Effects of Land Use Conversion on the Soil Microbial Community Composition and Functionality in the Urban Wetlands of North-Eastern China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Soil Sampling
2.2. Measurement of Soil Physicochemical Properties
2.3. DNA Extraction and Sequencing
2.4. Statistical Analysis
3. Results
3.1. Effect of Land-Use Type on Soil Physicochemical Properties
3.2. Diversity of Soil Microbial Communities under Different Land-Use Types
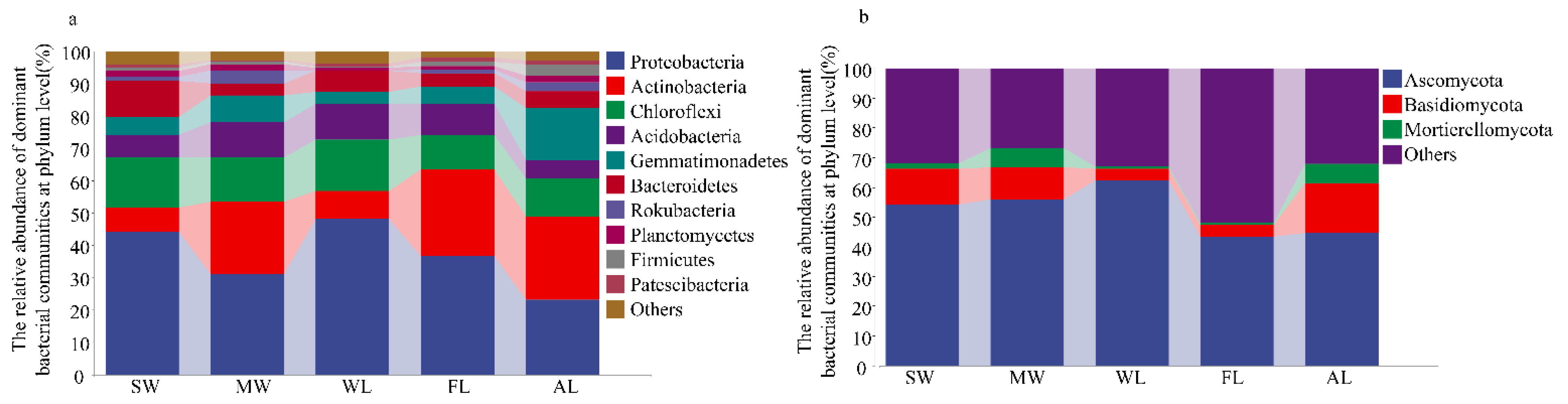
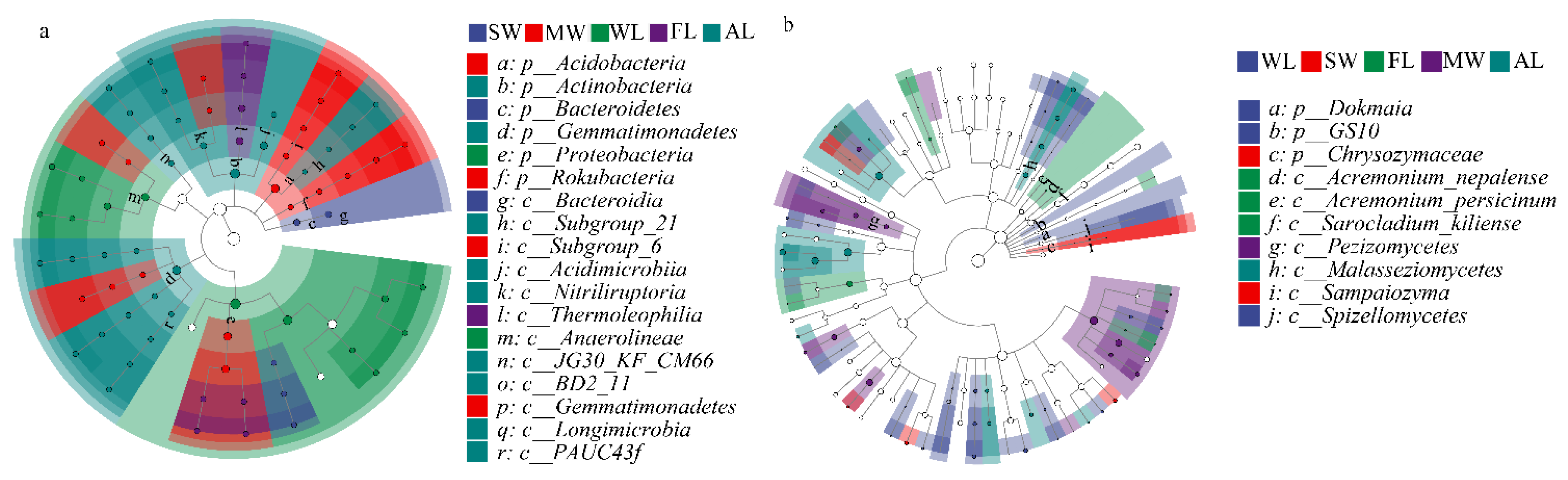
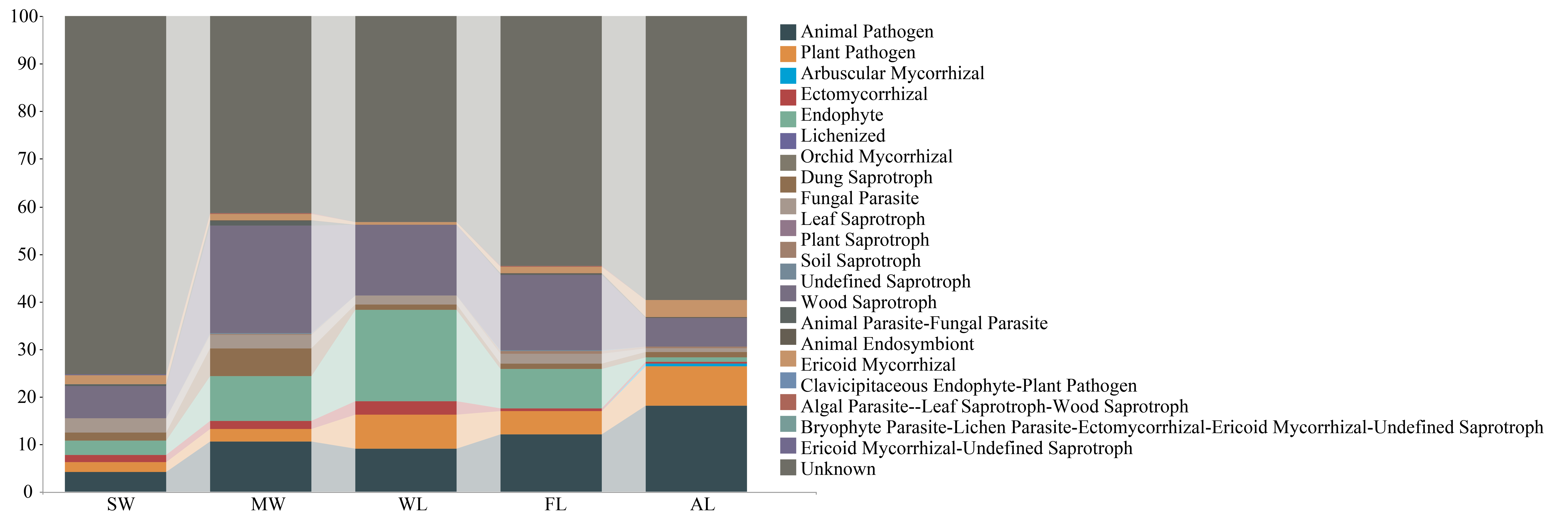
3.3. Composition of Soil Microbial Community under Different Land-Use Types
3.4. Associations between Microbial Communities and Soil Physicochemical Properties under Different Land-Use Types
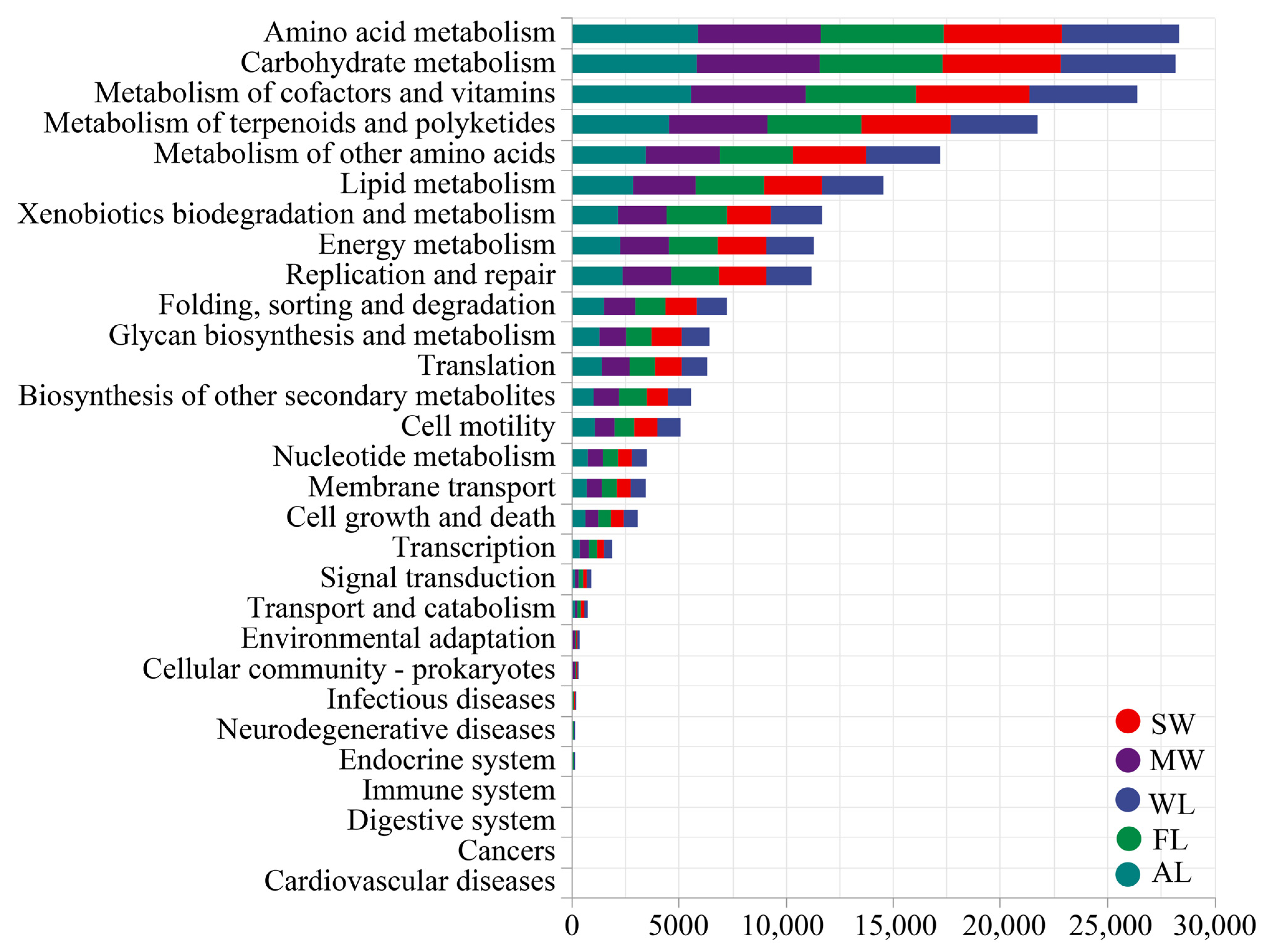
3.5. Microbial Functional Responses to Different Land-Use Types
4. Discussion
4.1. Land-Use Type Influence Soil Physicochemical Properties
4.2. Land-Use Type Influence on the Composition and Structure of Soil Microbial Community
4.3. Correlation between Soil Microbial Communities and Soil Variables under Different Land-Use Types
4.4. Prediction of Metabolic Functions of Soil Bacteria under Different Land-Use Types
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smith, P.; House, J.; Bustamante, M.; Sobocká, J.; Harper, R.; Pan, G.-X.; West, P.; Clark, J.; Adhya, T.; Rumpel, C.; et al. Global change pressures on soils from land use and management. Glob. Chang. Biol. 2016, 22, 1008–1028. [Google Scholar] [CrossRef] [PubMed]
- Keesstra, S.; Mol, G.; De Leeuw, J.; Okx, J.; Molenaar, C.; Cleen, M.; Visser, S. Soil-related sustainable development goals: Four concepts to make land degradation neutrality and restoration work. Land 2018, 7, 133. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Jia, Y.; Li, Z.; Tao, J.; Lin, L.; Chen, K.; Zhenyuan, L.; Tan, X.; Zhang, Q. Trophic structure in response to land use in subtropical streams. Ecol. Indic. 2021, 127, 107746. [Google Scholar] [CrossRef]
- Mganga, K.; Razavi, B.; Kuzyakov, Y. Land use affects soil biochemical properties in Mt. Kilimanjaro region. Catena 2016, 141, 22–29. [Google Scholar] [CrossRef]
- Bai, Y.; Wong, C.; Jiang, B.; Hughes, A.; Wang, M.; Wang, Q. Developing China’s ecological redline policy using ecosystem services assessments for land use planning. Nat. Commun. 2018, 9, 3034. [Google Scholar] [CrossRef] [Green Version]
- Zedler, J.; Kercher, S. Wetland resources: Status, trends, ecosystem services, and restorability. Annu. Rev. Environ. Resour. 2005, 15, 39–74. [Google Scholar] [CrossRef] [Green Version]
- Camacho, V.; Tello-Alcaide, E.; Wootton, A.; Valencia-Barrera, E. Land use change and urban ecosystem services: A case study of urban wetlands in a rapidly sprawling city in the highlands of Chiapas, Mexico. J. Manag. Sustain. 2019, 9, 67. [Google Scholar] [CrossRef]
- Wang, S.; Sun, L.; Ling, N.; Zhu, C.; Chi, F.; Li, W.; Hao, X.; Zhang, W.; Bian, J.; Chen, L.; et al. Exploring soil factors determining composition and structure of the bacterial communities in saline-alkali soils of Songnen Plain. Front. Microbiol. 2020, 10, 2902. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Liu, X.; Li, X.; Tian, C. Soil organic carbon changes following wetland restoration: A global meta-analysis. Geoderma 2019, 353, 89–96. [Google Scholar] [CrossRef]
- An, J.; Liu, C.; Wang, Q.; Yao, M.; Rui, J.; Zhang, S.; Li, X. Soil bacterial community structure in Chinese wetlands. Geoderma 2019, 337, 290–299. [Google Scholar] [CrossRef]
- Guan, Y.; Jiang, N.; Wu, Y.; Yang, Z.; Bello, A.; Yang, W. Disentangling the role of salinity-sodicity in shaping soil microbiome along a natural saline-sodic gradient. Sci. Total Environ. 2020, 765, 142738. [Google Scholar] [CrossRef] [PubMed]
- Bacani, V.; Sakamoto, Y.; Quénol, H.; Vannier, C.; Corgne, S. Markov chains–cellular automata modeling and multicriteria analysis of land cover change in the Lower Nhecolândia subregion of the Brazilian Pantanal wetland. J. Appl. Remote Sens. 2016, 10, 016004. [Google Scholar] [CrossRef]
- Wu, Y.C.; Tan, L.; Liu, W.; Wang, B.; Cai, Y.; Lin, X. Profiling bacterial diversity in a limestone cave of the western Loess Plateau of China. Front. Microbiol. 2015, 6, 244. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.; Li, Y.; Zhang, X.; Yan, Z.; Wu, H.; Li, M.; Yan, L.; Zhang, K.; Wang, J.; Kang, X. Soil pH and nutrients shape the vertical distribution of microbial communities in an alpine wetland. Sci. Total Environ. 2021, 774, 145780. [Google Scholar] [CrossRef]
- Bardgett, R.; Putten, W. Belowground biodiversity and ecosystem functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef]
- Sheng, Y.; Wang, H.; Wang, M.; Li, H.; Li, X.; Pan, F.; Chen, X.; Shen, X.; Mao, Z. Effects of soil texture on the growth of young apple trees and soil microbial community structure under replanted conditions. Hortic. Plant J. 2020, 6, 123–131. [Google Scholar] [CrossRef]
- Sleator, R.D.; Shortall, C.; Hill, C. Metagenomics. Lett. Appl. Microbiol. 2008, 47, 361–366. [Google Scholar] [CrossRef]
- Tiwari, S.; Singh, C.; Boudh, S.; Rai, P.K.; Gupta, V.K.; Singh, J.S. Land use change: A key ecological disturbance declines soil microbial biomass in dry tropical uplands. J. Environ. Manag. 2019, 242, 1–10. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, X.; Wu, Y.; Zhang, L.; Cheng, J.; Wei, G.; Lin, Y. Natural revegetation of a semiarid habitat alters taxonomic and functional diversity of soil microbial communities. Sci. Total Environ. 2018, 635, 598–606. [Google Scholar] [CrossRef]
- Lauber, C.; Strickland, M.; Bradford, M.; Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 2008, 40, 2407–2415. [Google Scholar] [CrossRef]
- Guan, X.; Wang, J.; Zhao, H.; Wang, J.; Luo, X.; Liu, F.; Zhao, F. Soil bacterial communities shaped by geochemical factors and land use in a less-explored area, Tibetan Plateau. BMC Genom. 2013, 14, 820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Ren, Z.; Qu, X.; Zhang, M.; Yu, Y.; Zhang, Y.; Peng, W. Microbial community structure and functional properties in permanently and seasonally flooded areas in Poyang Lake. Sci. Rep. 2020, 10, 4819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.H.; Wang, Y.F.; Li, H.; Gu, J.D. Niche specificity of ammonia-oxidizing archaeal and bacterial communities in a freshwater wetland receiving municipal wastewater in Daqing, Northeast China. Ecotoxicology 2014, 23, 2081–2091. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zheng, X.; Zhang, H.; Yu, X.; Lian, Y.; Yang, X.; Yu, H.; Hu, R.; He, Z.; Xiao, F.; et al. Metagenomic insights into the effects of submerged plants on functional potential of microbial communities in wetland sediments. Mar. Life Sci. Technol. 2021, 3, 405–415. [Google Scholar] [CrossRef]
- Yi, X.; Ning, C.; Feng, S.; Gao, H.; Zhao, J.; Liao, J.; Peng, Y.; Zhao, S.; Liu, S. Urbanization-induced environmental changes strongly affect wetland soil bacterial community composition and diversity. Environ. Res. Lett. 2022, 17, 014027. [Google Scholar] [CrossRef]
- Sui, X.; Zhang, R.; Frey, B.; Yang, L.; Li, M.H.; Ni, H. Land use change effects on diversity of soil bacterial, Acidobacterial and fungal communities in wetlands of the Sanjiang Plain, northeastern China. Sci. Rep. 2019, 9, 18535. [Google Scholar] [CrossRef]
- Dong, C.; Shi, Y.; Wang, J.; Zeng, H.; Wang, W. Divergent responses of the soil bacteria community to multi-level nitrogen enrichment in temperate grasslands under different degrees of degradation. Land Degrad. Dev. 2021, 32, 3524–3535. [Google Scholar] [CrossRef]
- Rao, D.; Meng, F.; Yan, X.; Zhang, M.; Yao, X.; Kim, K.S.; Zhao, J.; Qiu, Q.; Xie, F.; Zhang, W. Changes in soil microbial activity, bacterial community composition and function in a long-term continuous soybean cropping system after corn insertion and fertilization. Front. Microbiol. 2021, 12, 638326. [Google Scholar] [CrossRef]
- Lu, R. Soil and Agro-Chemical Analytical Methods; China Agricultural Science and Technology Press: Beijing, China, 1999. [Google Scholar]
- Rayment, G.; Higginson, F. The Australian Handbook of Soil and Water Chemical Methods; Inkata Press: Melbourne, Australia, 1992; Volume 63. [Google Scholar]
- Jackson, M. Soil Chemical Analysis; Prentice-Hall: New York, NY, USA, 1958; Volume 46. [Google Scholar]
- Adeloju, S.; Bond, A.; Briggs, M. Critical evaluation of some wet digestion methods for the stripping voltammetric determination of selenium in biological materials. Anal. Chem. 1984, 56, 2397–2401. [Google Scholar] [CrossRef]
- Paliwal, K.V.; Maliwal, G.L. Prediction of exchangeable sodium percentage from cation exchange equilibria. Geoderma 1971, 6, 75–78. [Google Scholar] [CrossRef]
- Kemp, P.; Aller, J. Bacterial diversity in aquatic and other environments: What 16S rDNA libraries can tell us. FEMS Microbiol. Ecol. 2004, 47, 161–177. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Ryberg, M.; Kristiansson, E.; Abarenkov, K.; Larsson, K.-H.; Kõljalg, U. Taxonomic reliability of DNA sequences in public sequence databases: A Fungal Perspective. PLoS ONE 2006, 1, e59. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2012. [Google Scholar]
- Zuur, A.F.; Ieno, E.N.; Smith, G.M. Principal coordinate analysis and non-metric multidimensional scaling. In Analysing Ecological Data; Springer: New York, NY, USA, 2007; pp. 259–264. [Google Scholar]
- Morris, C. Multivariate analysis of ecological data using Canoco 5, 2nd Edition. Afr. J. Range Forage Sci. 2015, 32, 1–2. [Google Scholar] [CrossRef]
- Nguyen, N.; Song, Z.; Bates, S.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.; Kennedy, P. FUN-Guild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2015, 20, 241–248. [Google Scholar] [CrossRef]
- Douglas, G.; Maffei, V.; Zaneveld, J.; Yurgel, S.; Brown, J.; Taylor, C.; Huttenhower, C.; Langille, M. PICRUSt2: An improved and extensible approach for metagenome inference. bioRxiv 2019, 672295. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Xu, N.; Wang, H.; Li, J.; Zhong, H.; Zong, C. Variations in the diversity of soil microbial community and structure under various categories of degradation wetland in Sanjiang Plain, northeastern China. Land Degrad. Dev. 2021, 32, 2143–2156. [Google Scholar] [CrossRef]
- Lu, M.; Ren, Y.; Wang, S.; Tian, K.; Sun, X.; Peng, S. Contribution of soil variables to bacterial community composition following land use change in Napahai plateau wetlands. J. Environ. Manag. 2019, 246, 77–84. [Google Scholar] [CrossRef]
- Ma, Q.; Cui, L.; Song, H.; Gao, C.; Hao, Y.; Luan, J.; Wang, Y.; Li, W. Aboveground and belowground biomass relationships in the Zoige peatland, Eastern Qinghai–Tibetan Plateau. Wetlands 2017, 37, 461–469. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Y.; Sun, L.; Sun, T. Effects of grassland degradation on vegetation and soil properties in the middle of the Songnen Plain. Pratacultural Sci. 2018, 35, 2347–2353. (In Chinese) [Google Scholar]
- Pan, C.; Li, Y.; Xiaoya, Y.; Ren, S. Drivers of soil bacterial diversity in sandy grasslands in China. Sci. Rep. 2021. [Google Scholar] [CrossRef]
- McKinley, V.L. Effects of land use and restoration on soil microbial communities. In Understanding Terrestrial Microbial Communities; Springer: Cham, Switzerland, 2019; pp. 173–242. [Google Scholar]
- Lauber, C.L.; Ramirez, K.S.; Aanderud, Z.; Lennon, J.; Fierer, N. Temporal variability in soil microbial communities across land-use types. ISME J. 2013, 7, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.; Kellner, E.; Martin, G.; Freedman, Z.; Hubbart, J.; Stephan, K.; Kelly, C.; Morrissey, E. Land use intensification destabilizes stream microbial biodiversity and decreases metabolic efficiency. Sci. Total Environ. 2021, 767, 145440. [Google Scholar] [CrossRef] [PubMed]
- Asmelash, F.; Bekele, T.; Belay, Z.; Kebede, F. Soil physicochemical property and arbuscular mycorrhizal fungi resilience to degradation and deforestation of a dry evergreen Afromontane forest in central Ethiopia. Land Degrad. Dev. 2021, 32, 3338–3350. [Google Scholar] [CrossRef]
- Feng, H.; Wang, S.; Gao, Z.; Wang, Z.; Ren, X.; Hu, S.; Pan, H. Effect of land use on the composition of bacterial and fungal communities in saline–sodic soils. Land Degrad. Dev. 2019, 30, 1851–1860. [Google Scholar] [CrossRef]
- Kraychenko, A.N.; Guber, A.K.; Razavi, B.S.; Koestel, J.; Quigley, M.Y.; Robertson, G.P.; Kuzyakov, Y. Microbial spatial footprint as a driver of soil carbon stabilization. Nat. Commun. 2019, 10, 10. [Google Scholar] [CrossRef] [Green Version]
- Zhao, F.; Ren, C.; Zhang, L.; HHan, X.; HYang, G.; Wang, J. Changes in soil microbial community are linked to soil carbon fractions after afforestation: Soil microbial community affects carbon fractions. Eur. J. Soil Sci. 2018, 69, 370–379. [Google Scholar] [CrossRef]
- Hanson, C.; Allison, S.; Bradford, M.; Wallenstein, M.; Treseder, K. Fungal taxa target different carbon sources in forest soil. Ecosystems 2008, 11, 1157–1167. [Google Scholar] [CrossRef]
- Dong, H.; Kong, C.; Wang, P.; Huang, Q. Temporal variation of soil friedelin and microbial community under different land uses in a long-term agroecosystem. Soil Biol. Biochem. 2014, 69, 275–281. [Google Scholar] [CrossRef]
- Bradley, D.; Martiny, J. Patterns of fungal diversity and composition along a salinity gradient. ISME J. 2010, 5, 379–388. [Google Scholar] [CrossRef]
- Gao, W.; Gao, K.; Guo, Z.; Liu, Y.; Jiang, L.; Liu, C.; Liu, X.; Wang, G. Different responses of soil bacterial and fungal communities to 3 years of biochar amendment in an alkaline soybean soil. Front. Microbiol. 2021, 12, 630418. [Google Scholar] [CrossRef]
- Shahariar, S.; Helgason, B.; Soolanayakanahally, R.; Bedard-Haughn, A. Soil enzyme activity as affected by land-use, salinity, and groundwater fluctuations in wetland soils of the prairie pothole region. Wetlands 2021, 41, 31. [Google Scholar] [CrossRef]
- Rousk, J.; Bååth, E.; Brookes, P.; LLauber, C.; Lozupone, C.; Gregory Caporaso, J.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; DBardgett, R.; MVitousek, P.; Maestre, F.; AWilliams, M.; JEldridge, D.; Lambers, H.; Neuhauser, S.; Gallardo, A.; García-Velázquez, L.; et al. Changes in belowground biodiversity during ecosystem development. Proc. Natl. Acad. Sci. USA 2019, 116, 201818400. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Waldrop, M.; Holloway, J.; Smith, D.; Goldhaber, M.; Drenovsky, R.; Scow, K.; Dick, R.; Howard, D.; Wylie, B.; Grace, J. The interacting roles of climate, soils, and plant production on soil microbial communities at a continental scale. Ecology 2017, 98, 1957–1967. [Google Scholar] [CrossRef] [Green Version]
- Ait Barka, E.; Vatsa, P.; Sanchez, L.; Vaillant-Gaveau, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; Wezel, G. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2015, 80, 1–43. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Miao, Y.; Gan, Y.; Wei, S.; Tan, S.; Rask, K.A.; Wang, L.; Dai, J.; Chen, W.; Ekelund, F. Soil bacterial community response to long-term land use conversion in Yellow River Delta. Appl. Soil Ecol. 2020, 156, 103709. [Google Scholar] [CrossRef]
- Grandy, S.; Neff, J. Molecular C dynamics downstream: The biochemical decomposition sequence and its impact on soil organic matter structure and function. Sci. Total Environ. 2008, 404, 297–307. [Google Scholar] [CrossRef]
- Rath, K.; Maheshwari, A.; Bengtson, P.; Rousk, J. Comparative toxicities of salts on microbial processes in soil. Appl. Environ. Microbiol. 2016, 82, 2012–2020. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.M.; Wang, S.P.; Jiang, L.L.; Zhang, L.R.; Cui, S.J.; Meng, F.D.; Wang, Q.; Li, X.N.; Zhou, Y. Changes of soil microbial community under different degraded gradients of alpine meadow. Agric. Ecosyst. Environ. 2016, 222, 213–222. [Google Scholar] [CrossRef]
- Che, R.; Wang, Y.; Li, K.; Xu, Z.; Hu, J.; Wang, F.; Rui, Y.; Li, L.; Pang, Z.; Cui, X. Degraded patch formation significantly changed microbial community composition in alpine meadow soils. Soil Tillage Res. 2019, 195, 104426. [Google Scholar] [CrossRef]
- Urbanová, M.; Šnajdr, J.; Baldrian, P. Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 2015, 84, 53–64. [Google Scholar] [CrossRef]
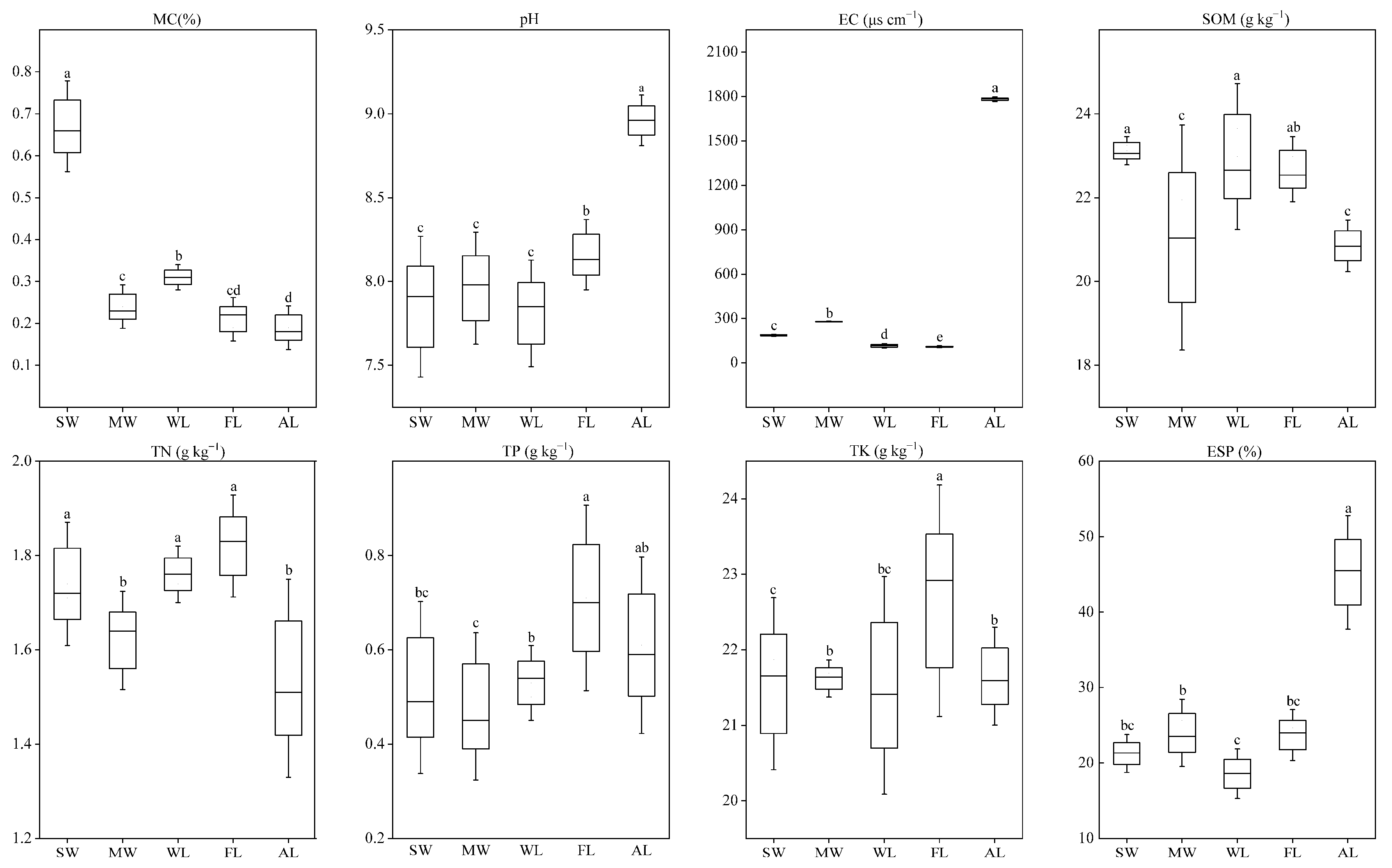


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Gao, W.; Zou, Y.; Dong, H.; Yu, F.; Wang, H.; Zong, C. Effects of Land Use Conversion on the Soil Microbial Community Composition and Functionality in the Urban Wetlands of North-Eastern China. Forests 2022, 13, 1148. https://doi.org/10.3390/f13071148
Wu Y, Gao W, Zou Y, Dong H, Yu F, Wang H, Zong C. Effects of Land Use Conversion on the Soil Microbial Community Composition and Functionality in the Urban Wetlands of North-Eastern China. Forests. 2022; 13(7):1148. https://doi.org/10.3390/f13071148
Chicago/Turabian StyleWu, Yining, Weifeng Gao, Yu Zou, Haiyan Dong, Fei Yu, He Wang, and Cheng Zong. 2022. "Effects of Land Use Conversion on the Soil Microbial Community Composition and Functionality in the Urban Wetlands of North-Eastern China" Forests 13, no. 7: 1148. https://doi.org/10.3390/f13071148
APA StyleWu, Y., Gao, W., Zou, Y., Dong, H., Yu, F., Wang, H., & Zong, C. (2022). Effects of Land Use Conversion on the Soil Microbial Community Composition and Functionality in the Urban Wetlands of North-Eastern China. Forests, 13(7), 1148. https://doi.org/10.3390/f13071148




