Xylem Transcriptome Analysis in Contrasting Wood Phenotypes of Eucalyptus urophylla × tereticornis Hybrids
Abstract
1. Introduction
2. Results
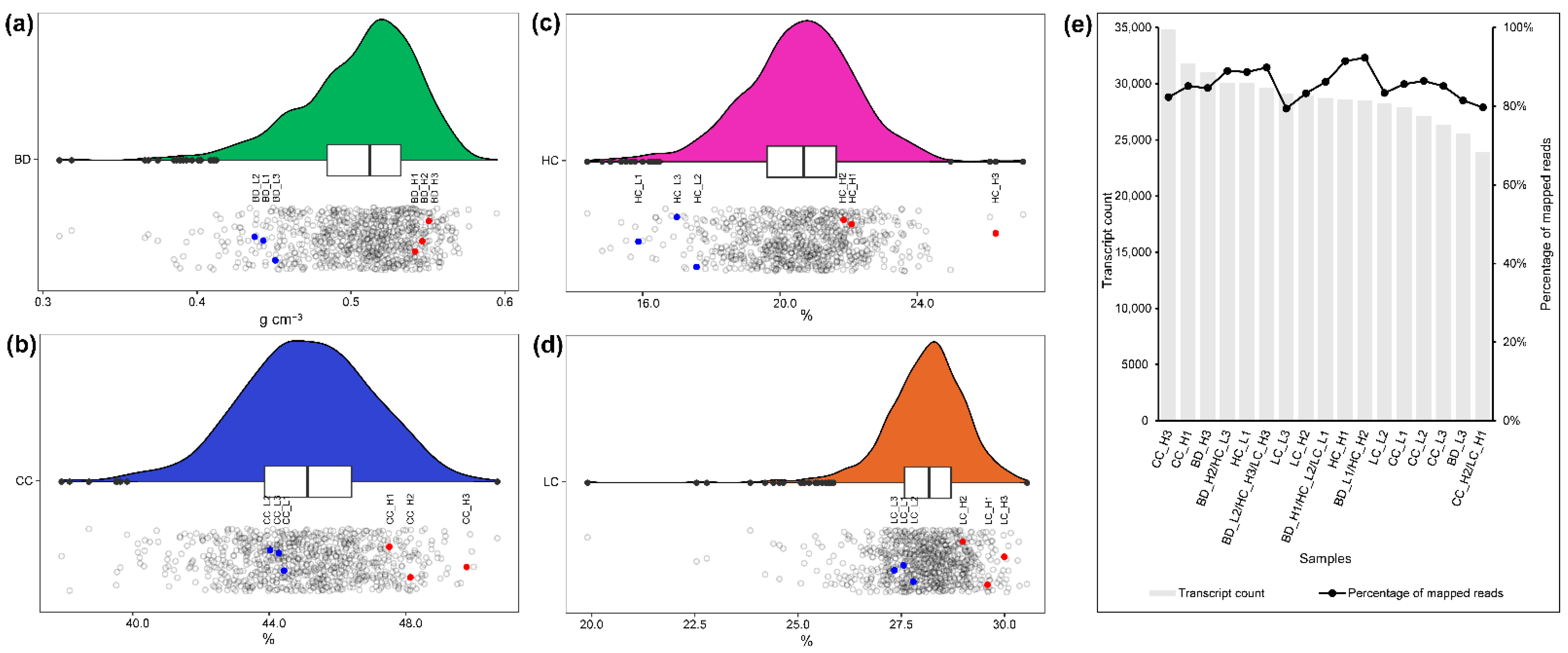
2.1. Transcriptome Sequencing Quality
2.2. Analysis of Differentially Expressed Genes
2.3. Alternative Splicing Identification and Differentially Spliced Gene Analysis
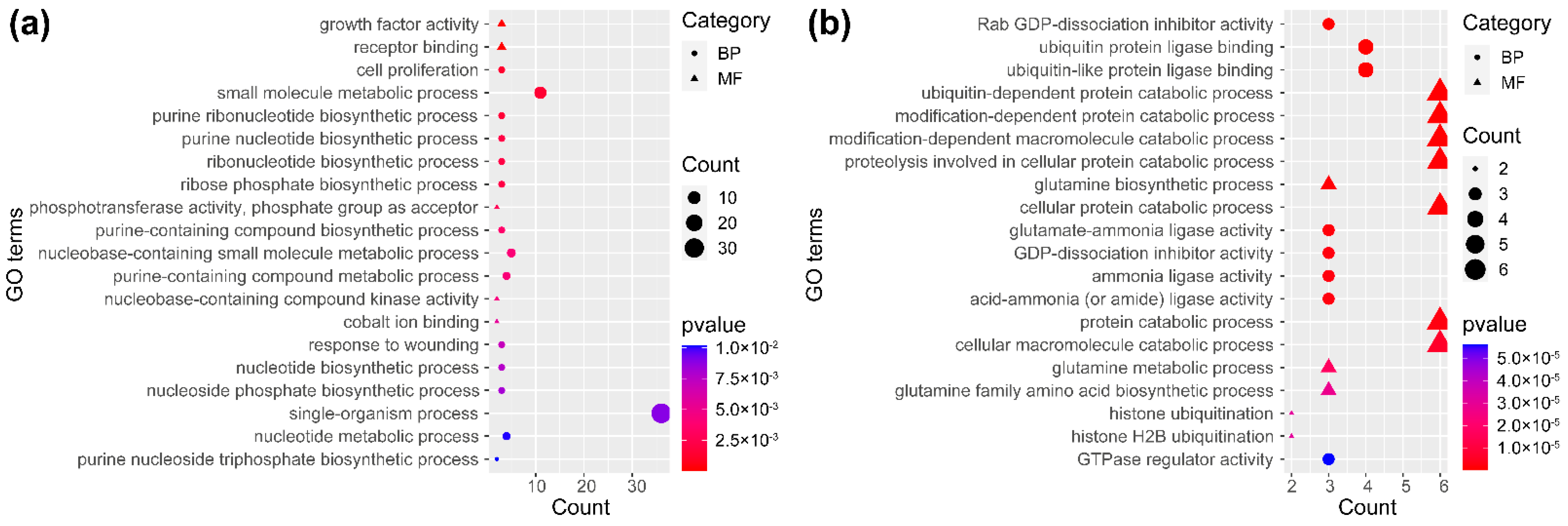
2.4. Functional Comparison of Key DEGs and DSGs
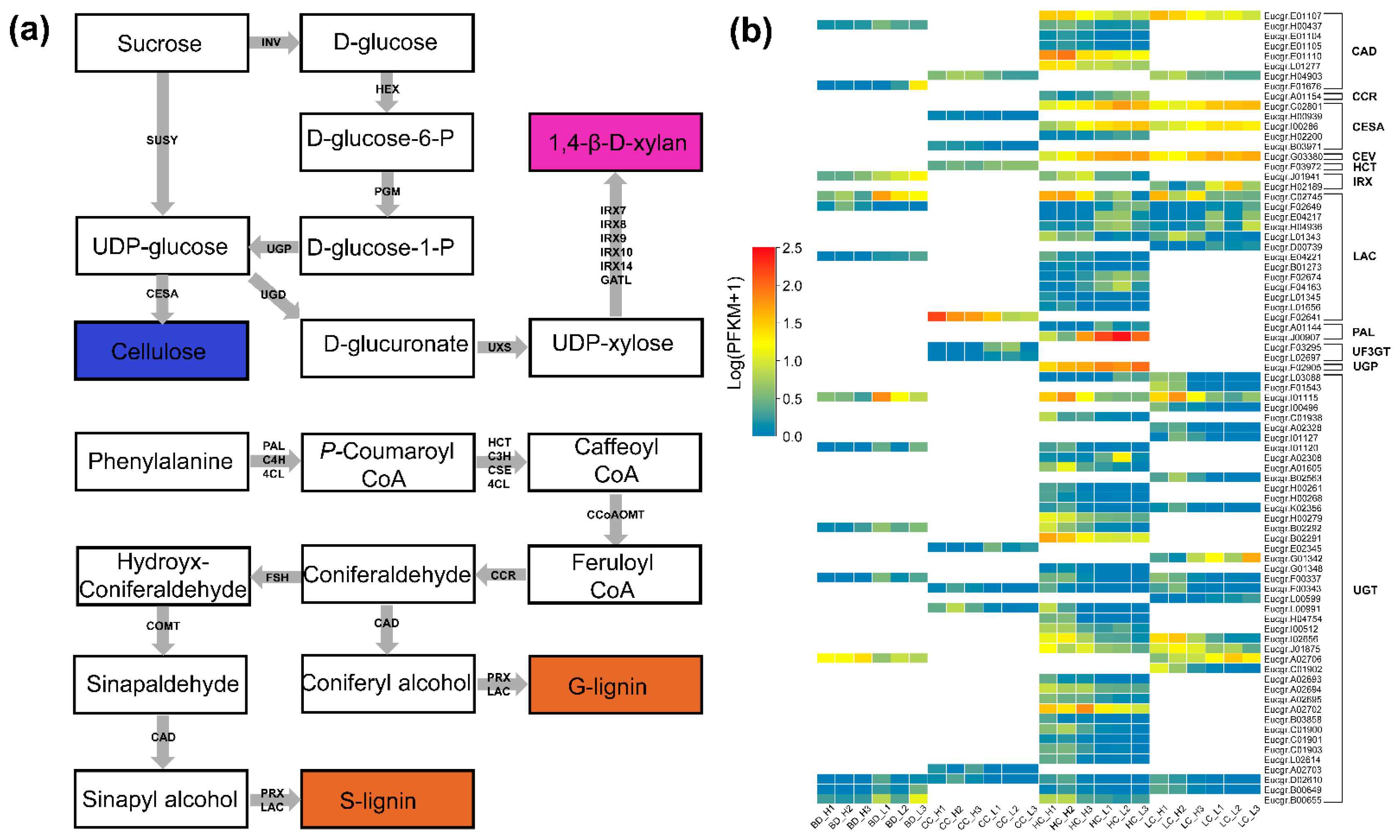
2.5. Wood Biosynthetic Pathway Genes
3. Discussion
3.1. DEGs and DSGs in Individuals with Contrasting Wood Properties
3.2. The Relationship between DEGs and DSGs
3.3. Transcriptional Levels of Wood Biosynthetic Pathway Genes
3.4. Further Research Arising from This Study
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Extraction, Library Construction and RNA Sequencing
4.3. Quality Control of Sequences and Reference-Guided Transcriptome Mapping
4.4. Differentially Expressed Gene and Differentially Spliced Gene Detection
4.5. Analysis of Gene Ontology and Biological Pathways and Transcription Factor Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ladiges, P.Y.; Udovicic, F.; Nelson, G. Australian Biogeographical Connections and the Phylogeny of Large Genera in the Plant Family Myrtaceae: Australian Biogeographical Connections. J. Biogeogr. 2003, 30, 989–998. [Google Scholar] [CrossRef]
- Denis, M.; Favreau, B.; Ueno, S.; Camus-Kulandaivelu, L.; Chaix, G.; Gion, J.-M.; Nourrisier-Mountou, S.; Polidori, J.; Bouvet, J.-M. Genetic Variation of Wood Chemical Traits and Association with Underlying Genes in Eucalyptus urophylla. Tree Genet. Genomes 2013, 9, 927–942. [Google Scholar] [CrossRef]
- Gan, S.; Shi, J.; Li, M.; Wu, K.; Wu, J.; Bai, J. Moderate-Density Molecular Maps of Eucalyptus urophylla S. T. Blake and E. tereticornis Smith Genomes Based on RAPD Markers. Genetica 2003, 118, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Varghese, M.; Harwood, C.E.; Hegde, R.; Ravi, N. Evaluation of Provenances of Eucalyptus camaldulensis and Clones of E. camaldulensis and E. tereticornis at Contrasting Sites in Southern India. Silvae Genet. 2008, 57, 170–179. [Google Scholar] [CrossRef]
- Chen, S.; Weng, Q.; Li, F.; Li, M.; Zhou, C.; Gan, S. Genetic Parameters for Growth and Wood Chemical Properties in Eucalyptus urophylla × E. tereticornis Hybrids. Ann. For. Sci. 2018, 75, 16. [Google Scholar] [CrossRef]
- Yang, H.; Weng, Q.; Li, F.; Zhou, C.; Li, M.; Chen, S.; Ji, H.; Gan, S. Genotypic Variation and Genotype-by-Environment Interactions in Growth and Wood Properties in a Cloned Eucalyptus urophylla × E. tereticornis Family in Southern China. For. Sci. 2018, 64, 225–232. [Google Scholar] [CrossRef]
- He, X.; Li, F.; Li, M.; Weng, Q.; Shi, J.; Mo, X.; Gan, S. Quantitative Genetics of Cold Hardiness and Growth in Eucalyptus as Estimated from E. urophylla × E. tereticornis Hybrids. New For. 2012, 43, 383–394. [Google Scholar] [CrossRef]
- Li, F.; Zhou, C.; Weng, Q.; Li, M.; Yu, X.; Guo, Y.; Wang, Y.; Zhang, X.; Gan, S. Comparative Genomics Analyses Reveal Extensive Chromosome Colinearity and Novel Quantitative Trait Loci in Eucalyptus. PLoS ONE 2015, 10, e0145144. [Google Scholar] [CrossRef][Green Version]
- Marco de Lima, B.; Cappa, E.P.; Silva-Junior, O.B.; Garcia, C.; Mansfield, S.D.; Grattapaglia, D. Quantitative Genetic Parameters for Growth and Wood Properties in Eucalyptus “Urograndis” Hybrid Using near-Infrared Phenotyping and Genome-Wide SNP-Based Relationships. PLoS ONE 2019, 14, e0218747. [Google Scholar] [CrossRef]
- Thomas, L.H.; Martel, A.; Grillo, I.; Jarvis, M.C. Hemicellulose Binding and the Spacing of Cellulose Microfibrils in Spruce Wood. Cellulose 2020, 27, 4249–4254. [Google Scholar] [CrossRef]
- Soares, B.C.D.; Lima, J.T.; de Assis, C.O. Influence of Density and Lignin Content on Cleavage Strength of Eucalyptus grandis Wood. Trees 2022. [Google Scholar] [CrossRef]
- Xu, P.; Kong, Y.; Song, D.; Huang, C.; Li, X.; Li, L. Conservation and Functional Influence of Alternative Splicing in Wood Formation of Populus and Eucalyptus. BMC Genom. 2014, 15, 780. [Google Scholar] [CrossRef] [PubMed]
- Lerouxel, O.; Cavalier, D.M.; Liepman, A.H.; Keegstra, K. Biosynthesis of Plant Cell Wall Polysaccharides—A Complex Process. Curr. Opin. Plant Biol. 2006, 9, 621–630. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Li, W.; Sun, Y.-H.; Kumari, S.; Wei, H.; Li, Q.; Tunlaya-Anukit, S.; Sederoff, R.R.; Chiang, V.L. SND1 Transcription Factor-Directed Quantitative Functional Hierarchical Genetic Regulatory Network in Wood Formation in Populus trichocarpa. Plant Cell 2013, 25, 4324–4341. [Google Scholar] [CrossRef]
- Shinya, T.; Iwata, E.; Nakahama, K.; Fukuda, Y.; Hayashi, K.; Nanto, K.; Rosa, A.C.; Kawaoka, A. Transcriptional Profiles of Hybrid Eucalyptus Genotypes with Contrasting Lignin Content Reveal That Monolignol Biosynthesis-Related Genes Regulate Wood Composition. Front. Plant Sci. 2016, 7, 443. [Google Scholar] [CrossRef]
- Nakahama, K.; Urata, N.; Shinya, T.; Hayashi, K.; Nanto, K.; Rosa, A.C.; Kawaoka, A. RNA-Seq Analysis of Lignocellulose-Related Genes in Hybrid Eucalyptus with Contrasting Wood Basic Density. BMC Plant Biol. 2018, 18, 156. [Google Scholar] [CrossRef] [PubMed]
- Bedre, R.; Irigoyen, S.; Petrillo, E.; Mandadi, K.K. New Era in Plant Alternative Splicing Analysis Enabled by Advances in High-Throughput Sequencing (HTS) Technologies. Front. Plant Sci. 2019, 10, 740. [Google Scholar] [CrossRef]
- Marquez, Y.; Brown, J.W.S.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome Survey Reveals Increased Complexity of the Alternative Splicing Landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef]
- Dong, C.; He, F.; Berkowitz, O.; Liu, J.; Cao, P.; Tang, M.; Shi, H.; Wang, W.; Li, Q.; Shen, Z.; et al. Alternative Splicing Plays a Critical Role in Maintaining Mineral Nutrient Homeostasis in Rice (Oryza Sativa). Plant Cell 2018, 30, 2267–2285. [Google Scholar] [CrossRef]
- Bao, H.; Li, E.; Mansfield, S.D.; Cronk, Q.C.; El-Kassaby, Y.A.; Douglas, C.J. The Developing Xylem Transcriptome and Genome-Wide Analysis of Alternative Splicing in Populus trichocarpa (Black Cottonwood) Populations. BMC Genom. 2013, 14, 359. [Google Scholar] [CrossRef]
- Noble, J.D.; Balmant, K.M.; Dervinis, C.; de los Campos, G.; Resende, M.F.R.; Kirst, M.; Barbazuk, W.B. The Genetic Regulation of Alternative Splicing in Populus deltoides. Front. Plant Sci. 2020, 11, 590. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, G.; Liu, X.; Yu, Z.; Peng, S. Integrated Analysis of Seed MicroRNA and MRNA Transcriptome Reveals Important Functional Genes and MicroRNA-Targets in the Process of Walnut (Juglans Regia) Seed Oil Accumulation. IJMS 2020, 21, 9093. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, Y.; Jin, Y.; Kan, L.; Shen, C.; Malladi, A.; Nambeesan, S.; Xu, Y.; Dong, C. Transcriptome Analysis of Pyrus Betulaefolia Seedling Root Responses to Short-Term Potassium Deficiency. IJMS 2020, 21, 8857. [Google Scholar] [CrossRef] [PubMed]
- Myburg, A.A.; Grattapaglia, D.; Tuskan, G.A.; Hellsten, U.; Hayes, R.D.; Grimwood, J.; Jenkins, J.; Lindquist, E.; Tice, H.; Bauer, D.; et al. The Genome of Eucalyptus grandis. Nature 2014, 510, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Elissetche, J.P.; Valenzuela, S.; García, R.; Norambuena, M.; Iturra, C.; Rodríguez, J.; Mendonça, R.T.; Balocchi, C. Transcript Abundance of Enzymes Involved in Lignin Biosynthesis of Eucalyptus globulus Genotypes with Contrasting Levels of Pulp Yield and Wood Density. Tree Genet. Genomes 2011, 7, 697–705. [Google Scholar] [CrossRef]
- Thavamanikumar, S.; Southerton, S.; Thumma, B. RNA-Seq Using Two Populations Reveals Genes and Alleles Controlling Wood Traits and Growth in Eucalyptus nitens. PLoS ONE 2014, 9, e101104. [Google Scholar] [CrossRef]
- Li, X.; Wu, H.X.; Southerton, S.G. Identification of Putative Candidate Genes for Juvenile Wood Density in Pinus radiata. Tree Physiol. 2012, 32, 1046–1057. [Google Scholar] [CrossRef][Green Version]
- Dharanishanthi, V.; Dasgupta, M.G. Construction of Co-Expression Network Based on Natural Expression Variation of Xylogenesis-Related Transcripts in Eucalyptus tereticornis. Mol. Biol. Rep. 2016, 43, 1129–1146. [Google Scholar] [CrossRef]
- Li, Y.I.; Knowles, D.A.; Humphrey, J.; Barbeira, A.N.; Dickinson, S.P.; Im, H.K.; Pritchard, J.K. Annotation-Free Quantification of RNA Splicing Using LeafCutter. Nat. Genet. 2018, 50, 151–158. [Google Scholar] [CrossRef]
- Zhang, G.; Guo, G.; Hu, X.; Zhang, Y.; Li, Q.; Li, R.; Zhuang, R.; Lu, Z.; He, Z.; Fang, X.; et al. Deep RNA Sequencing at Single Base-Pair Resolution Reveals High Complexity of the Rice Transcriptome. Genome Res. 2010, 20, 646–654. [Google Scholar] [CrossRef]
- Iida, K. Genome-Wide Analysis of Alternative Pre-MRNA Splicing in Arabidopsis Thaliana Based on Full-Length CDNA Sequences. Nucleic Acids Res. 2004, 32, 5096–5103. [Google Scholar] [CrossRef] [PubMed]
- Nishida, S.; Kakei, Y.; Shimada, Y.; Fujiwara, T. Genome-Wide Analysis of Specific Alterations in Transcript Structure and Accumulation Caused by Nutrient Deficiencies in Arabidopsis thaliana. Plant J. 2017, 91, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Han, X.; Sang, J.; He, X.; Liu, M.; Qiao, G.; Zhuo, R.; He, G.; Hu, J. Transcriptome Analysis of Immature Xylem in the Chinese Fir at Different Developmental Phases. PeerJ 2016, 4, e2097. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Patton, M.F.; Arena, G.D.; Salminen, J.-P.; Steinbauer, M.J.; Casteel, C.L. Transcriptome and Defence Response in Eucalyptus camaldulensis Leaves to Feeding by Glycaspis brimblecombei Moore (Hemiptera: Aphalaridae): A Stealthy Psyllid Does Not Go Unnoticed: Eucalyptus-Glycaspis Psyllid Interactions. Austral Entomol. 2018, 57, 247–254. [Google Scholar] [CrossRef]
- Morreale, F.E.; Walden, H. Types of Ubiquitin Ligases. Cell 2016, 165, 248–248.e1. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF Family Transcription Factors in Plant Abiotic Stress Responses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.-L.; Xing, G.-M.; Liu, J.-X.; Duan, A.-Q.; Xu, Z.-S.; Li, M.-Y.; Zhuang, J.; Xiong, A.-S. Advances in AP2/ERF Super-Family Transcription Factors in Plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, Z.; Li, D.; Wang, F.; Zhang, R.; Wang, J. Molecular Characterization of the ERF Family in Susceptible Poplar Infected by Virulent Melampsora Larici-Populina. Physiol. Mol. Plant Pathol. 2019, 108, 101437. [Google Scholar] [CrossRef]
- Seyfferth, C.; Wessels, B.; Jokipii-Lukkari, S.; Sundberg, B.; Delhomme, N.; Felten, J.; Tuominen, H. Ethylene-Related Gene Expression Networks in Wood Formation. Front. Plant Sci. 2018, 9, 272. [Google Scholar] [CrossRef]
- Berget, S.M.; Moore, C.; Sharp, P.A. Spliced Segments at the 5′ Terminus of Adenovirus 2 Late MRNA. Proc. Natl. Acad. Sci. USA 1977, 74, 3171–3175. [Google Scholar] [CrossRef]
- Xie, T.; Liu, Z.; Wang, G. Structural Basis for Monolignol Oxidation by a Maize Laccase. Nat. Plants 2020, 6, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Bryan, A.C.; Jawdy, S.; Gunter, L.; Gjersing, E.; Sykes, R.; Hinchee, M.A.W.; Winkeler, K.A.; Collins, C.M.; Engle, N.; Tschaplinski, T.J.; et al. Knockdown of a Laccase in Populus deltoides Confers Altered Cell Wall Chemistry and Increased Sugar Release. Plant Biotechnol. J. 2016, 14, 2010–2020. [Google Scholar] [CrossRef] [PubMed]
- Gion, J.-M.; Carouché, A.; Deweer, S.; Bedon, F.; Pichavant, F.; Charpentier, J.-P.; Baillères, H.; Rozenberg, P.; Carocha, V.; Ognouabi, N.; et al. Comprehensive Genetic Dissection of Wood Properties in a Widely-Grown Tropical Tree: Eucalyptus. BMC Genom. 2011, 12, 301. [Google Scholar] [CrossRef] [PubMed]
- Grubb, C.D.; Zipp, B.J.; Kopycki, J.; Schubert, M.; Quint, M.; Lim, E.-K.; Bowles, D.J.; Pedras, M.S.C.; Abel, S. Comparative Analysis of Arabidopsis UGT74 Glucosyltransferases Reveals a Special Role of UGT74C1 in Glucosinolate Biosynthesis. Plant J. 2014, 79, 92–105. [Google Scholar] [CrossRef]
- De Micco, V.; Carrer, M.; Rathgeber, C.B.K.; Julio Camarero, J.; Voltas, J.; Cherubini, P.; Battipaglia, G. From Xylogenesis to Tree Rings: Wood Traits to Investigate Tree Response to Environmental Changes. IAWA 2019, 40, 155–182. [Google Scholar] [CrossRef]
- Solomon, O.L.; Berger, D.K.; Myburg, A.A. Diurnal and Circadian Patterns of Gene Expression in the Developing Xylem of Eucalyptus Trees. S. Afr. J. Bot. 2010, 76, 425–439. [Google Scholar] [CrossRef][Green Version]
- Chao, Q.; Gao, Z.-F.; Zhang, D.; Zhao, B.-G.; Dong, F.-Q.; Fu, C.-X.; Liu, L.-J.; Wang, B.-C. The Developmental Dynamics of the Populus Stem Transcriptome. Plant Biotechnol. J. 2019, 17, 206–219. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-Level Expression Analysis of RNA-Seq Experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome. Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Molecular Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Gaete-Loyola, J.; Lagos, C.; Beltrán, M.F.; Valenzuela, S.; Emhart, V.; Fernández, M. Transcriptome Profiling of Eucalyptus nitens Reveals Deeper Insight into the Molecular Mechanism of Cold Acclimation and Deacclimation Process. Tree Genet. Genomes 2017, 13, 37. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Jin, J.; Gao, G. PlantRegMap: Charting Functional Regulatory Maps in Plants. Nucleic Acids Res. 2019, 48, D1104–D1113. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A Web Server for Annotation and Identification of Enriched Pathways and Diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a Central Hub for Transcription Factors and Regulatory Interactions in Plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; He, J.; Zhou, C.; Weng, Q.; Chen, S.; Bush, D.; Li, F. Xylem Transcriptome Analysis in Contrasting Wood Phenotypes of Eucalyptus urophylla × tereticornis Hybrids. Forests 2022, 13, 1102. https://doi.org/10.3390/f13071102
Zhu X, He J, Zhou C, Weng Q, Chen S, Bush D, Li F. Xylem Transcriptome Analysis in Contrasting Wood Phenotypes of Eucalyptus urophylla × tereticornis Hybrids. Forests. 2022; 13(7):1102. https://doi.org/10.3390/f13071102
Chicago/Turabian StyleZhu, Xianliang, Jiayue He, Changpin Zhou, Qijie Weng, Shengkan Chen, David Bush, and Fagen Li. 2022. "Xylem Transcriptome Analysis in Contrasting Wood Phenotypes of Eucalyptus urophylla × tereticornis Hybrids" Forests 13, no. 7: 1102. https://doi.org/10.3390/f13071102
APA StyleZhu, X., He, J., Zhou, C., Weng, Q., Chen, S., Bush, D., & Li, F. (2022). Xylem Transcriptome Analysis in Contrasting Wood Phenotypes of Eucalyptus urophylla × tereticornis Hybrids. Forests, 13(7), 1102. https://doi.org/10.3390/f13071102








