Abstract
Halotolerant plant-growth-promoting rhizobacteria (PGPR) could not only promote plant growth, but also help in counteracting the detrimental effects of salt stress. In the present study, a total of 76 bacteria were isolated from the rhizosphere, non-rhizospheric soil and endophytes of the halophyte Salsola tetrandra, collected from natural saline soils in Algeria. Phylogenetic analysis based on the 16S rDNA sequence of Gram-negative bacteria (n = 51) identified, showed seventeen representative isolates grouped into four genera (Pseudomonas, Acinetobacter, Enterobacter, and Providencia). These bacterial isolates that exhibited different PGPR traits were selected and tested for their ability to tolerate different abiotic stress (NaCl, PEG8000, and pH). The majority of isolates were drought tolerant (60% of PEG8000) and had an optimal growth at high pH values (pH 9 and 11) and some strains tolerated 2 M of NaCl. Strains identified as Enterobacter xiangfangensis BE1, Providencia rettgeri BR5 and Pseudomonas stutzeri MLR6 showed high capacity of adaptation on their PGP traits. The salt-tolerant isolates were finally chosen to promote growth and enhance salt tolerance, separately or combined, of Arabidopsis thaliana (Col-0) exposed or not to 0.1 M NaCl, by following fresh and root weight, primary root elongation and lateral root number. The best bacterial effect was recorded for the MLR6 strain in increasing shoot fresh weight and for BE1 in terms of root fresh weight in the absence of salt stress. At stressed conditions, all growth parameters declined. However, inoculation of Arabidopsis thaliana with the three bacterial strains (MLR6, BE1 and BR5), single or in co-culture, conferred an increase in the shoot weight, primary root length and lateral root number. The use of these strains separately or combined as biofertilizers seems to be a powerful tool in the development of sustainable agriculture in saline soils.
1. Introduction
Salt stress is one of the most important threats to agricultural production and food security. For more than 3000 years, it has been a threat to agriculture in many regions of the world [1,2,3,4,5,6]. Globally, 22% of the total arable land and 33% of the total irrigated agricultural area are under salt stress. Therefore, 50% of the arable land area will be affected by salinity in the next few years [7]. According to the Food and Agriculture Organization of the United Nations (FAO) report on “The State of the World’s Soil Resources”, in more than 100 countries, approximately 1 billion hectares of soil are affected by salt problems [8]. Osmotic pressure leads to nutritional imbalance, morphological damage and reduced photosynthesis. This leads to a decline in growth and productivity, which can lead to cell death [3,9]. Like many other abiotic stresses, salt stress restricts plant growth by increasing soil osmotic potential and reducing water uptake through the roots. High salt concentration disturbs the steady state of water potential and the distribution of ions at the cellular level and throughout the plant. To cope with these limitations, plants have established several physiological and biochemical mechanisms to adapt [3,10]. In order to achieve salt tolerance, the damage must be mitigated; then, homeostatic conditions should be restored under the new stress conditions and, finally, growth must start again, but at a slower rate [3,11].
Halophytes account for about 1% of the world’s flora and can survive and reproduce naturally in saline soil. These plants have developed various mechanisms to respond to soil salinity, such as osmotic regulation, ion absorption and transport regulation [12,13]. Another mechanism for plants to alleviate salt stress is related to the beneficial effects of rhizosphere salt-tolerant microorganisms [2,4,14]. Salt-tolerant plant rhizosphere growth promoters (ST-PGPR) are becoming an effective biological tool to reduce the toxic effects of high salt concentration and promote plant growth, while repairing degraded saline soil [7,15]. ST-PGPR uses a series of direct or indirect mechanisms involved in improving salt stress in crop plants [4,16,17]. Studies have confirmed that ST-PGPR produces various types of plant hormones, such as auxin, gibberellin, cytokinin [18], synthesizes ACC deaminase [19,20], and produces extracellular polysaccharides [21,22] and osmoprotectants (proline, trehalose, and glycine betaine) [23,24]. They regulate plant defense systems and activate plant antioxidant enzymes [17]. In addition, different new genes in plants, such as SOS1, are regulated by PGPR, which activates the plant defense system to resist salt [7].
Several bacterial genera are recognized as PGPR growing under saline conditions. In addition to the genus Bacillus, which exists in a wide range of natural habitats [25,26], in accordance with its degree of physiological and genetic adaptability, other researchers have isolated other bacteria from saline environments, belonging to Gram-negative bacteria, such as Pseudomonas, Enterobacter, Providencia and Acenitobacter, and have been used to improve the productivity of various crops under saline conditions [16]. Pseudomonas is one of the most diverse bacterial genera found in soil, water, plants, etc. The rhizosphere of the plant is the most important ecological niche for Pseudomonas compared to bulk soil. Plant root exudates preferentially interact with Gram-negative bacteria over Gram-positive bacteria due to the composition of the cell wall and the composition of root exudates [27]. For the past two decades, Pseudomonas has been widely used in sustainable agriculture, as agents for promoting plant growth and in the management of biotic and abiotic stress.
However, in order to successfully apply PGPR in saline areas, the bio-inoculant must be isolated from the affected soil. The rhizosphere of halophytes is suitable for searching for bacteria capable of stimulating plant growth [4,28]. A salt-tolerant strain Pseudomonas sp. M30-35, isolated from the rhizosphere of Haloxylon ammodendron, a perennial C4 succulent xero-halophyte with exceptional drought and salt-tolerance abilities, was found to contain 34 genes possessing homology with genes associated with PGP traits and abiotic stress tolerance [29]. Recently, the genomes of many ST-PGPRs have also been sequenced, providing information on their salt tolerance and plant-growth-promoting attributes [30]. However, only a few reports have investigated the promoting effect of PGPRs isolated from halophytes on glycophyte growth under salt stress. Therefore, may halophyte-associated bacteria ameliorate salt tolerance in glycophytes?
In this study, we aimed to use ST-PGPR from halophytes collected in different arid and semi-arid regions in Algeria, as a future inoculant for crop production, in order to induce salt tolerance and augment crop performance in salt-dominated land. We managed to isolate endophytic, rhizospheric and non-rhizospheric bacteria from salsola tetrandra and to evaluate the PGP potential of the cultivated bacterial population. The selected bacteria, which tolerate different abiotic stress, were tested for their PGPR potential under NaCl stress. The salt-tolerant PGPR selected were qualified to improve the growth and resistance of Arabidopsis thaliana under salt stress conditions.
2. Materials and Methods
2.1. Site Location and Sampling of Plants
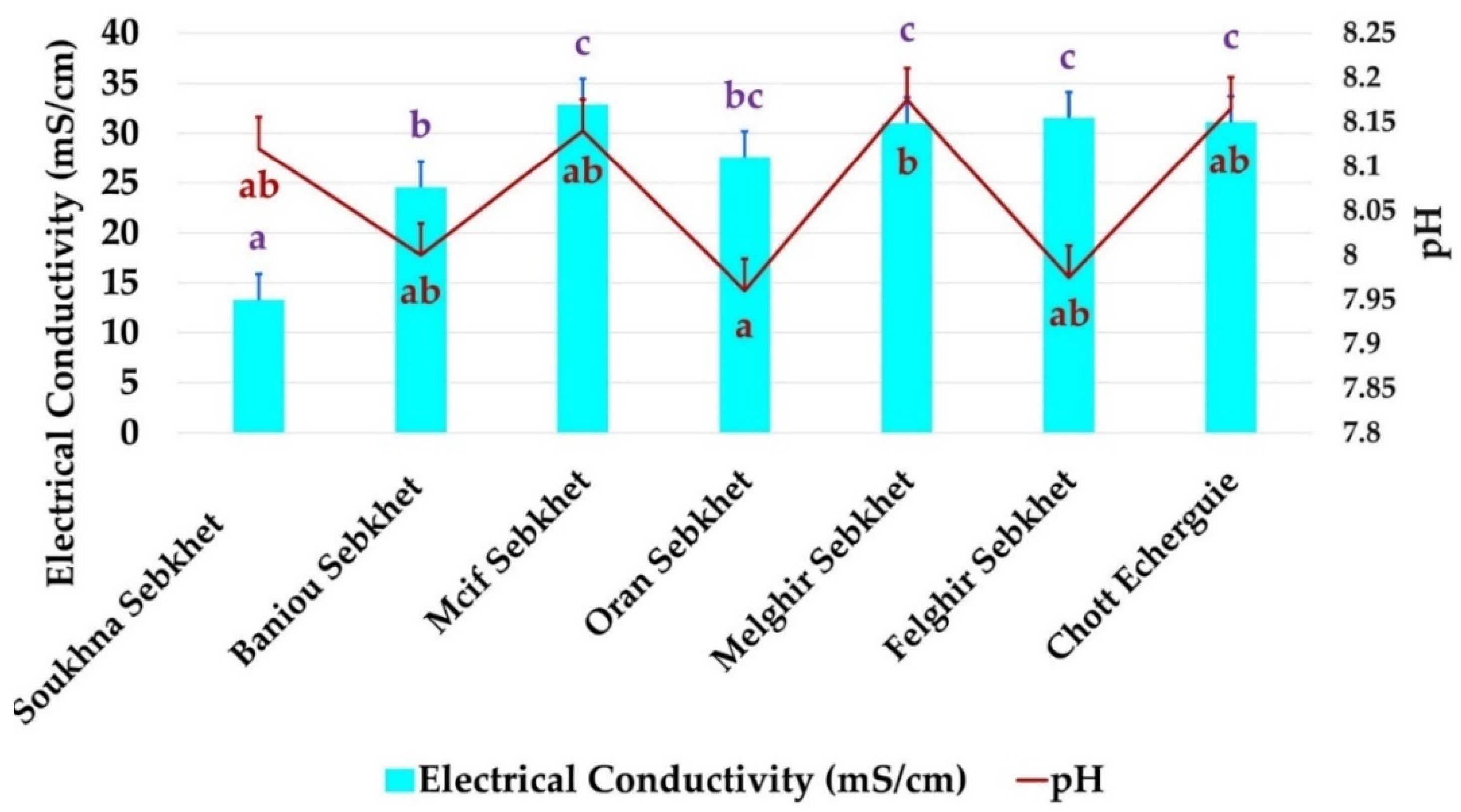
Soil samples were collected from diverse arid and semi-arid regions in Algeria (Figure 1). Based on their predominance in each physiographic region, the halophyte plants belonging to Salsola tetrandra species were used for the isolation of rhizospheric and endophytic bacteria. Two independent plants at last two meters away from each other were taken randomly and bulked to obtain a representative composite sample. Seven sites were sampled: Soukhna Sebkhet (SS) (N 35°55′, E 05°45′), Baniou Sebkhet (BS), (N 35°22.000′, E 4°32.513′), Mcif Sebkhet (SM), (N 35°22.000′, E 4°32.513′), Oran Sebkhet (OS), (N 35°, W 0°), Felghir Sebkhet (FS), (N 34°10.631′, E 6°17.322′), Melghir Sebkhet (MLS), (N 34°10.631′, E 6°17.322′) and Chott Echerguie (CE), (N 34°03.376′, W 0°05.164′). Soils and plants were placed into sterile bags and transported to the laboratory under controlled conditions. The chemical and physical properties of examined soil are given in Figure 2.
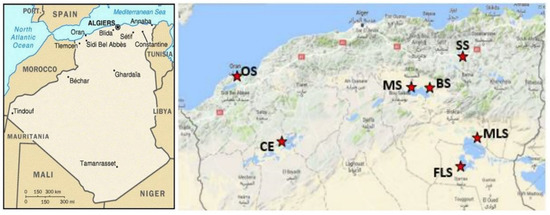
Figure 1.
Map of sampling locations. SS, Soukhna sebkhet; BS, Baniou sebkhet; MS, Mcif sebkhet; OS, Oran sebkhet; MLS, Melghir sebkhet; FLS: Felghir sebkhet; CE, Chott Echerguie.
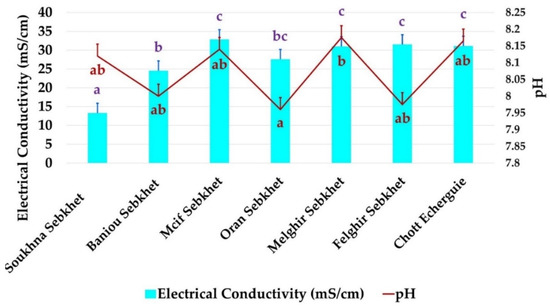
Figure 2.
Soil chemical and physical properties of the seven Sebkhet and Chott sampled. Data present mean ± standard error. Bars labelled with different letters are significantly different among the treatments at p < 0.05 using Tukey’s honest significant difference (HSD) test. In each bar groups, bars labelled with the same letter are not significantly different from each other according to Tukey’s HSD at p < 0.05. (Letters with Blue and red colors belong to EC and pH, respectively.)
2.2. Isolation of Cultivable Bacteria
Bacterial isolation from soil samples was performed according to the method described by Cherif-Silini et al. [2]. Ten grams of each soil sample (rhizospheric and non-rhizospheric soil) was transferred into 250 mL Erlenmeyer flasks containing 90 mL of sterile NaCl (9 g/L) solution. The mixture was then shaken for 30 min at approximately 150 rpm. Serial dilutions of each suspension were made. Aliquots of 0.1 mL from each dilution were spread in triplicate on Petri dishes containing King’s B and Tryptic soy agar media. To isolate the endophytes, the roots were washed through a series of sterile distilled water to remove all rhizospheric particles. They were then disinfected by immersion in ethanol 70% for 5 min and in sodium hypochlorite solution (2%) for 30 min followed by several washes with sterile distilled water. To check the efficiency of root disinfection, the final washing water was spread on nutrient agar and the plates were incubated at 30 °C for 48 h. Surface disinfected roots (1 g) were then ground in 10 mL sterile NaCl (9 g/L) solution. Appropriate serial dilutions of root homogenate samples were made and 0.1 mL from each dilution was spread in triplicate on Petri dishes containing the King’s B (KB) and Tryptic soy agar (TSA) media. Numbers of colony-forming units (CFU) per gram of soil/root were determined after 48 h at 28 ± 2 °C. A total of 76 isolates were collected and the colonies showing morphological differences were selected and spread on agar media to obtain pure colonies. Pure isolates were named according to the sampling sites (Figure 1) and sampling compartment (N: Non-rhizospheric, R: Rhizospheric and E: Endophytes) followed by a code number. These isolates were subjected to morphological examination based on macroscopic and microscopic characteristics (cell form, arrangement, Gram). All isolates were stored at −80 °C in 20% glycerol.
2.3. Plan-Growth-Promoting Traits of the Bacterial Isolates
2.3.1. Quantification of Indole-3-Acetic Acid (IAA)
IAA activity was quantified with colorimetric method as described in [31]. For this, isolates were cultured in 10 mL of Luria–Bertani (LB) broth supplemented with L-tryptophan (5 mM) in triplicate and incubated at 28 °C/48 h. The bacterial cultures were then centrifuged for 15 min at 10,000 rpm. Next, 1 mL of the supernatant was mixed with 2 mL of Salkowski’s reagent (1 mL 0.5 M FeCl3 in 50 mL of 35% HClO4) and incubated in darkness for 30 min. Development of pink color indicated IAA production and the absorbance was read at 530 nm. IAA concentration in the supernatant was determined using a calibration standard curve of pure IAA (Sigma-Aldrich, St. Louis, MO, USA) [32].
2.3.2. Phosphate Solubilization
Solubilization of tri-calcium phosphate was detected in Pikovskaya’s agar medium [33]. Each isolate was spotted in triplicate on the surface medium and phosphate solubilizing activity was estimated after seven days at 28 °C. The development of the clear zone around bacterial colony indicated positive phosphate solubilization activity.
2.3.3. Siderophores Production
Siderophores activity was detected according to the method of Pérez-Miranda et al. [34]. The isolates grown in LB broth medium were spotted in KB agar medium in triplicate at 28 °C/24 h. Then, 15 mL of CAS agar medium described by Schwyn and Neilands [35] was applied over the KB agar medium containing cultivated isolates. The development of orange halo around bacterial colonies indicated siderophores production.
2.3.4. NH3 Production
Bacterial isolates were tested for the production of ammonia by inoculation of fresh cutlers in 10 mL of peptone water (Peptone: 10 g/L, NaCl: 5 g/L, Na2HPO4: 3.5 g/L, K2HPO4: 1.5 g/L) in triplicate. A drop (0.5 mL) of Nessler’s reagent was added in each tube after 48 h of incubation at 28 °C. Development of brown to yellow color indicated a positive test for ammonia production [36].
2.3.5. Hydrogen Cyanide Production
Hydrogen cyanide (HCN) production from glycine was detected according to the method of Bakker and Schippers [37]. The bacterial isolates were streaked in TSA medium supplemented with glycine (4.4 g/L) in triplicate. A Whatman filter paper No. 3 soaked in picric acid and sodium carbonate Na2CO3 (0.5 and 2%, respectively) was placed in the lid of the plate. Plates were sealed with parafilm and incubated at 28 °C for 4 days. Cyanogenesis activity was revealed by observing change in filter paper colors from yellow to orange-brown.
2.3.6. Nitrogen Fixation
N-fixation ability was detected by observing the growth on N-free semi-solid Jensen’s medium in triplicate. Plates were incubated at 28 °C for 7 days and then isolates were re-inoculated in the same medium. Bacterial growth on medium surface indicated positive activity for atmospheric nitrogen fixation.
2.3.7. Cellulase Production
The ability of isolates to degrade cellulose was performed in carboxymethyl cellulose (CMC) medium [38]. In brief, 5 µL of each isolate was inoculated in plates with the medium, in triplicate. After 48 h of incubation at 28 °C, plates were stained with iodine for 5 min and observed for clear zone around the culture.
2.3.8. Chitinase Production
The isolates were characterized for chitinase activity on colloidal chitin agar medium following the method of Murthy and Bleakley [39]. Chitin agar plates were inoculated with freshly grown bacteria and incubated at 28 °C for 7 days in triplicate. Chitinase activity was detected by formation of a clear zone.
2.3.9. Protease Production
Protease activity was assessed on skim milk (10%) agar plate. After inoculation with 5 µL of each isolate the plates were incubated at 28 °C for 48 h in triplicate. Protease activity was detected as a clear zone around bacterial colony [40].
2.3.10. Antagonistic Effects
Thirty-six isolates (36) reported positive for siderophore; HCN, NH3, cellulase, chitinase and protease production were tested for antagonistic activity against Fusarium oxysporum, Alternaria alternata, Phytophthora infestens and Aspergillus niger provided by the Laboratory of Applied Microbiology of Setif University. The pathogens were grown on potato dextrose agar (PDA) medium at 28 °C for dual culture bioassay. The Selected isolates grown in LB broth were inoculated in PDA plates. A 5-mm five-day-old disc for each fungal pathogen was placed in the center of the plate inoculated with bacterial isolate. Plates were incubated at 28 °C for 7 days and pathogen inhibition percentage was calculated using formula percentage of inhibition = (R1 − R2)/R1 × 100, where R1 = radial growth of pathogen in control; R2 = radial growth of pathogen in dual culture experiments with antagonists.
2.4. DNA Extraction, PCR Amplification of ITS-rDNA Region, 16S-rDNA and Sequencing
The isolated bacterial cultures were tested for their reaction to Gram staining. Among 76 bacterial isolates, 51 were Gram-negative and were selected for further culture independent analysis. Genomic DNA extraction of selected isolates was performed by suspending isolated colonies in TE buffer (1 mM of Tris-HCl and EDTA 10 mM pH 7.4) in a micro-centrifuge tube. These cells were heated for 10 min at 100 °C and were centrifuged at 13,000 rpm for 5 min at 4 °C. The supernatant was used as template DNA for the amplification of intergenic transcribed spacer (ITS) region and 16S-rRNA gene. The bacterial collection was de-replicated by fingerprinting analysis of the rRNA 16S–23S intergenic transcribed spacer (ITS) region, using universal primers ITSR (5′…CAAGGCATCCACGT…3′), ITSF (5′…GTCGTAACAAGGTAGCCGTA…3′). Each 25 mL reaction contained 100 ng template DNA, 25 mM/L concentrations of each dNTPs, 25 mM/L MgCl2, 5 U/μL of Taq DNA polymerase and 25 mM/L concentrations of each primer. The PCR thermal cycling conditions were set as initial denaturation at 94 °C for 3 min and 35 cycles of 94 °C for 45 s (DNA denaturation), 55 °C for 1 min (primer annealing) and 72 °C for 2 min (DNA extension), with a final extension at 72 °C for 7 min in a DNA thermal cycler (Bio-Rad). PCR products (2 µL) were checked by electrophoresis in 1.5% agarose gel and stained with ethidium bromide. Gel images were captured using Gel Doc 2000 system (Bio-Rad, Tunis, Tunisia), and bacteria redundancy was reduced by evaluating the different ITS profiles. Strains exhibiting the same band patterns were grouped in the same ITS haplotype [41]. One representative strain from each group has been selected for subsequent identification using 16S-rRNA genes sequencing. Seventeen representative isolates of all detected haplotypes were further characterized by 16S-rRNA gene sequencing and phylogenetic analysis.
The amplification of the 16S-rRNA gene was performed with the primers 16SR (5′…CTACGGCTACCTTGTTACGA…3′) and 16SF (5′…AGAGTTTGATCCTGGCTCAG…3′) using the same PCR protocol described above. The PCR products were separated by electrophoresis in 1.5% agarose gel, stained with ethidium bromide (0.5 µg/mL) and visualized in UV light. Next, 60 ng of each product was used as template for sequencing reactions, which were performed by the LMBA laboratory of the University of El Mannar, Tunis. Nucleotide sequences were analyzed by BLAST search and compared against bacterial 16S-rDNA sequences available in the Gene bank data base [42]. The sequences were aligned and subjected for construction of neighbor joining phylogenetic tree using (MEGA) software, version 6. The nucleotide sequences were submitted in the GenBank data base (https://www.ncbi.nlm.nih.gov/genbank/, accessed on 10 December 2021) under accession numbers.
2.5. Stress Tolerance of Gram-Negative Isolates
Seventeen Gram-negative representative isolates were selected to study their growth ability under NaCl, PEG8000 and pH stress. The tolerance of the selected strains was examined on microplate using TSB medium supplemented with NaCl (0 to 2.2 M), PEG8000 (from 0 to 70%) and having different pH values (4, 7, 9 and 11), respectively. The microplate wells were inoculated with bacterial culture prepared in TSB medium. The tests were performed in duplicate. The growth of the bacteria was estimated after 48 h of incubation at 28 °C with a microplate reader (Multiskan GO, version 1.00.40).
2.6. Selection for Salt-Tolerant PGPR
Five selected isolates that tolerated various studied abiotic stresses were screened to evaluate their ability to produce PGP traits under salt stress. The selected strains were tested quantitatively for phosphate solubilization according to the method of Nautiyal [33] modified by adding the various NaCl concentrations (0–0.1–0.2–0.3 M of NaCl). After 7 days of growth, culture was centrifuged and supernatant was used to determine P-solubilization through the colorimetric method of Fiske and Subbarow [43].
Siderophores production was determined in KB liquid medium supplemented with the same NaCl concentrations incubated at 28 ± 2 °C for 72 h and the resulting cultures were centrifuged at 5000 rpm for 30 min. Following this, 500 μL of the supernatant was mixed with 500 μL of Chrom Azurol S liquid (CAS) medium [35]. The OD was measured at 630 nm after 20 min of incubation and siderophore units were calculated as follows:
% siderophore unit = (Absorbance of reference (Ar) − Absorbance of test (As))/(Absorbance of reference (Ar)) × 100
The ability of the isolates to produce IAA was determined by the method of Bric et al. [31] described above but modified by adding graded series of NaCl concentrations. All tests were performed in duplicate.
2.7. In Vitro Inoculation Assays of Arabidopsis thaliana Seedlings under Salt Stress
2.7.1. Bacterial Strains
Three salt-tolerant PGPRs (BE1, BR5 and MLR6) were selected based on the results of their growth performance under salt stress. The isolates were further evaluated for their PGP effect on Arabidopsis thaliana seedlings.
2.7.2. Plant Material and Plant Growth Conditions
Seeds of Arabidopsis thaliana (ecotype Col-0)/Rédei-L211-497) (LEHLE seeds Round Rock, TX, USA) were surface-sterilized by soaking in 70% ethanol for 2 min followed by soaking in 1% sodium hypochlorite for 20 min, after those seeds were rinsed 4 times in sterile distilled water. The sterilized seeds were placed in Petri plates containing half-strength Murashige and Skoog (MS) salt medium [44] supplemented with 1.5% sucrose and solidified with 0.8% agar. Seeds were vernalized for 2 days at 4 °C in the absence of light and then incubated in a plant growth chamber (16 h light/8 h darkness photoperiod and 23 ± 1 °C). Germinated seedlings were transferred after 6 days to plates for the experimental uses described below.
2.7.3. Plant Inoculation
Germinated seedlings were transferred to Petri plates (6 seedlings per plate) containing MS agar with or not 100 mM NaCl. Then, 100 μL of 108 CFU/mL bacterial culture suspensions was applied on TSA medium below the seedlings. Five treatments were applied (Control, MLR6, BE1, BR5 and MLR6 + BE1 + BR5) under 0 and 100 mM NaCl and each treatment was performed in triplicate. Plates were incubated for 14 days (23 ± 1 °C, 16 h/8 h light/darkness).
2.7.4. Measurement of Morphological Parameters
Arabidopsis thaliana seedlings were collected and their growth evaluated by estimation of root and shoot fresh weight (g), primary root length (cm), and lateral root number.
2.8. Statistical Analysis
The statistical analysis of the data was performed using analysis of variance (ANOVA) and when significant effects were detected, the groups were compared using a post-hoc Tukey’s HSD test. The level of significance used for all statistical tests is 5% (p < 0.05). The statistical program used was IBM SPSS Statistics v.22.
3. Results
3.1. Soil Chemical and Physical Properties
All the sites chosen were considered hyper saline. The average values of electrical conductivity (Ec) varied from 11.29 ds/m in sebkhet Soukhna to 32.5 ds/m in sebkhet Mcif. The soils of sebkhet Mcif, Melghir, Felghir, Chott Echerguie were the most saline. The salinity of the soil of sebkhet Baniou and Oran seemed close (Figure 2). All soils were alkaline, varying from pH 7.94 to 8.20. The soils of sebkhet soukhna, Baniou, Mcif, felghir and Chott Echerguie had similar pH values. Oran sebkha had the lowest pH value, 7.94, while Melghir sebkhet had the highest pH value, 8.20.
3.2. Bacterial Enumeration
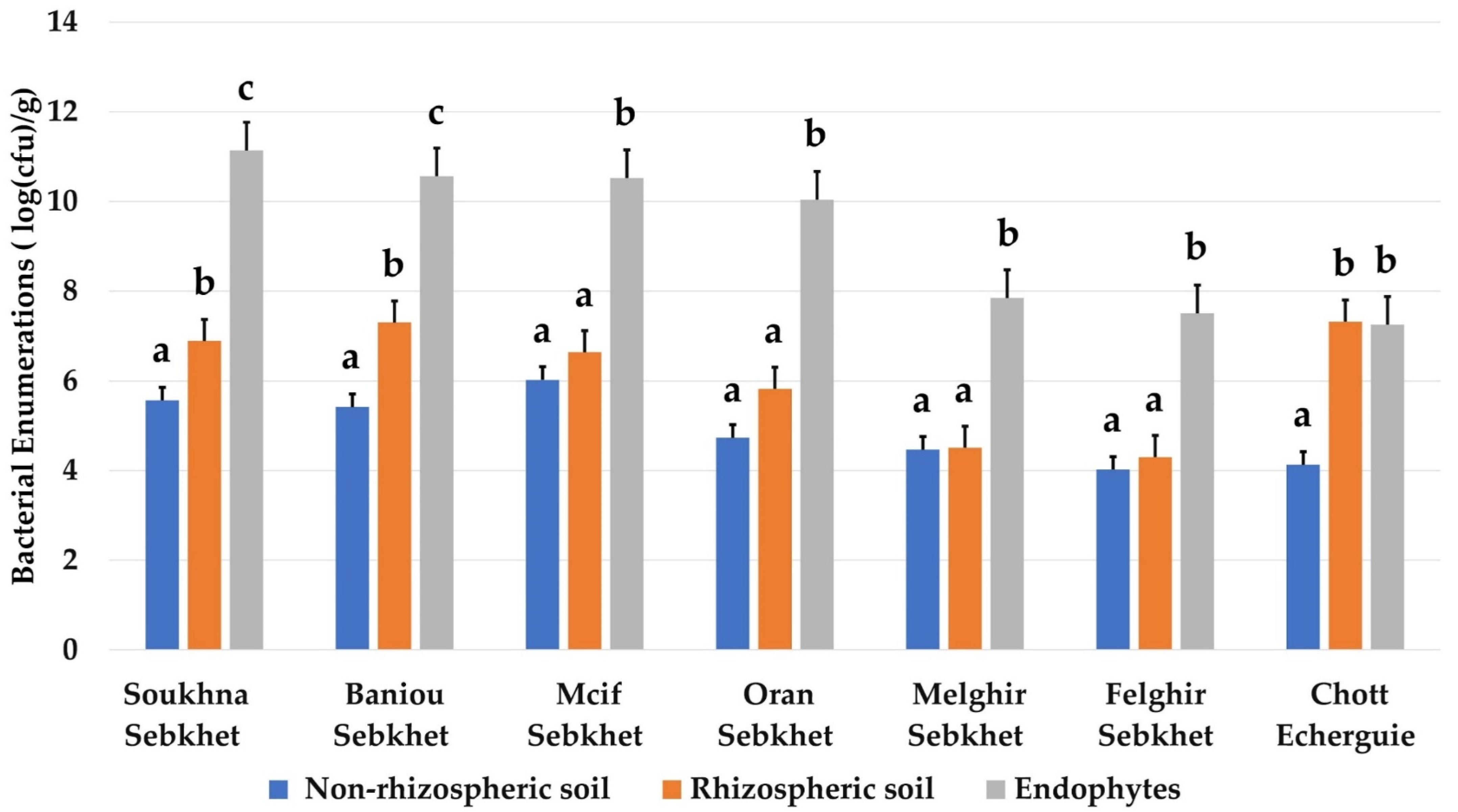
The average densities of endophytic bacteria were significantly higher compared to those of rhizospheric and non-rhizospheric soil in all studied sites (Figure 3). The number of endophytic bacteria was remarkably higher in Soukhna and Baniou sebkhet, which were characterized by the lowest values of Ec (11.29–23.5 ds/m). The sebkha soils of Mcif, Oran, Felghir and Melghir presented the same profile of isolated bacteria in endophytic, rhizospheric and non-rhizospheric samples. However, the extreme site of Chott Echerguie presented a remarkable number of rhizospheric bacteria, despite its high salinity (32.5 ds/m). It seemed that there was no relation between soil salinity and bacterial population across the studied areas.
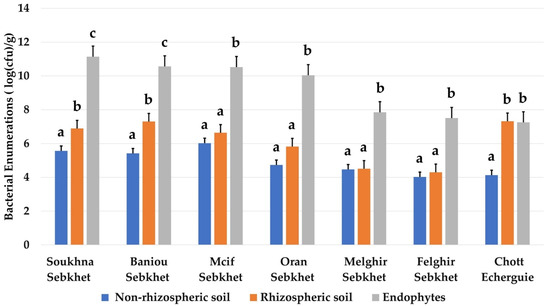
Figure 3.
Bacterial enumerations (log(cfu)/g) from different habitats (non-rhizospheric soil, rhizospheric soil, and endophytes) in the seven locations. Data present mean ± standard error. Bars labelled with different letters are significantly different among the treatments at p < 0.05 using Tukey’s HSD test. In each bar groups, bars labelled with the same letter are not significantly different from each other according to Tukey’s HSD at p < 0.05.
3.3. Plant-Growth-Promoting Traits
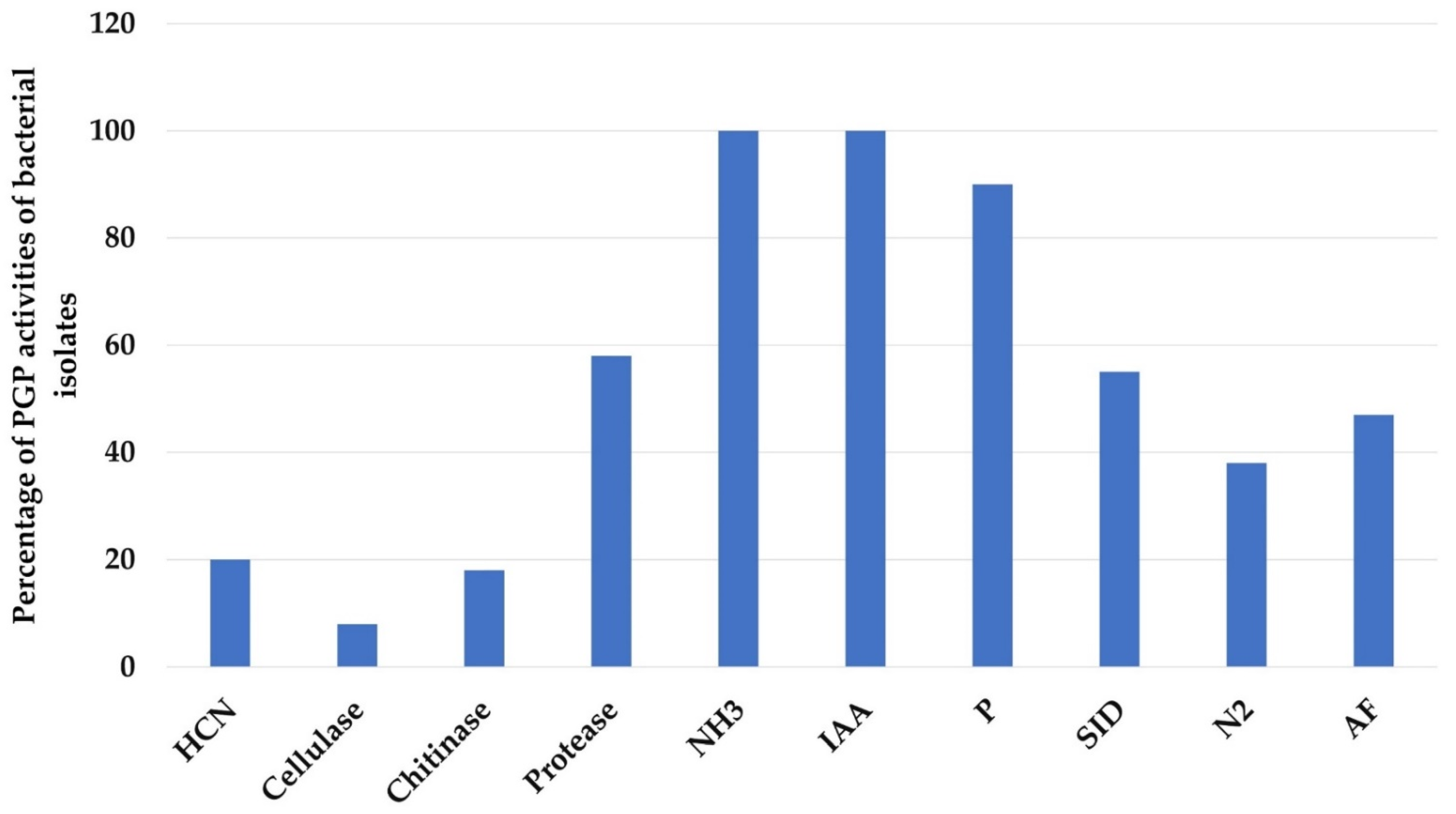
The percentage of isolates exhibiting the different PGP activities is shown in Figure 4. All 76 isolates (100%) were IAA and NH3 producers. Further, 62 strains (82%) were phosphate solubilizers and 49 strains (64%) were siderophores producers. Of the 76 isolates, only 29 (38%) were able to fix atmospheric N2. Among those, 14 (18%) and 15 (20%) of the isolates produced chitinase and HCN, respectively, but only 6 isolates (8%) exhibited cellulolytic activity. In contrast, more than half of the isolates, 44 (58%), appeared to be protease producers and 47% of the isolates exhibited antagonistic activity.
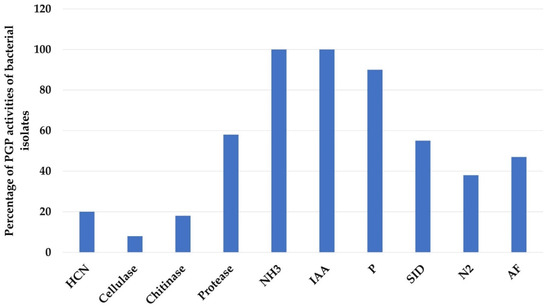
Figure 4.
Percentage of plant growth promoting (PGP) activities of bacterial isolates.
3.4. Identification of Strains and Phylogenetic Analysis
Phylogenetic analysis based on the 16S rDNA sequence showed that the 17 isolates were grouped into 4 genera of Pseudomonas, Acinetobacter, Enterobacter and Providencia. Isolates belonging to the genera Pseudomonas fell into twelve species: Pseudomonas azotoformans P8 (KX611495), Pseudomonas koreensis SN5 (KU577964), Pseudomonas straminea MR2 (KU577961), Pseudomonas sp. MN8 (KU577945), Pseudomonas sp. BR4 (KU577953), Pseudomonas plecoglossicida MR1 (KU577960), Pseudomonas stutzeri MLR6 (KU577948), Pseudomonas stutzeri CR6 (KU577968), Pseudomonas lini SE1 (KU577951), Pseudomonas sp. CR5 (KX611491), Pseudomonas psychrophila ON6 (KU577957) and Pseudomonas trivialis SR1 (KU577950). Isolates belonging to Acinetobacter were assigned to two species, namely, Acinetobacter sp. FLR3 (KU577963) and Acinetobacter johnsonii MN6 (KU577959). Two species of Providencia were identified as Providencia vermicola ME1 (KU577949) and Providencia rettgeri BR5 (KU577954). One isolate of Enterobacter was identified as Enterobacter xiangfangensis BE1 (KU577955). The nucleotide sequences derived from 16S-rDNA have been submitted to GenBank (Figure 5).

Figure 5.
Maximum likelihood phylogenetic tree of seventeen Gram-negative bacteria. Xanthomonas floridensis WHRI 8848 was used as outgroup. Supports for branches were assessed by bootstrap resampling of the data set with 1000 replications.
3.5. Effect of NaCl, PEG8000 and pH on Bacterial Growth
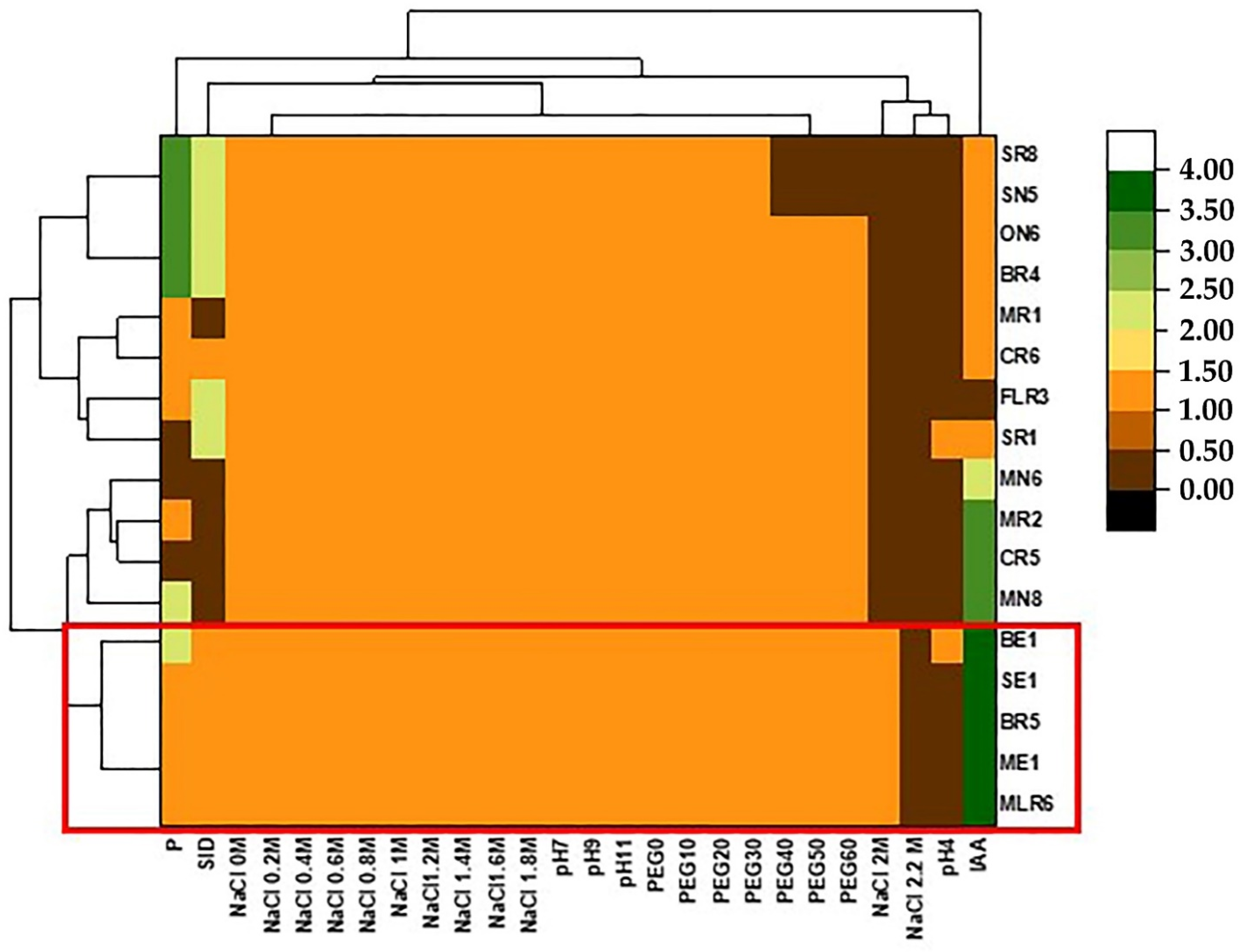
Bacterial growth at different NaCl, PEG8000 and pH concentrations is shown in Figure 6. Almost all of the isolates were able to grow at NaCl range (0–1.8 M). However, four strains (MLR6, SR1, BE1, and SE1) were tolerant at 2 M. The majority of isolates were drought tolerant and grew at the range of 0–60% of PEG8000. Only three strains (SN5, SE1, SR8) could not tolerate up to 30% PEG8000. All studied isolates showed optimal growth at high pH values (pH 9 and 11) and among them, two strains (ME1 and BE1) were able to grow at pH 4. In order to select the performant bacteria, the high amounts of stress tolerance and the results of the PGP characteristics (phosphate solubilization, production of IAA, and siderophore) of the strains were considered. Thus, the strains BE1, ME1, MLR6, BR5, and SE1 (Figure 6) were chosen to study whether their PGP activities remained high under different NaCl concentrations.
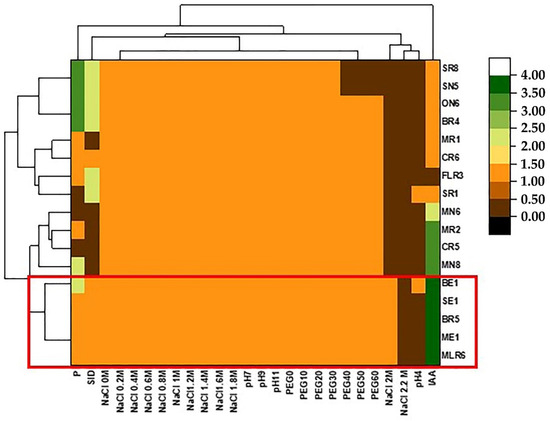
Figure 6.
Heat map showing the response of the isolates to different stress tolerance (NaCl, pH, and PEG) and the expression of their PGP activities (IAA, P, and SID). The bacterial isolates (BE1, BR5, ME,1 SE1, and MLR6) having the best capacities are highlighted with the same colors.
3.6. Effect of NaCl on Plant-Growth-Promoting Traits
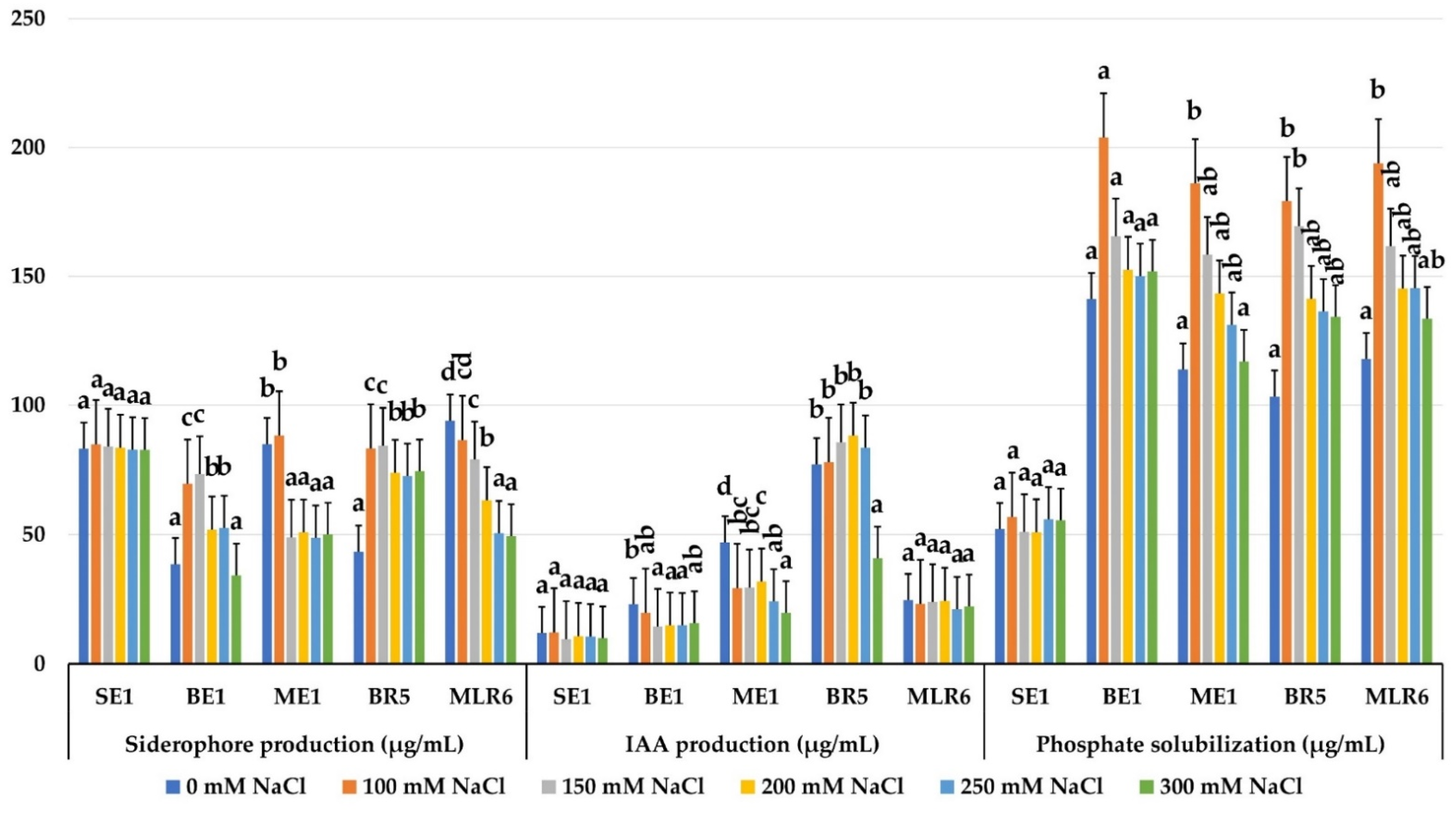
The five selected strains (BE1, ME1, MLR6, BR5 and SE1) were able to produce IAA, siderophores and solubilize phosphate in the absence and presence of salt stress. The amount of Ca3(PO4)2 solubilized by the strains was not affected by NaCl concentrations. The amount of soluble phosphate ranged from 52.00 ± 3.50 to 193.90 ± 21.16 μg/mL. The BE1, MLR6, ME1 and BR5 strains showed a great ability to solubilize inorganic phosphate, with an average amount of 162.52–149.61–141.64, and 144 μg/mL, respectively. A low solubilization was observed for the SE1 strain (53.62 μg/mL) (Figure 7). All selected strains produced IAA in significant amounts under salt stress. The amount produced by BR5 (73.88 μg/mL) was the highest, followed by ME1 (30.15 μg/mL) and MLR6 (23.12 μg/mL) (Figure 7) and the production rate was not modified compared to the control without salt. The SE1 strain produced the best rate of siderophores, with an average of 83.51%, and this activity remained stable at all salt concentrations tested (Figure 7). The BR5 and MLR6 strains produce 71.94% and 70.41%, respectively, and remain productive, despite high NaCl concentrations. However, the activity of ME1 and BE1 strains was affected by salinity. Their production decreases in the presence of salt. Based on this result, the BE1, BR5 and MLR6 strains were chosen for further inoculation testing.
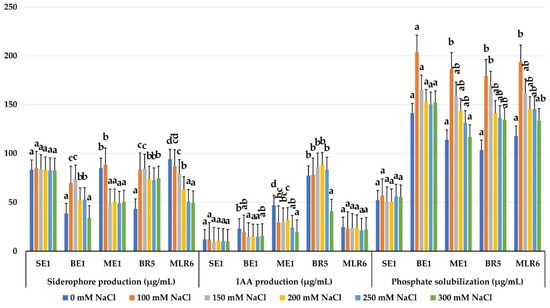
Figure 7.
Effect of salinity on phosphate solubilization, IAA production, and siderophore production of selected bacterial isolates. Data present mean ± standard error. Bars labelled with different letters are significantly different among the treatments at p < 0.05 using Tukey’s HSD test. In each bar groups, bars labelled with the same letter are not significantly different from each other according to Tukey’s HSD at p < 0.05.
3.7. Arabidopsis Thaliana Inoculation
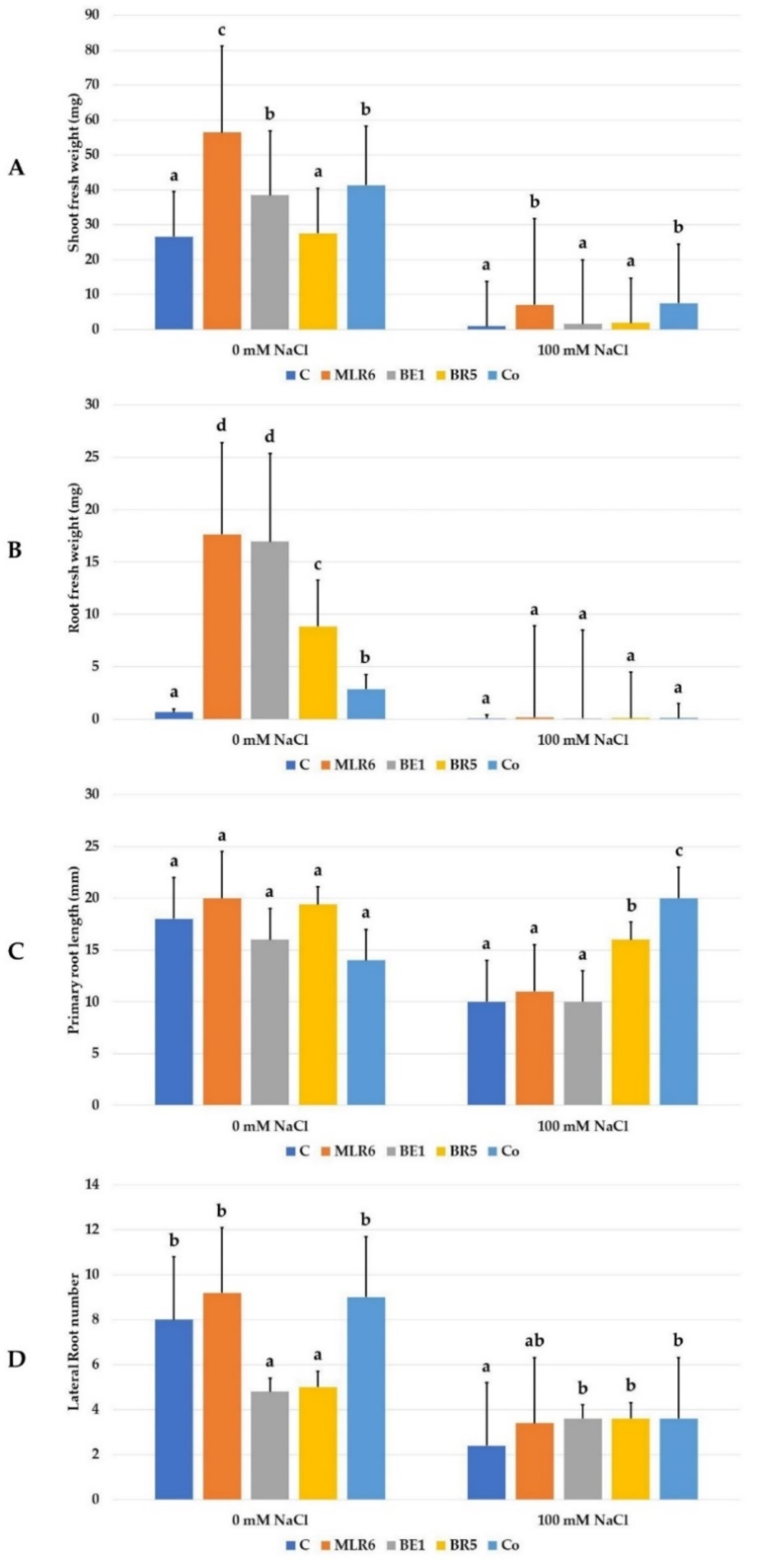
The inoculation of A. thaliana with the three selected strains and their coculture (MLR6, BE1, BR5, and MLR6 + BE1 + BR5) was used to assess their capacity to improve growth under 0 and 100 mM of NaCl (Figure 8). Analysis of morphological parameters of the plants, fresh weight, root weight, primary root elongation, and lateral root number (Figure 9) showed a significant decrease in these different growth parameters under salinity stress. Bacterial inoculation significantly improved the shoot and root fresh weight (Figure 9) of A. thaliana in the absence of NaCl stress compared to non-inoculated plants. The best bacteria effect was recorded to MLR6 for increases in shoot fresh weight (Figure 9) and both MLR6 and BE1 for root fresh weight (Figure 9). Any significant enhancement was observed on root length and lateral root number in inoculated plants compared to control. However a negative effect from the bacteria was observed in plants inoculated with BE1 and BR5 on lateral root number (Figure 9). Under saline conditions, the plants inoculated with the MLR6 strain and the co-culture showed significant enhancement in shoot weight, as compared to control plants (Figure 9). A similar increase in primary root length was observed in plants inoculated with both BR5 and the co-culture, which noted the better response (Figure 9). The MLR6, BR5 and BE1 and their co-culture improved lateral root number in inoculated plants compared to control plants (Figure 9). However, they did not have any significative enhancement in root fresh weight.

Figure 8.
In vitro effect of bacterial isolates on Arabidopsis seedlings growth exposed to NaCl (0, 100 mM) concentrations.
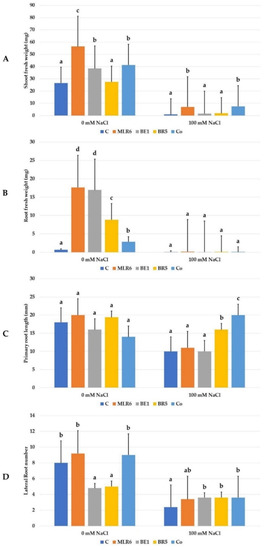
Figure 9.
Effect of bacterial isolates on (A) shoot fresh weight (mg), (B) root fresh weight (mg), (C) primary root length (mm), and (D) lateral root number of Arabidopsis seedlings exposed to NaCl (0, 100 Mm) concentrations. Data present mean ± standard error. Bars labelled with different letters are significantly different among the treatments at p < 0.05 using Tukey’s HSD test. In each bar groups, bars labelled with the same letter are not significantly different from each other according to Tukey’s HSD at p < 0.05.
4. Discussion
Soil salinization is a serious stress condition and also land-degradation problem in arid and semi-arid regions, causing major problems for crop productivity [45]. Salt induces osmotic and ionic stress in plants, leading to nutrient imbalance, morphological damage and reduced photosynthesis, resulting in decreased growth/productivity and death [9]. Abnormal physiological functions in plants affected by salt can be reprogrammed through plant-growth-promoting rhizobacteria (PGPR) interactions, thereby increasing the rate of photosynthesis, plant growth regulators, antioxidants and compatible solutes, and regulating the expression of stress-responsive genes and proteins [46]. Inoculating seeds of various crops with PGPR can increase root and shoot growth, dry weight, fruit and seed yield, and enhance plant tolerance to salt stress [16]. However, in order to successfully apply PGPR in saline areas, the inoculant should be isolated from indigenous soil affected by salt. Microorganisms in the rhizosphere of halophytes in saline soils may provide valuable resources for improving the salt tolerance of crops [47]. In the present study, seventy-six (76) strains of endophytic, rhizospheric and non-rhizospheric bacteria associated to the Salsola tetrandra halophyte were isolated from seven sebkha in Algeria. The physico-chemical characteristics of the sampling sites showed that these soils were highly saline, with Ec > 4 dS/m ranging from 11.29 to 35.5 dS/m. Based on salinity norms, the studied soils are saline. They have basic pH values (7.97 to 8.20), which are compatible with that of natural saline environments.
We showed that halophytic root samples presented the highest abundance of bacteria compared to rhizospheric and non-rhizospheric samples. The abundance of the endophytic bacteria population might be due to its location inside the roots, protecting them from salt constraints. Forchetti et al. [48] revealed that the endophytic bacteria benefits by sheltering them from environmental stress and competition with other microbes.
Most of our isolates have the ability to dissolve phosphate, produce IAA, siderophore, and fix N2. IAA produced by PGPR isolates is an important attribute for improving plant growth. This hormone helps to promote root growth, which could explain the adaptation of plants to extremely hard soils, which is conducive to root anchoring and acquisition of water and minerals [49]. IAA-producing bacteria are also involved in alleviating salinity-induced dormancy, showing a highly stimulating effect on root and shoot length [50].
One of the main factors limiting plant growth is the low bioavailability of phosphorus and iron in soils (Gamalero et al. [51]). Therefore, bacteria that can dissolve and mineralize phosphor and synthesize siderophores have great potential as biological fertilizers. In our work, most of the strains (82%) were able to solubilize inorganic phosphate in media, where production of siderophores was observed in 64% of total strains. The ability to produce siderophores is a PGPR property and is also considered a biocontrol property, as iron chelation helps restrict the amount available to pathogens.
About 18% of the isolates appear to possess chitinolytic activity. In contrast, only 8% exhibited cellulolytic activity and more than half of the isolates (58%) seem to be protease producers. All selected isolates proved antifungal, according to their inhibitory effect. These lytic enzymes are another trait associated with PGPR, enabling them to limit fungal pathogen growth. In vitro studies showed that the exposure of selected plant pathogenic fungi to lytic enzymes, such as chitinase, protease, gluconase or cellulase, can result in degradation of the structural matrix of the fungal cell wall [52]. It has been demonstrated that indigenous desert microorganisms promote plant health in desert agro-ecosystems via an antagonist potential towards phytopathogens [53].
In this study, phylogenetic analysis showed the abundance of Gram-negative bacteria in the sampled sites. This result could be explained by the culture conditions chosen in this research; in particular, the King B culture medium is recommended for the isolation of Pseudomonas. In addition, soil salinity reduces soil diversity and microbial activity due to the decrease in organic matter. Thus, soils containing intermediate levels of salinity harbor higher amounts of bacteria than fungi. Studies have reported that total bacteria populations and Gram-negative ones have not changed significantly in salinity, while total fluorescent pseudomonad populations apparently increased. The literature suggested that salt is one of the major problems in agriculture and affects the growth of soil microflora [54]. The abundance of Pseudomonas species is remarkable in our study, similarly to what has been reported by many cases, in both the soil environment and inside the plant [55]. Pseudomonas species found in saline soil include P. aeruginosa, P. fluorescens, P. putida, P. stutzeri, P. mendocina, P. mallei, and P. diminuta [56]. Hu et al. [57] reported that the majority of bacterial species belong to Gram-negative bacteria, especially to Pseudomonas spp., accounted for most of the microorganisms detected that can tolerate saline soil. P. putida was shown to reduce salt-stress-induced damage in citrus plants through several PGP traits, such as lowering abscisic acid (ABA) and salicylic acid (SA) levels, increasing accumulation of IAA in the leaf and inhibiting accumulation of root chloride and proline and reducing photosystem II (Fv/Fm) efficiency during salt stress [58]. Acinetobacter strains also show potential to act as PGPRs, particularly under stressed environmental conditions, where the iron, phosphate, or zinc sources may be limited [59]. Providencia vermicola strains were also found to be promising in phosphate solubilization and auxin biosynthesis, in addition to their considerable acetylene reduction capacity [60]. Bacteria of the genus Enterobacter has been associated with numerous biological models. The Enterobacter spp. strain EJ01, isolated from Dianthus japonicus thunb (China sea rose), was described as a bacterium capable of aiding vegetative growth, besides alleviating salt stress in tomato and Arabidopsis [61].
The abundance of these taxonomic groups over others in our study may be related to their ability to tolerate salinity. Further, these isolated bacteria could be less dependent on other factors determining their abundance, such as oxygen availability [50], mineralization, ionic type or hydrocarbon content [56,62,63].
Culture media containing different NaCl, PEG and pH concentration were performed to select the best stress-tolerant strains. The five strains showed high capacity for tolerance to the various tested stress. The high level of tolerance may be explained by the fact that they were isolated directly from a soil region known for its salinity. MLR6, SR1, BE1 and SE1 strains were tolerant at 2 M. Bacteria isolated from a saline environment are more likely to survive inhibitory salt concentrations than other bacteria inhabiting non-saline regions. The growth of these isolates with a high tolerance to salt stress can be explained by their cellular machinery, which is adapted to such conditions [64].
An important factor to take into account when selecting new tolerable isolates is to test their PGPR activities under the same conditions in which they will be applied. Our results revealed that the five selected strains can produce IAA with significant amounts across the NaCl range tested, where the BR5 strain (73.88 μg/mL) recorded the highest level. On the other hand, the strains BE1 162.52 μg/mL, MLR6 149.61 μg/mL, ME1 41.64 μg/mL, BR5 144 μg/mL showed a high ability to solubilize inorganic phosphate, while the SE1 strain was the most siderophore productive, with an average of 83.51%, and its activity remained stable in all tested salt concentrations. The ability of these strains to exhibit their PGP activities under high levels of NaCl could be an important characteristic for them to improve plants growth under similar saline conditions [6]. Therefore, they can be recommended as biological fertilizers or biocontrol agents to promote plant growth and to reduce production costs and pollution with chemical fertilizers [4].
In our work, three selected PGP strains (MLR6, BE1, BR5) were used to study the effect of stress alleviation and growth promotion of the A. thaliana plant under salt stress. The results showed that all growth parameters were severely decreased in uninoculated plants under stressed conditions. This reduction is in line with the results of many authors working on different plants [2,6,65,66]. It is well known that salt stress induces metabolic perturbation in plants, involving a broad spectrum of metabolic pathways, in both primary and secondary metabolism. In addition to reducing plant growth, salt stress can also adversely affect the plant productivity by damaging the photosynthetic machinery [67].
In inoculated Arabidopsis plants, with single or co-culture, the effect of stress treatment is slight and significantly increases in shoot weight, primary root length lateral root number and was detected compared to uninoculated ones. The maintaining of root growth constitutes an adaptive response of inoculated Arabidopsis plants to stressed conditions. As reported by Albacete et al. [68], plants inoculated with bacterial strain-produced IAA displayed higher root and leaf growth and they considered this effect as an adaptive response to salinity.
The production of IAA is beneficial for plant growth and development. It plays a very important role in cell division, root elongation, proliferation of root hairs and in the plant tolerance mechanism [69,70]. Such results of growth improvement with PGPR inoculation under salinity were previously reported by several authors [6,9,71,72,73,74,75].
Our work confirms that halophytes are a source of salt-tolerant plant-growth-promoting bacteria, which could successfully improve the performance of glycophytes under saline conditions. The application of salt-tolerant PGPR strains, as both biofertilizers and biopesticides in agriculture, may be a low-cost and environmentally friendly option that can be used to manage saline soil to increase crop productivity [76].
5. Conclusions
The results of this study reveal that the rhizosphere of the halophyte Salsola tetrandra represents excellent niches for salt-tolerant rhizobacteria, with an interesting potential for PGP activity, even at high salt concentrations. Harnessing the PGPR potential of ME1, BR5 and MLR6 strains is an alternative in improving plant growth under salt stress. These bacteria and their combinations were able to alleviate salt stress and promote the growth parameters of A. thaliana at 100 mM NaCl. The selection of these strains as phyto-stimulators under salt stress seems to be a promising strategy for enhancing plant growth in arid areas.
Indeed, this study extended the range of halotolerant PGPR strains, with promising results, showing they can be used as biofertilizers to promote plant growth and combat biotic and abiotic stresses. These results encourage further research on their application to other strategic crops. Further research should focus on the use of these HT-PGPRs in salt-affected agricultural fields as commercial biofertilizers, to improve salinity tolerance and crop productivity. The plant growth stimulants market size is booming but is expected to further improve in the near future. In the future, biostimulants involving HT-PGPR and their metabolites are expected to take off and enter the market. Therefore, the development of bioformulations from these PGPRs can be crucial to combat salinity and improve the yield of salt-affected agro-ecosystems, leading to better productivity and sustainability.
Author Contributions
Conceptualization, N.E.H.R., H.C.-S., A.S., F.N.A., A.C.B. and L.B.; methodology, N.E.H.R., H.C.-S., A.S. and L.B.; software, A.C.B. and L.B.; validation, N.E.H.R., H.C.-S., A.S., F.N.A., A.C.B. and L.B.; formal analysis, A.C.B. and L.B.; investigation, N.E.H.R., H.C.-S., A.S., F.N.A., A.C.B. and L.B.; resources, F.N.A., T.O. and L.B.; data curation, A.C.B. and L.B.; writing—original draft preparation, N.E.H.R., H.C.-S. and A.S.; writing—review and editing, A.C.B. and L.B.; visualization, H.C.-S. and L.B.; supervision, H.C.-S. and L.B.; project administration, H.C.-S. and L.B.; funding acquisition, F.N.A. and L.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Flowers, T. Preface. J. Exp. Bot. 2006, 57, iv. [Google Scholar] [CrossRef]
- Cherif-Silini, H.; Thissera, B.; Bouket, A.C.; Saadaoui, N.; Silini, A.; Eshelli, M.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Yahiaoui, B.; et al. Durum Wheat Stress Tolerance Induced by Endophyte Pantoea agglomerans with Genes Contributing to Plant Functions and Secondary Metabolite Arsenal. Int. J. Mol. Sci. 2019, 20, 3989. [Google Scholar] [CrossRef] [PubMed]
- Bouhouch, S.; Eshelli, M.; Slama, H.B.; Bouket, A.C.; Oszako, T.; Okorski, A.; Rateb, M.E.; Belbahri, L. Morphological, Biochemical, and Metabolomic Strategies of the Date Palm (Phoenix dactylifera L., cv. Deglet Nour) Roots Response to Salt Stress. Agronomy 2021, 11, 2389. [Google Scholar] [CrossRef]
- Cherif-Silini, H.; Silini, A.; Chenari Bouket, A.; Alenezi, F.N.; Luptakova, L.; Bouremani, N.; Nowakowska, J.A.; Oszako, T.; Belbahri, L. Tailoring Next Generation Plant Growth Promoting Microorganisms as Versatile Tools beyond Soil Desalinization: A Road Map towards Field Application. Sustainability 2021, 13, 4422. [Google Scholar] [CrossRef]
- Saidi, S.; Cherif-Silini, H.; Bouket, A.C.; Silini, A.; Eshelli, M.; Luptakova, L.; Alenezi, F.N.; Belbahri, L. Improvement of Medicago sativa Crops Productivity by the Co-inoculation of Sinorhizobium meliloti–Actinobacteria Under Salt Stress. Curr. Microbiol. 2021, 78, 1344–1357. [Google Scholar] [CrossRef]
- Kerbab, S.; Silini, A.; Chenari Bouket, A.; Cherif-Silini, H.; Eshelli, M.; El Houda Rabhi, N.; Belbahri, L. Mitigation of NaCl Stress in Wheat by Rhizosphere Engineering Using Salt Habitat Adapted PGPR Halotolerant Bacteria. Appl. Sci. 2021, 11, 1034. [Google Scholar] [CrossRef]
- Abbas, R.; Rasul, S.; Aslam, K.; Baber, M.; Shahid, M.; Mubeen, F.; Naqqash, T. Halotolerant PGPR: A hope for cultivation of saline soils. J. King Saud Univ. Sci. 2019, 4, 1195–1201. [Google Scholar] [CrossRef]
- FAO and ITPS. Status of the World’s Soil Resources (SWSR)—Main Report. Rome: Food and Agriculture Organization of the United Nations and Intergovernmental Technical Panel on Soils. 2015. Available online: https://www.fao.org/3/bc590e/bc590e.pdf (accessed on 10 December 2021).
- Panwar, M.; Tewari, R.; Nayyar, H. Native halo-tolerant plant growth promoting rhizobacteria Enterococcus and Pantoea sp. improve seed yield of Mungbean (Vigna radiata L.) under soil salinity by reducing sodium uptake and stress injury. Physiol. Mol. Biol. Plants 2016, 22, 445–459. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, G.; Zhang, J.; Zhang, Y.; Xie, Q.; Zhao, Z.; Pan, Y.; Hu, Z. The abiotic stress-responsive NAC-type transcription factor SlNAC4 regulates salt and drought tolerance and stress-related genes in tomato (Solanum lycopersicum). Plant Cell Rep. 2014, 33, 1851–1863. [Google Scholar] [CrossRef]
- Rekik, I.; Chaabane, Z.; Missaoui, A.; Chenari Bouket, A.; Luptakova, L.; Elleuch, A.; Belbahri, L. Effects of untreated and treated wastewater at the morphological, physiological and biochemical levels on seed germination and development of sorghum (Sorghum bicolor (L.) Moench), alfalfa (Medicago sativa L.) and fescue (Festuca arundinacea Schreb.). J. Hazard. Mater. 2017, 326, 165–176. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Meng, Y.; Li, B.; Ma, X.; Lai, Y.; Si, E.; Yang, K.; Xu, X.; Shang, X.; Wang, H.; et al. Physiological and proteomic analyses of salt stress response in the halophyte Halogeton glomeratus. Plant Cell Environ. 2015, 38, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Szymańska, S.; Plociniczak, T.; Piotrowska-Seget, Z.; Zloch, M.; Ruppel, S.; Hrynkiewicz, K. Metabolic potential and community structure of endophytic and rhizosphere bacteria associated with the roots of the halophyte Aster tripolium L. Microbiol. Res. 2016, 182, 68–79. [Google Scholar] [CrossRef]
- Slama, H.B.; Triki, M.A.; Chenari Bouket, A.; Ben Mefteh, F.; Alenezi, F.N.; Luptakova, L.; Cherif-Silini, H.; Vallat, A.; Oszako, T.; Gharsallah, N.; et al. Screening of the High-Rhizosphere Competent Limoniastrum monopetalum’ Culturable Endophyte Microbiota Allows the Recovery of Multifaceted and Versatile Biocontrol Agents. Microorganisms 2019, 7, 249. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.; Bellingrath-Kimura, S.D.; Mishra, J.; Arora, N.K. Salt-Tolerant Plant Growth Promoting Rhizobacteria for Enhancing Crop Productivity of Saline Soils. Front. Microbiol. 2019, 10, 2791. [Google Scholar] [CrossRef]
- Hashem, A.; Abd_Allah, E.F.; Alqarawi, A.A.; Al-Huqail, A.A.; Wirth, S.; Egamberdieva, D. The interaction between arbuscular mycorrhizal fungi and endophytic bacteria enhances plant growth of Acacia gerrardii under salt stress. Front. Microbiol. 2016, 7, 1089. [Google Scholar] [CrossRef]
- Arkhipova, T.N.; Evseeva, N.V.; Tkachenko, O.V.; Burygin, G.L.; Vysotskaya, L.B.; Akhtyamova, Z.A.; Kudoyarova, G.R. Rhizobacteria Inoculation Effects on Phytohormone Status of Potato Microclones Cultivated In Vitro under Osmotic Stress. Biomolecules 2020, 10, 1231. [Google Scholar] [CrossRef]
- Bomle, D.V.; Kiran, A.; Kumar, J.K.; Nagaraj, L.S.; Pradeep, C.K.; Ansari, M.A.; Alghamdi, S.; Kabrah, A.; Assaggaf, H.; Dablool, A.S.; et al. Plants Saline Environment in Perception with Rhizosphere Bacteria Containing 1-Aminocyclopropane-1-Carboxylate Deaminase. Int. J. Mol. Sci. 2021, 22, 11461. [Google Scholar] [CrossRef]
- Belbahri, L.; Chenari Bouket, A.; Rekik, I.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Petrovova, E.; Oszako, T.; Cherrad, S.; Vacher, S.; et al. Comparative Genomics of Bacillus amyloliquefaciens Strains Reveals a Core Genome with Traits for Habitat Adaptation and a Secondary Metabolites Rich Accessory Genome. Front. Microbiol. 2017, 8, 1438. [Google Scholar] [CrossRef]
- Sunita, K.; Mishra, I.; Mishra, J.; Prakash, J.; Arora, N.K. Secondary metabolites from halotolerant plant growth promoting rhizobacteria for ameliorating salinity stress in plants. Front. Microbiol. 2020, 11, 567768. [Google Scholar] [CrossRef]
- Timmusk, S.; El-Daim, I.A.A.; Copolovici, L.; Tanilas, T.; Kännaste, A.; Behers, L.; Nevo, E.; Seisenbaeva, G.; Stenstrom, E.; Niinemets, U. Drought-tolerance of wheat improved by rhizosphere bacteria from harsh environments: Enhanced biomass production and reduced emissions of stress volatiles. PLoS ONE 2014, 9, e96086. [Google Scholar] [CrossRef] [PubMed]
- Bano, A.; Fatima, M. Salt tolerance in Zea mays (L.) following inoculation with Rhizobium and Pseudomonas. Biol. Fertil. Soils 2009, 45, 405–413. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Singh, D.P. Effect of salt-tolerant plant growth promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biol. 2015, 17, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Brito, P.H.; Chevreux, B.; Serra, C.R.; Schyns, G.; Henriques, A.O.; Pereira-Leal, J.B. Genetic Competence Drives Genome Diversity in Bacillus subtilis. Genome Biol. Evol. 2018, 10, 108–124. [Google Scholar] [CrossRef] [PubMed]
- Alenezi, F.N.; Slama, H.B.; Bouket, A.C.; Cherif-Silini, H.; Silini, A.; Luptakova, L.; Nowakowska, J.A.; Oszako, T.; Belbahri, L. Bacillus velezensis: A Treasure House of Bioactive Compounds of Medicinal, Biocontrol and Environmental Importance. Forests 2021, 12, 1714. [Google Scholar] [CrossRef]
- Mavrodi, O.V.; McWilliams, J.R.; Peter, J.O.; Berim, A.; Hassan, K.A.; Elbourne, L.D.H.; LeTourneau, M.K.; Gang, D.R.; Paulsen, I.T.; Weller, D.M.; et al. Root Exudates Alter the Expression of Diverse Metabolic, Transport, Regulatory, and Stress Response Genes in Rhizosphere Pseudomonas. Front. Microbiol. 2021, 12, 651282. [Google Scholar] [CrossRef] [PubMed]
- Sáenz-Mata, J.; Palacio-Rodríguez, R.; Sánchez-Galván, H.; Balagurusamy, N. Plant Growth Promoting Rhizobacteria Associated to Halophytes. In Potential Applications in Agriculture Sabkha Ecosystems; Khan, M.A., Ed.; Springer: Cham, Switzerland, 2016; Volume 5. [Google Scholar]
- He, A.-L.; Niu, S.-Q.; Zhao, Q.; Li, Y.-S.; Gou, J.-Y.; Gao, H.-J.; Suo, S.-Z.; Zhang, J.-L. Induced Salt Tolerance of Perennial Ryegrass by a Novel Bacterium Strain from the Rhizosphere of a Desert Shrub Haloxylon ammodendron. Int. J. Mol. Sci. 2018, 19, 469. [Google Scholar] [CrossRef]
- Frikha-Dammak, D.; Ayadi, H.; Hakim-Rekik, I.; Belbahri, L.; Maalej, S. Genome analysis of the salt-resistant Paludifilum halophilum DSM 102817T reveals genes involved in flux-tuning of ectoines and unexplored bioactive secondary metabolites. World J. Microbiol. Biotechnol. 2021, 37, 178. [Google Scholar] [CrossRef]
- Bric, J.M.; Bostock, R.M.; Silverstone, S.E. Rapid In situ assay for indoleacetic Acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 1991, 57, 535–538. [Google Scholar] [CrossRef]
- Gangoir, M.; Rani, S.; Sharman, N. Diversity of endophytic Actinomycetes from wheat and its potential as plant growth promoting and biocontrol agents. J. Adv. Labor. Res. Biol. 2012, 3, 17–24. [Google Scholar]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Miranda, S.; Cabirol, N.; George-Téllez, R.; Zamudio-Rivera, L.S.; Fernández, F.J. O-CAS, a fast and universal method for siderophore detection. J. Microbiol. Methods 2007, 70, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Cappuccino, J.C.; Sherman, N. Microbiology: A Laboratory Manual, 3rd ed.; Benjamin/Cumming pub. Co.: New York, NY, USA, 1992; pp. 125–179. [Google Scholar]
- Bakker, A.W.; Schippers, B. Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas spp. mediated plant growth-stimulation. Soil Biol. Biochem. 1987, 19, 451–457. [Google Scholar] [CrossRef]
- Kasana, R.C.; Salwan, R.; Dhar, H.; Dutt, S.; Gulati, A. A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Curr. Microbiol. 2008, 57, 503–507. [Google Scholar] [CrossRef]
- Murthy, N.; Bleakley, B. Simplified method of preparing colloidal chitin used for screening of chitinase-producing microorganisms. Internet J. Microbiol. 2012, 10, e2bc3. [Google Scholar]
- Chang, W.T.; Hsieh, C.H.; Hsieh, H.S.; Chen, C. Conversion of crude chitosan to an anti-fungal protease by Bacillus cereus. World J. Microbiol. Biotechnol. 2009, 25, 375–382. [Google Scholar] [CrossRef]
- Ferjani, R.; Marasco, R.; Rolli, E.; Cherif, H.; Cherif, A.; Gtari, M.; Boudabous, A.; Daffonchio, D.; Ouzari, H. The date palm tree rhizosphere is a niche for plant growth promoting bacteria in the oasis ecosystem. BioMed Res. Int. 2015, 2015, 153851. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Fiske, C.H.; Subbarow, Y. The colorimetric determination of phosphorus. J. Biol. Chem. 1925, 66, 375–400. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Devkota, K.P.; Devkota, M.; Rezaei, M.; Oosterbaan, R. Managing salinity for sustainable agricultural production in salt-affected soils of irrigated drylands. Agri. Sys. 2022, 198, 103390. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Baek, K.H. Physiological and biochemical perspectives of non-salt tolerant plants during bacterial interaction against soil salinity. Plant Physiol. Biochem. 2017, 116, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Vaishnav, A.; Varma, A.; Tuteja, N.; Choudhary, D.K. PGPR-mediated amelioration of crops under salt stress. In Plant-Microbe Interaction: An Approach to Sustainable Agriculture; Choudhary, D.K., Ed.; Springer: Noida, India, 2016; pp. 205–226. [Google Scholar]
- Forchetti, G.; Masciarelli, O.; Alemano, S.; Alvarez, D.; Abdala, G. Endophytic bacteria in sunflower (Helianthus annuus L.): Isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl. Microbiol. Biotechnol. 2007, 76, 1145–1152. [Google Scholar] [CrossRef]
- Felestrino, É.B.; Santiago, I.F.; Freitas, L.D.S.; Rosa, L.H.; Ribeiro, S.P.; Moreira, L.M. Plant growth promoting bacteria associated with Langsdorffia hypogaea-rhizosphere-host biological interface: A neglected model of bacterial prospection. Front. Microbiol. 2017, 8, 172. [Google Scholar] [CrossRef]
- Marasco, R.; Mapelli, F.; Rolli, E.; Mosqueira, M.J.; Fusi, M.; Bariselli, P.; Reddy, M.; Cherif, A.; Tsiamis, G.; Borin, S.; et al. Salicornia strobilacea (synonym of Halocnemum strobilaceum) grown under different tidal regimes selects rhizosphere bacteria capable of promoting plant growth. Front. Microbiol. 2016, 7, 1–11. [Google Scholar] [CrossRef]
- Gamalero, E.; Glick, B.R. Plant growth-promoting bacteria in agriculture and stress environments. In Modern Soil Microbiology, 3rd ed.; Van Elsas, J.D., Trevors, J.T., Rosado, A.S., Nannipieri, P., Eds.; Springer: New Delhi, India, 2019; pp. 361–380. [Google Scholar]
- El-Sayed, W.S.; Akhkha, A.; El-Naggar, M.Y.; Elbadry, M. In vitro antagonistic activity, plant growth promoting traits and phylogenetic affiliation of rhizobacteria associated with wild plants grown in arid soil. Front. Microbiol. 2014, 5, 1–11. [Google Scholar] [CrossRef]
- Mukhtar, S.; Malik, K.A.; Mehnaz, S. Microbiome of halophytes: Diversity and importance for plant health and productivity. Microbiol. Biotechnol. Lett. 2019, 47, 1–10. [Google Scholar] [CrossRef]
- Deshwal, V.K.; Kumar, P. Effect of salinity on growth and PGPR activity of Pseudomonads. J. Acad. Ind. Res. 2013, 2, 353–356. [Google Scholar]
- Rangarajan, S.; Saleena, L.M.; Nair, S. Diversity of Pseudomonas spp. isolated from rice rhizosphere populations grown along a salinity gradient. Microb. Ecol. 2002, 43, 280–289. [Google Scholar] [CrossRef]
- Shi, Y.W.; Lou, K.; Li, C.; Wang, L.; Zhao, Z.Y.; Zhao, S.; Tian, C.Y. Illumina-based analysis of bacterial diversity related to halophytes Salicornia europaea and Sueada aralocaspica. J. Microbiol. 2015, 53, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wang, L.; Tang, Y.; Li, Y.; Chen, J.; Xi, X.; Zhang, Y.; Fu, X.; Wu, J.; Sun, Y. Variability in soil microbial community and activity between coastal and riparian wetlands in the Yangtze River estuary-Potential impacts on carbon sequestration. Soil Biol. Biochem. 2014, 70, 221–228. [Google Scholar] [CrossRef]
- Vives-Peris, V.; Gomez-Cadenas, A.; Perez-Clemente, R.M. Salt stress alleviation in citrus plants by plant growth-promoting rhizobacteria Pseudomonas putida and Novosphingobium sp. Plant Cell Rep. 2018, 37, 1557–1569. [Google Scholar] [CrossRef] [PubMed]
- Farokh, R.Z.; Sachdev, D.; Pour, N.K.; Engineer, A.; Pardesi, K.R.; Zinjarde, S.; Dhakephalkar, P.K.; Chopade, B.A. Characterization of plant-growth-promoting traits of Acinetobacter species isolated from rhizosphere of Pennisetum glaucum. J. Microbiol. Biotechnol. 2011, 21, 556–566. [Google Scholar]
- Hussain, K.; Hameed, S.; Shahid, M.; Ali, A.; Iqbal, J.; Hahn, D. First report of Providencia vermicola strains characterized for enhanced rapeseed growth attributing parameters. Int. J. Agric. Biol. 2015, 17, 1110–1116. [Google Scholar] [CrossRef]
- Kang, S.; Khan, A.L.; Waqas, M.; You, Y.; Kim, H.; Kim, J.; Hamayun, M.; Lee, I.; Khan, A.L.; Waqas, M. Plant growth-promoting rhizobacteria reduce adverse effects of salinity and osmotic stress by regulating phytohormones and antioxidants in Cucumis sativus. J. Plant Interact. 2014, 9, 673–682. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, C.; Feng, Q.; Liou, R.-M.; Lin, Y.-F.; Qiao, J.; Lu, Y.; Chang, Y. The Mechanisms of Sodium Chloride Stress Mitigation by Salt-Tolerant Plant Growth Promoting Rhizobacteria in Wheat. Agronomy 2022, 12, 543. [Google Scholar] [CrossRef]
- Xiao, M.; Sun, S.S.; Zhang, Z.Z.; Wang, J.M.; Qiu, L.-W.; Sun, H.-Y.; Song, Z.-Z.; Zhang, B.-Y.; Gao, D.-L.; Zhang, G.-Q.; et al. Analysis of bacterial diversity in two oil blocks from two low-permeability reservoirs with high salinities. Sci. Rep. 2016, 6, 19600. [Google Scholar] [CrossRef]
- Hajiabadi, A.A.; Arani, A.M.; Ghasemi, S.; Rad, M.H.; Etesami, H.; Manshadi, S.S.; Dolati, A. Mining the rhizosphere of halophytic rangeland plants for halotolerant bacteria to improve growth and yield of salinity-stressed wheat. Plant Physiol. Biochem. 2021, 163, 139–153. [Google Scholar] [CrossRef]
- Redondo-Gómez, S.; Mesa-Marín, J.; Pérez-Romero, J.A.; López-Jurado, J.; García-López, J.V.; Mariscal, V.; Molina-Heredia, F.P.; Pajuelo, E.; Rodríguez-Llorente, I.D.; Flowers, T.J.; et al. Consortia of Plant-Growth-Promoting Rhizobacteria Isolated from Halophytes Improve Response of Eight Crops to Soil Salinization and Climate Change Conditions. Agronomy 2021, 11, 1609. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Lugtenberg, B. Use of Plant Growth-Promoting Rhizobacteria to Alleviate Salinity Stress in Plants. In Use of Microbes for the Alleviation of Soil Stresses; Miransari, M., Ed.; Springer: New York, NY, USA, 2014; pp. 73–96. [Google Scholar]
- Flors, V.; Paradís, M.; García-Andrade, J.; Cerezo, M.; González-Bosch, C.; García-Agustín, P. A tolerant behavior in salt-sensitive tomato plants can be mimicked by chemical stimuli. Plant Signal Behav. 2007, 2, 50–57. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Albacete, A.; Ghanem, M.E.; Martinez-Andujar, C.; Acosta, M.; Sanchez-Bravo, J.; Martinez, V.; Lutts, S.; Dodd, I.C.; Perez-Alfocea, F. Hormonal changes in relation to biomass partitioning and shoot growth impairment in salinized tomato (Solanum lycopersicum L.) plants. J. Exp. Bot. 2008, 59, 4119–4131. [Google Scholar] [CrossRef] [PubMed]
- Marulanda, A.; Azcon, R.; Chaumont, F.; Ruiz-Lozano, J.M.; Aroca, R. Regulation of plasma membrane aquaporins by inoculation with a Bacillus megaterium strain in maize (Zea mays L.) plants under unstressed and salt-stressed conditions. Planta 2010, 232, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Sandhya, V.; Ali, S.Z.; Venkateswarlu, B.; Reddy, G.G.M. Effect of osmotic stress on plant growth promoting Pseudomonas spp. Arch. Microbiol. 2010, 192, 867–876. [Google Scholar] [CrossRef]
- Mayak, S.; Tirosh, T.; Glick, B.R. Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol. Biochem. 2004, 42, 565–572. [Google Scholar] [CrossRef]
- Paul, D.; Lade, H. Plant-growth-promoting rhizobacteria to improve crop growth in saline soils: A review. Agron. Sustain. Dev. 2014, 34, 737–752. [Google Scholar] [CrossRef]
- Liu, X.; Wei, W.; Zhu, W.; Su, L.; Xiong, Z.; Zhou, M.; Zheng, Y.; Zhou, D.X. Histone deacetylase AtSRT1 regulates metabolic flux and stress response in Arabidopsis. Mol. Plant 2017, 10, 1510–1522. [Google Scholar] [CrossRef]
- Poupin, M.J.; Timmermann, T.; Vega, A.; Zuñiga, A.; González, B. Effects of the plant growth-promoting bacterium Burkholderia phytofirmans PsJN throughout the life cycle of Arabidopsis thaliana. PLoS ONE 2013, 8, e69435. [Google Scholar] [CrossRef]
- Rabhi, N.E.H.; Silini, A.; Cherif-Silini, H.; Yahiaoui, B.; Lekired, A.; Robineau, M.; Esmaeel, Q.; Jacquard, C.; Vaillant-Gaveau, N.; Christophe Clément, C.; et al. Pseudomonas knackmussii MLR6, a rhizospheric strain isolated from halophyte, enhances salt tolerance in Arabidopsis thaliana. J. Appl. Microbiol. 2018, 125, 1836–1851. [Google Scholar] [CrossRef]
- Sharma, A.; Dev, K.; Sourirajan, A.; Choudhary, M. Isolation and characterization of salt-tolerant bacteria with plant growth-promoting activities from saline agricultural fields of Haryana, India. J. Genet. Eng. Biotechnol. 2021, 19, 99. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).