2.1. Data
The data for this work came from the Dali and Lanxia forest farms in Fujian province, China. During the exploratory analysis, some obvious data errors were removed (25 trees). A total of 340 trees, with crowns distributed evenly over 65 pure, even-aged, and unthinned temporary plots (30 m × 30 m) were selected to determine the crown shape estimate of Chinese fir trees. For each tree, we measured the sample crown height from its top to crown base (CH, m) and measured the crown radius (CR, m) at each sampling height. (see
Figure 1).
The crown data used in this study consisted of 1427 measured values, form 340 trees from Chinese fir stands with age ranging from 5 to 35 years. The tree factors used to establishment crown profile model should be selected according to the factors affecting the growth of trees. According to the relevant literature [
2,
3,
4,
6,
11,
34,
35,
37,
38,
44,
47,
51], the crown profile model is mainly affected by AGE, N, TH, DBH, LCL, CW, HCB, HLCR. In addition, in this study, we also defined the following composite tree factors: the tree crown length ratio, the HT to DBH ratio, and the tree crown fullness ratio. All the factors related to the crown and their descriptions are shown in
Table 1.
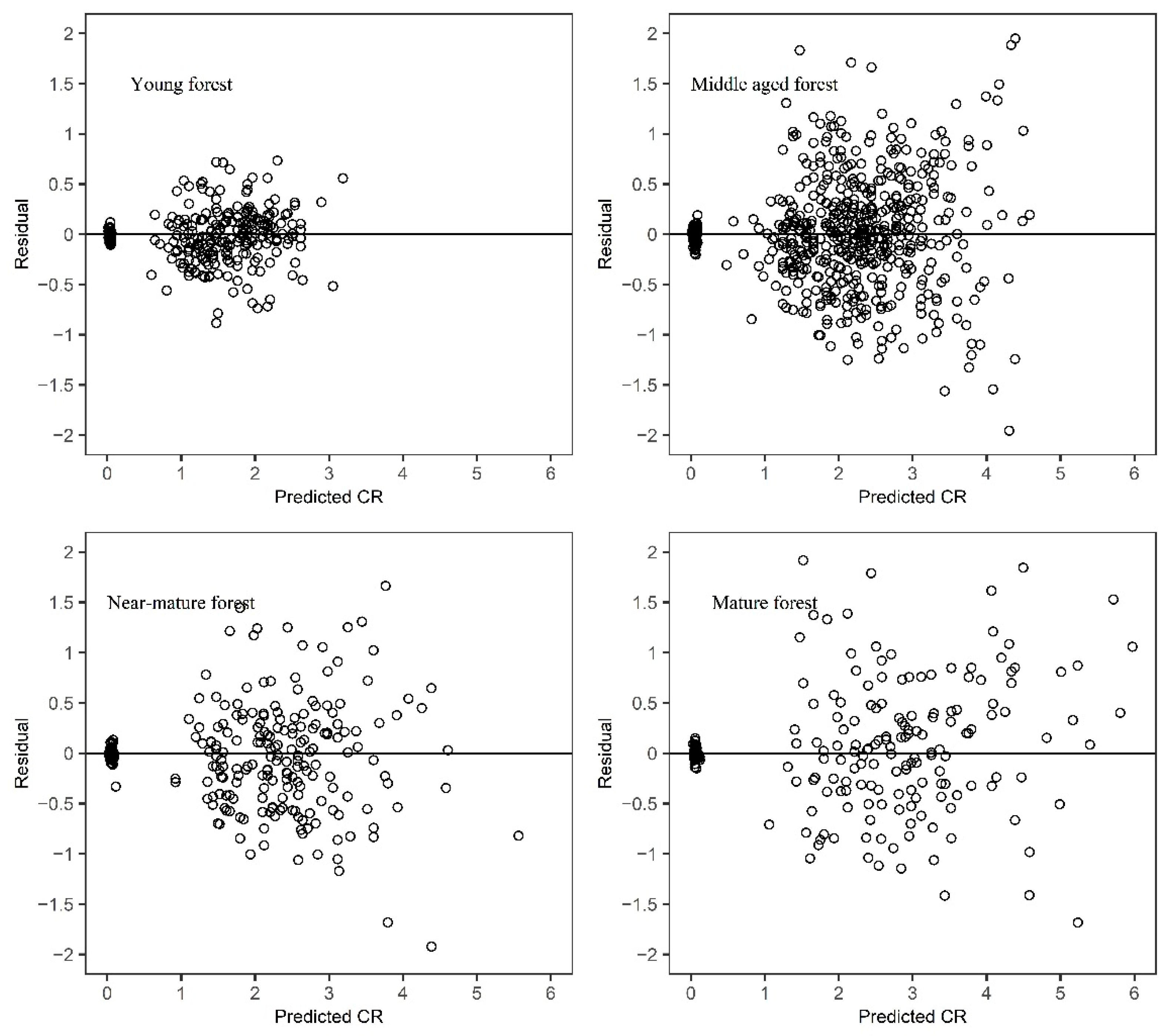
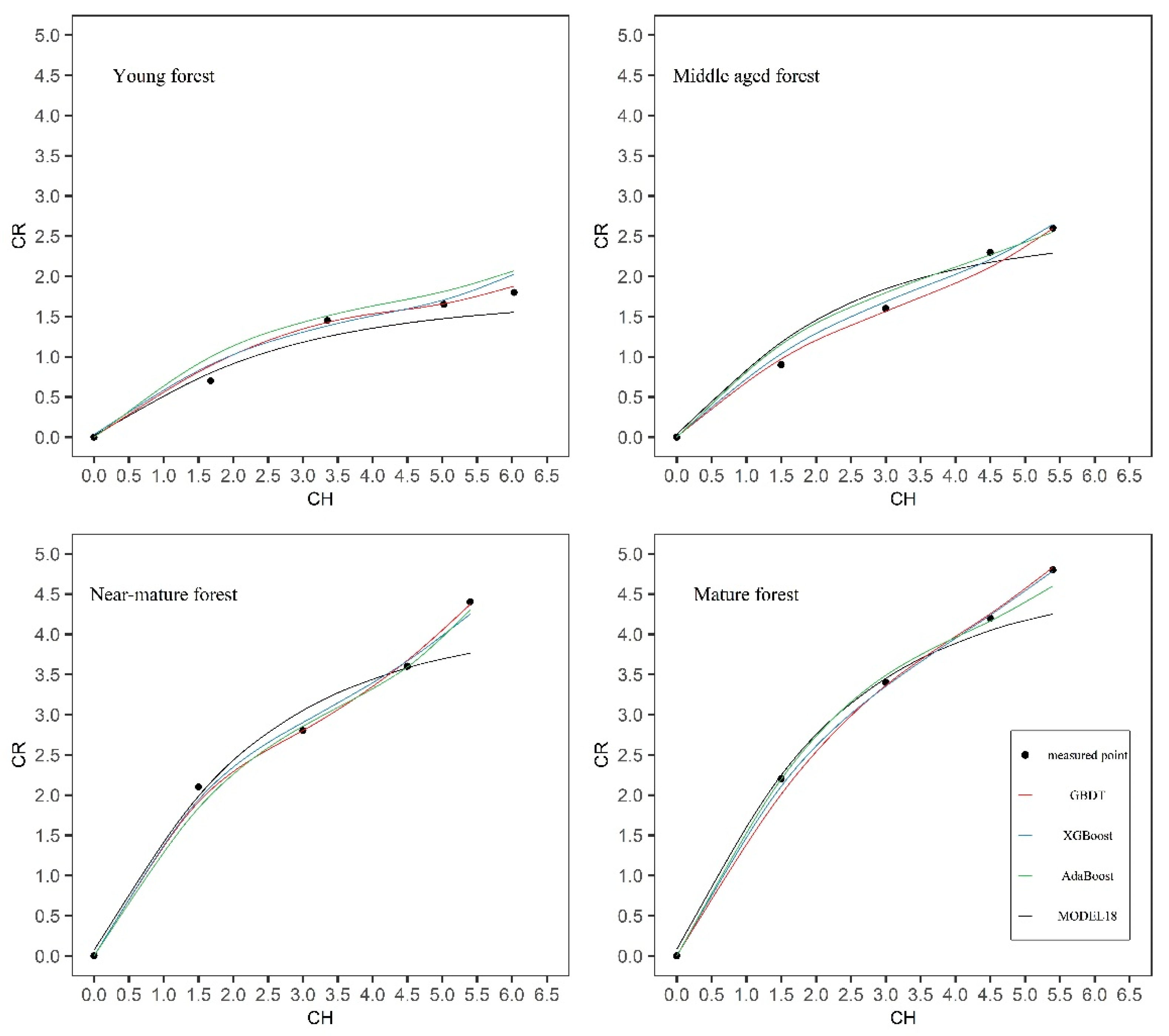
A detailed visual examination of the crown profiles suggested that sample trees from different age groups appeared to have a different crown form (
Figure 2). Further research is required to improve the prediction effect of adding age variables in the crown profile equations or algorithms.
2.2. Parametric Regression Approach for the Crown Profile Model
We initially selected 25 crown profile models proposed in the literature with different numbers of parameters that had previously shown good performance (
Table 2). The crown profile equations were estimated using nonlinear least-squares estimates through a Gauss–Newton algorithm, implemented in PROC MODEL (SAS Institute Inc., Cary, NC, USA, 2013) and we then compared the goodness-of-fit values. We screened for the parameters that passed
t-test, and which had R-squared (R
2) values greater than 60%, and selected these as the basic models.
Three different mean-covariance models were considered for estimation of means and covariances. The first is a saturated one in which all means and covariances are model parameters. The other two structured mean-covariance models use the same CR function to approximate means while characterizing covariances directly by using variance and correlation functions or indirectly via random effects within the framework of mixed-effects modeling [
60]. For the nonlinear marginal model, parameters estimates and the subsequent analysis of the residual variance-covariance matrix were performed using the SAS macro %NLINMIX (SAS Institute, Inc., Cary, NC, USA, 2004). The %NLINIMX macro extends the fitting algorithm used in the GENMOD procedure by using a quadratic instead of a simple method in the moment estimation equation for the correlation parameters [
61]. For the screened basic crown profile models, its parameter estimates and the subsequent analysis of the residual variance-covariance matrix were performed using the SAS macro %NLINMIX. To account for heteroscedastic, autocorrelated errors, the CAR(1) error structure was added to each CR equation for crown profile models (
type = SP(POW)(RCH) specifies the covariance structure in Equation (1)), and the residuals were used to calculate weights (weight = 1/sqrt(
)).
where,
represents variances;
represents correlation parameters;
the covariance between two observations depends on a distance metric; The
c-list contains the names of the numeric variables used as coordinates to determine distance.
2.3. Artificial Intelligence Procedures for the Crown Profile Model
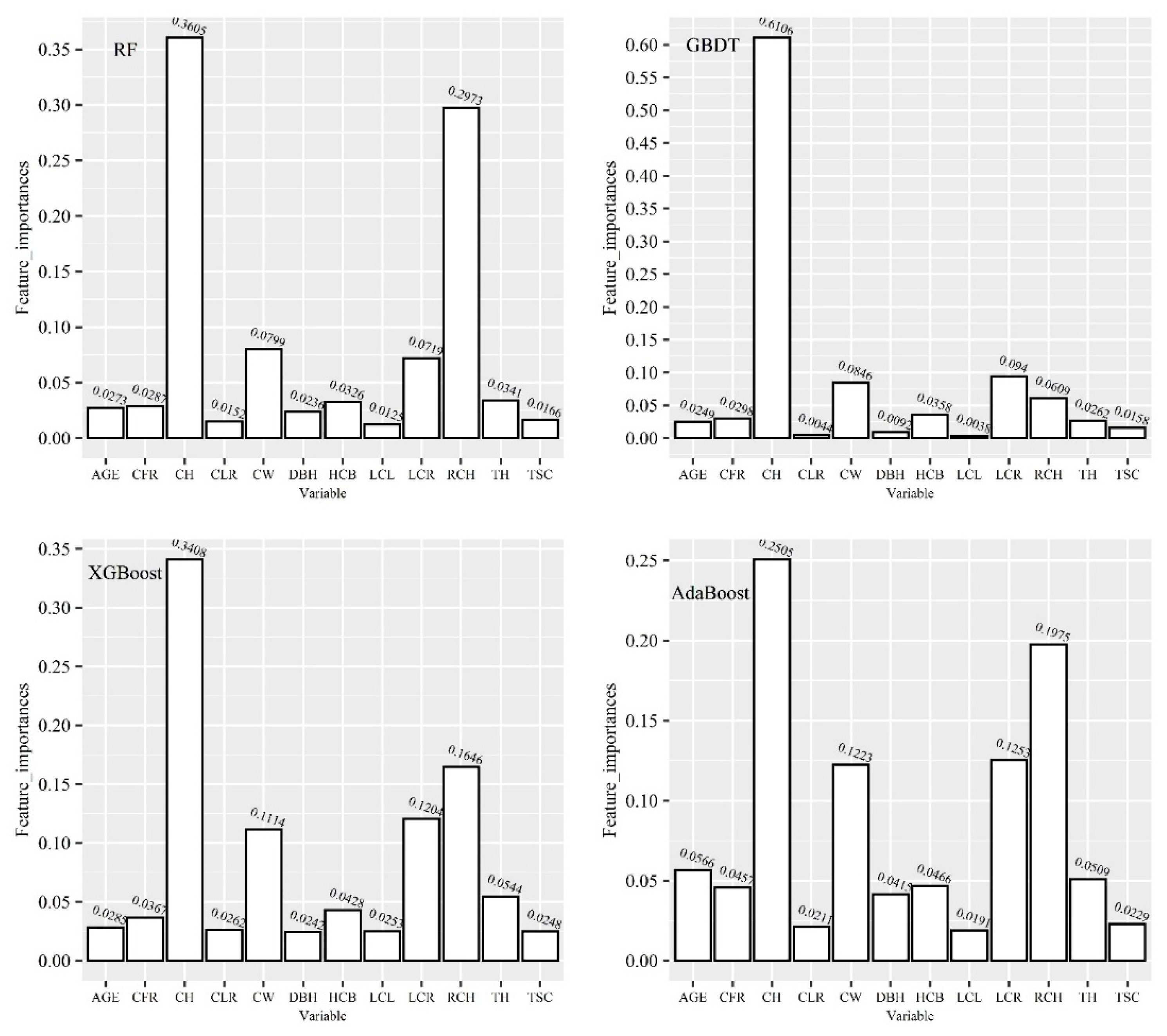
Six machine learning algorithms were used to construct the crown profile model of China fir trees in this paper, namely, artificial neural network (ANN), support vector regression (SVR), random forest (RF), adaptive boosting (AdaBoost), gradient boosting decision tree (GBDT) and extreme gradient boost (XGBoost) methods. Among these, RF, AdaBoost, GBDT and XGBoost belong to ensemble learning method. The objective is to predict
CR (crown radius) at different relative crown height
RCH.
where,
shows that some model or algorithm can depend on other variables
describing the tree.
From the model accuracy analysis, we should build the crown profile model that use all relevant variables ( DBH, TH, AGE, HCB, CW, LCL, LCR, TSC, CLR, CFR, CH) as input. From the model practicality analysis, we should train an AI algorithm that would use only standard forestry measures ( TH, DBH and AGE) as input. Considering different application scenarios, we trained these AI algorithms with two strategies for input variables: one is multivariable RCH, DBH, TH, AGE, HCB, CW, LCL, LCR, TSC, CLR, CFR, CH as inputs, and the other is only standard forestry measures as inputs.
ANN: ANN, also called a Multi-Layer Perceptron (MLP), achieves the goal of prediction through building a multilayer network. A typical neural network usually consists of an input layer, hidden layer and output layer. Based on different learning tasks, the number of hidden layers can reach any depth, and the output of each hidden layer is transformed by the activation function [
62,
63].
SVR: Support vector regression assumes that the allowable deviation between f(x) and y is at most ε, and the loss is calculated only when the absolute value of the difference between f(x) and y is greater than ε. At this time, it is equivalent to building an interval band with a width of 2ε with f(x) as the center. If the training sample falls into the interval band, the prediction is considered correct. It specifies the epsilon-tube within which no penalty is associated in the training loss function with points predicted within a distance εfrom the actual value.
RF: RF is an ensemble learner consisting of a collection of weak base learners (decision trees). By combining several weak base learners, the result can be achieved through voting or taking the mean value, which makes the results of the overall model have high accuracy and generalization performance [
64].
AdaBoost: AdaBoost is also an ensemble learning method that combines several weak learners into one strong learner to improve regression accuracy [
65].
Table 2.
Crown profile equations.
Table 2.
Crown profile equations.
| Model | Equation | Variable | Parameter | Reference |
|---|
| 1 | | RCH, LCR | | [34] |
| 2 | | RCH, LCR | | [35] |
| 3 | | RCH | | [37] |
| 4 | | RCH | | [23] |
| 5 | | CH, DBH, TH, LCL | | [2] |
| 6 | | RCH, LCR | | [3] |
| 7 | | RCH, LCR | | [4] |
| 8 | | RCH, LCR | | [48] |
| 9 | | RCH, LCR | | [4] |
| 10 | | RCH | | [51] |
| 11 | | RCH | | [51] |
| 12 | | RCH | | [45] |
| 13 | | RCH, DBH, CLR, TSC | | [45] |
| 14 | | RCH, DBH, CLR, TSC | | [45] |
| 15 | | RCH, DBH, CLR | | [45] |
| 16 | | RCH, DBH, CLR, TSC | | [11] |
| 17 | | RCH, LCR | | [66] |
| 18 | | RCH, LCR | | [66] |
| 19 | | RCH, LCR, HCB | | [66] |
| 20 | | RCH, LCR, HCB | | [66] |
| 21 | | RCH, LCR, AGE | | [66] |
| 22 | | RCH, LCR | | [6] |
| 23 | | RCH, LCR | | [6] |
| 24 | | RCH, LCR | | [6] |
| 25 | | RCH, LCR | | [6] |
GBDT: GBDT is a boosting ensemble learning algorithm, but its the specific process is different from that of AdaBoost. The goal of GBDT is to continuously reduce the loss of each iteration [
66].
XGBoost: XGBoost is an optimized distributed gradient boosting machine learning algorithms under the Gradient Boosting framework. A regularization technique is used in the XGBoost regression model. In the iterative optimization process, XGBoost uses the second-order approximation of the Taylor expansion of the objective function. Support parallelism is the flash point of XGBoost [
67].
In terms of hyperparameter optimization, the grid search and random search are two generic approaches to parameter search, which are provided in scikit-learn by GridSearchCV and RandomizedSearchCV. Given the finite set of values for each hyperparameter, GridSearchCV exhaustively considers all hyperparameter sets combinations, whereas RandomizedSearchCV can sample a given number of candidates from a hyperparameter space with a specified distribution [
68]. Considering the characteristics of different methods, in this paper we chose the strategy of combining the random and network search to simplify the required number of experimental iterations. Firstly, a random search was used to obtain the general direction in a large range of parameter space. Then, the results of random parameter selection were used as the basis for the following grid search. Different parameters and different preprocessing schemes were repeatedly compared, and the optimal hyperparameter set was finally screened out.
2.4. Model Evaluation and Validation
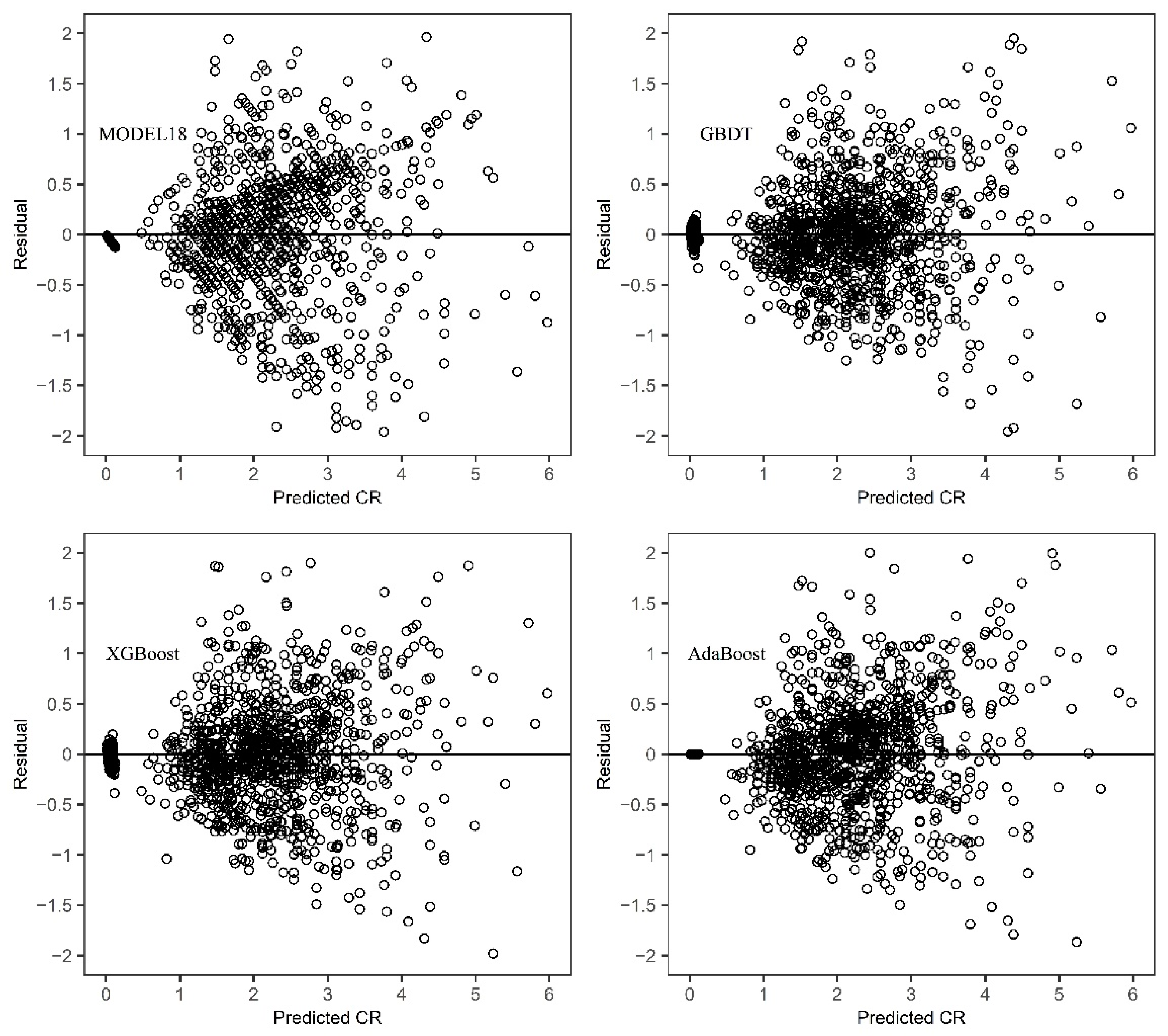
The way of dividing training set and test set judged whether the model has over fitting or under fitting problems. To assess the predictive accuracy of the estimated crown profile models, the leave-one-out approach based on tree level was selected for model validation. For N sample trees, each model was fitted N times. Each time, all the data points of one tree were removed from the fitting process, and predicted the values of CR were obtained for these using the coefficients estimated from the remaining data.
The performance indicators were calculated for both training and validation datasets. The evaluation criteria included the mean deviation (
ME, Equation (3)), the mean absolute deviation (
MAE, Equation (4)) [
53] and the mean squared error (
MSE, Equation (5)). We compared the goodness-of-fits using the root mean squared error (
RMSE, Equation (6)), the R-squared value (
R2, Equation (7)), the Akaike Information Criterion (
AIC, Equation (8)) and the Bayesian Information Criterion (
BIC, Equation (9)).
where
represents the observed value;
is the predicted value;
n is the number of tree points,
is the mean value for the observed, and is the number of parameters in the model.