Identification and Analysis of the EIN3/EIL Gene Family in Populus × xiaohei T. S. Hwang et Liang: Expression Profiling during Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Cloning of PsnEIN3/EIL Genes of Populus × xiaohei
2.3. Bioinformatic Analysis
2.4. Semi-Quantitative Reverse Transcription PCR, (sqRT-PCR)
3. Results and Analysis
3.1. Cloning and Identification of EIN3/EIL Gene Sequences of Populus × xiaohei
3.2. Bioinformatic Analysis of the EIN3/EIL Gene of Populus × xiaohei
3.2.1. Analysis of the Physicochemical Properties of PsnEIN3/EIL Gene in Populus × xiaohei
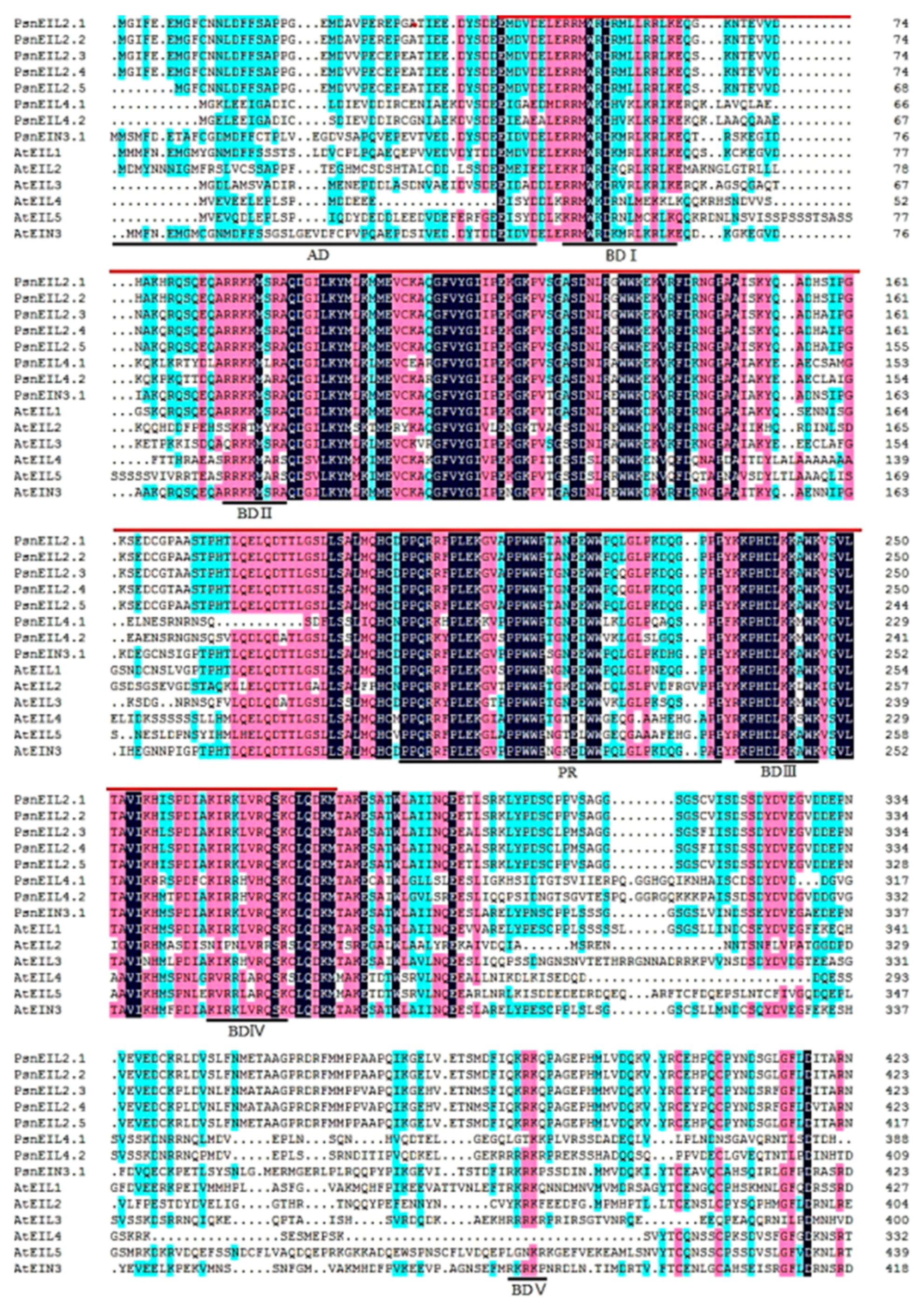
3.2.2. Multiple Sequence Alignment of PsnEIN3/EIL Proteins
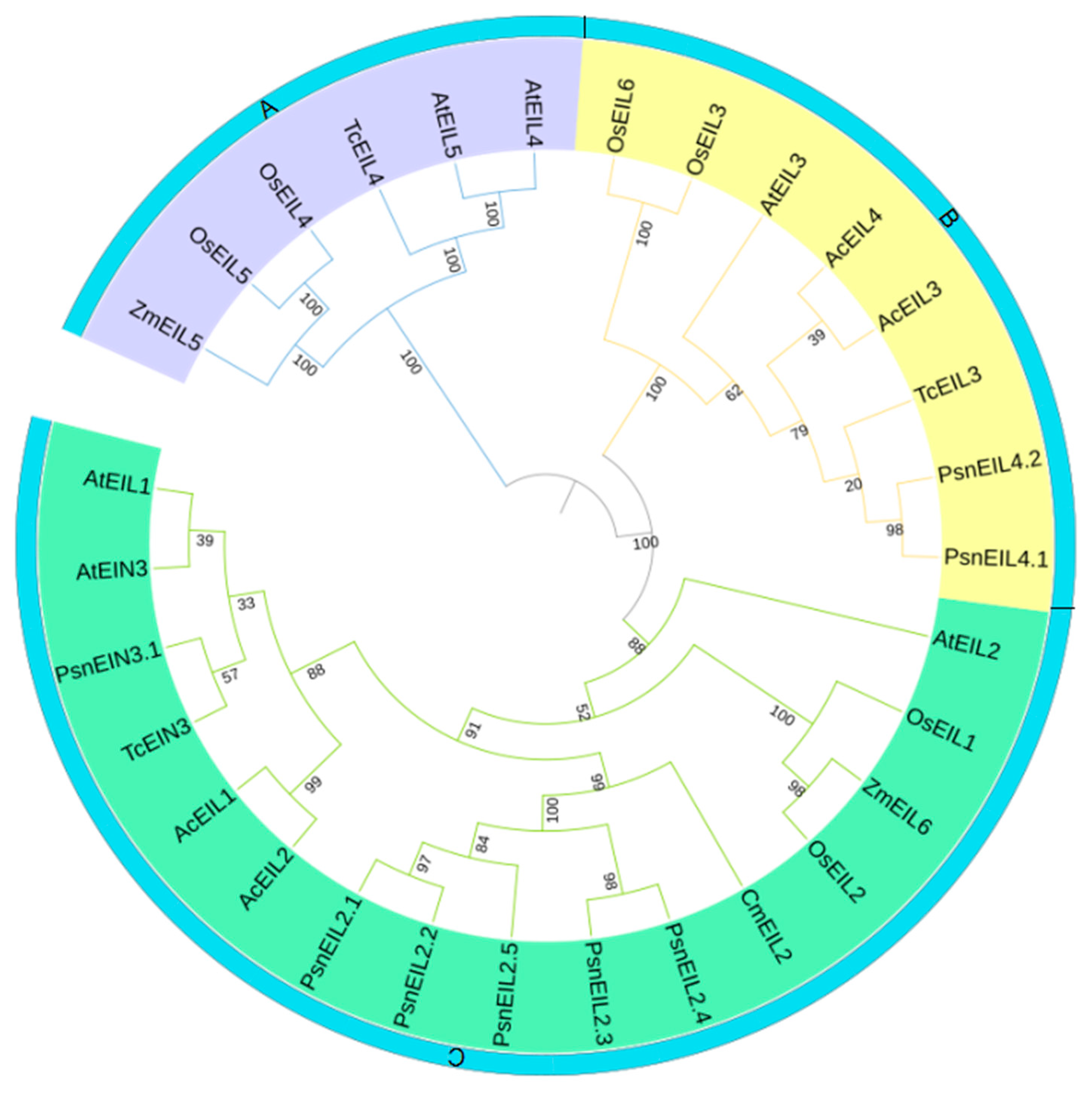
3.2.3. Phylogenetic Analysis of PsnEIN3/EIL Genes in Populus × xiaohei
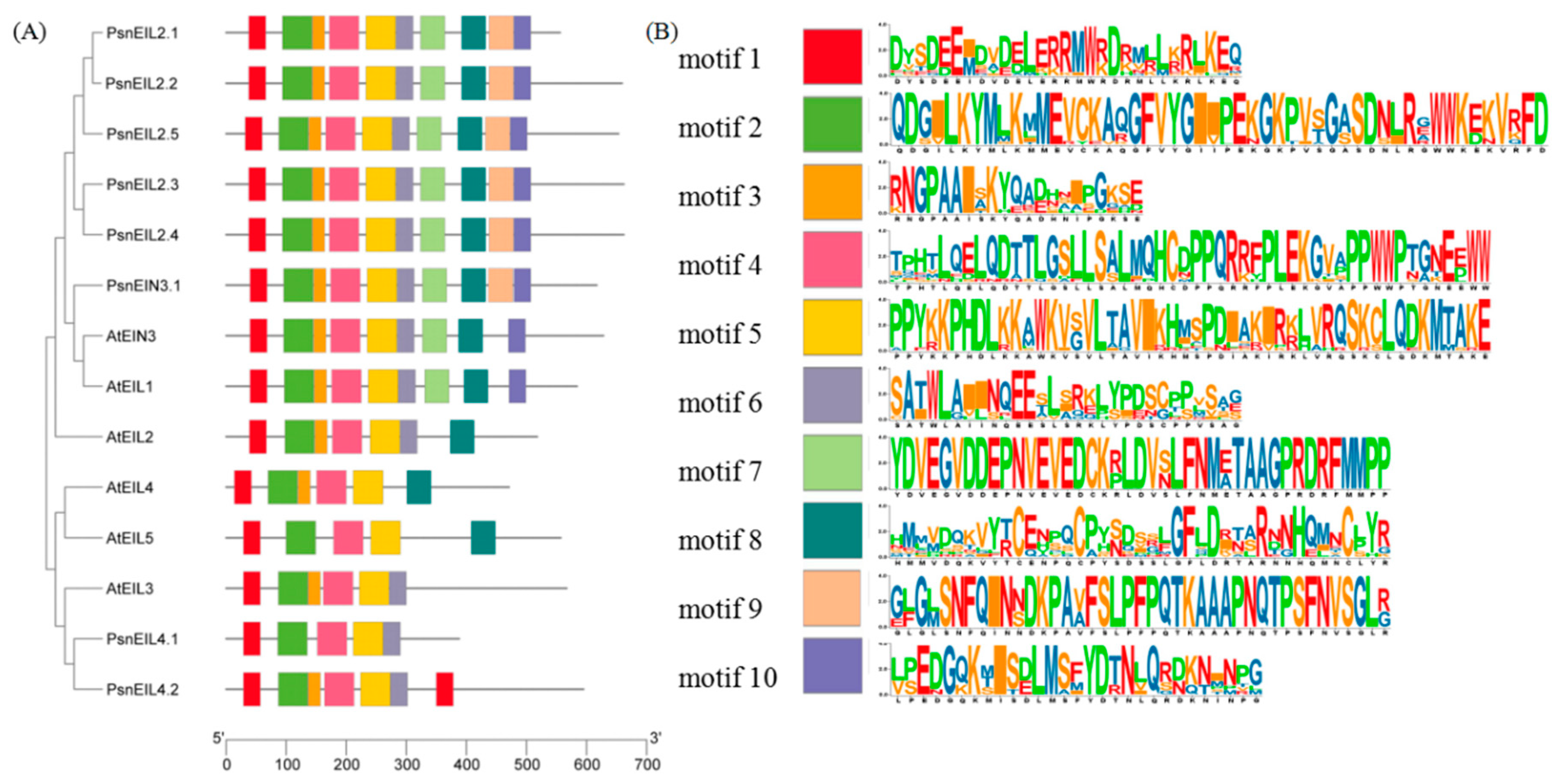
3.2.4. Analysis of Conserved Motifs Analysis of EIN3/EIL Proteins
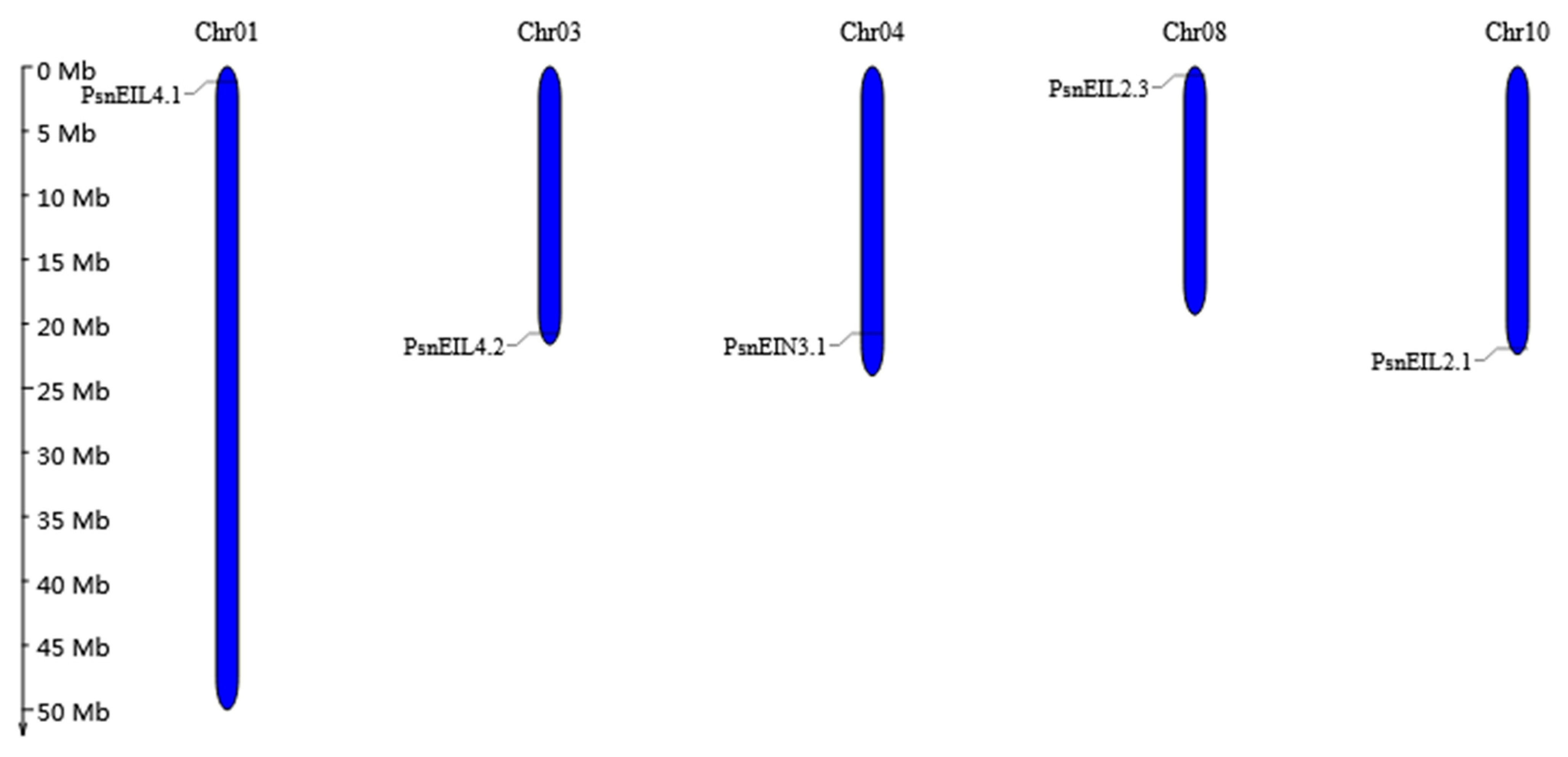
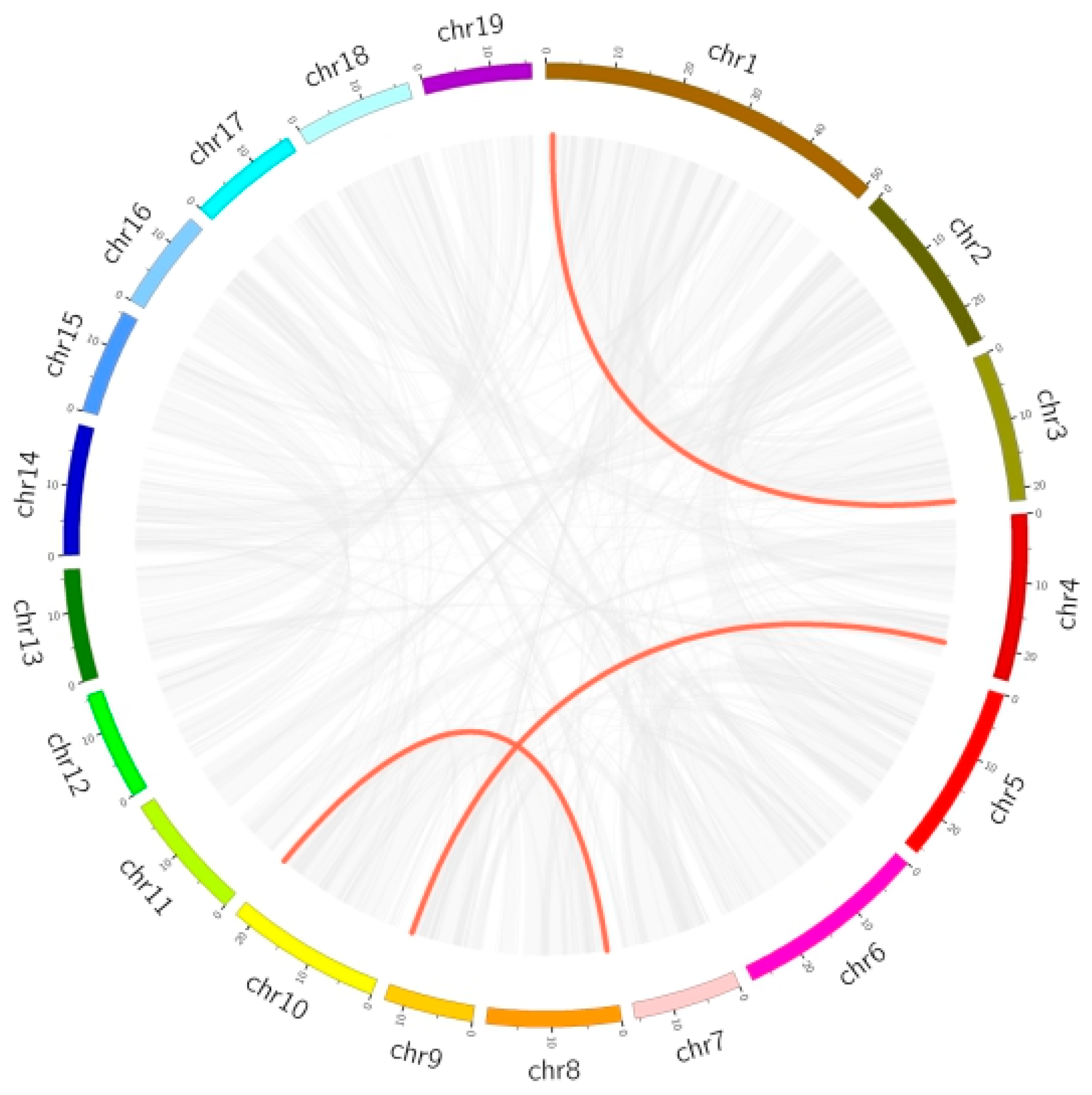
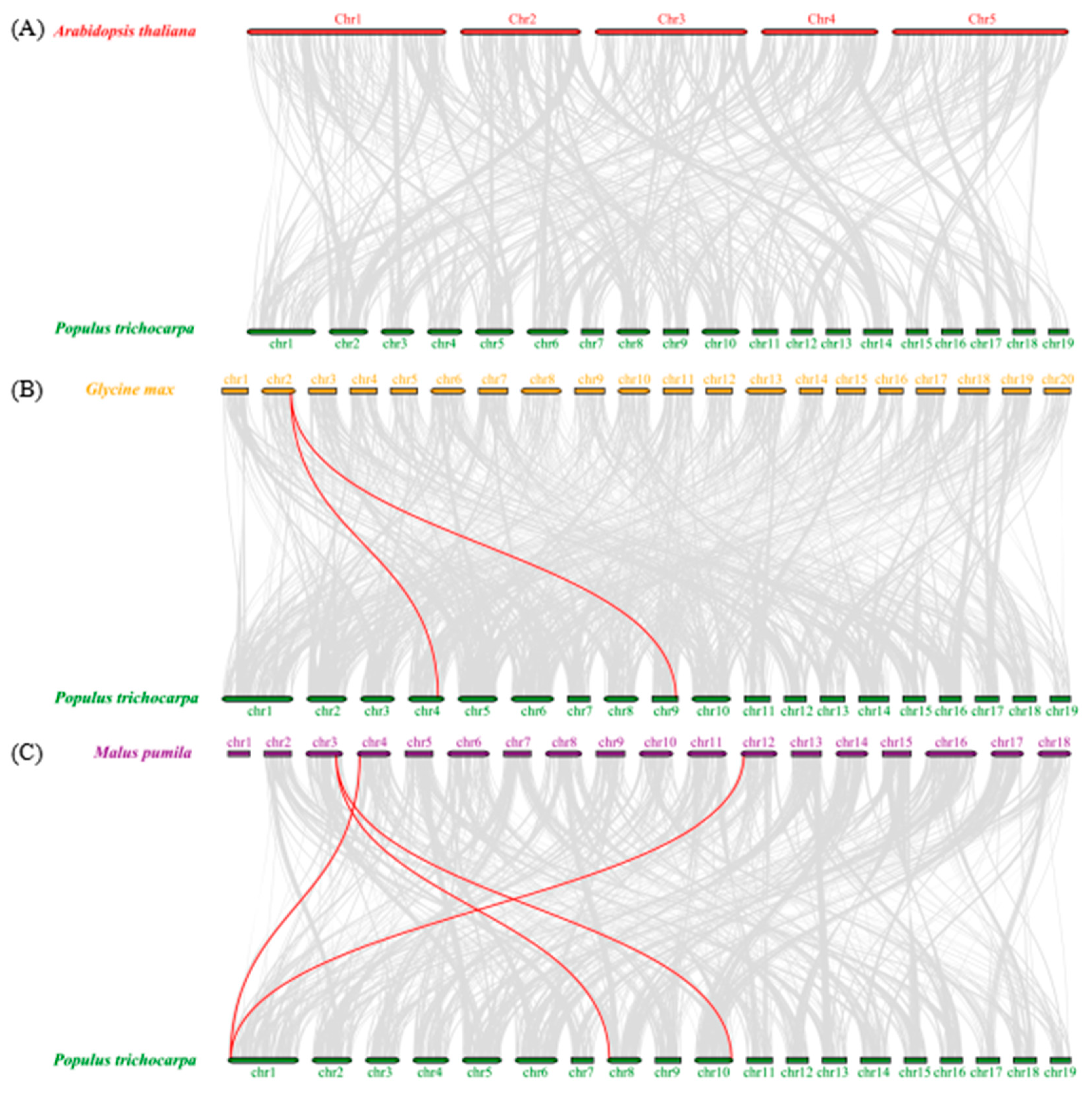
3.2.5. Chromosomal Location, Duplication Analysis, and Ka/Ks Calculation
3.3. Gene Expression Pattern Analysis of EIN3/EIL Genes
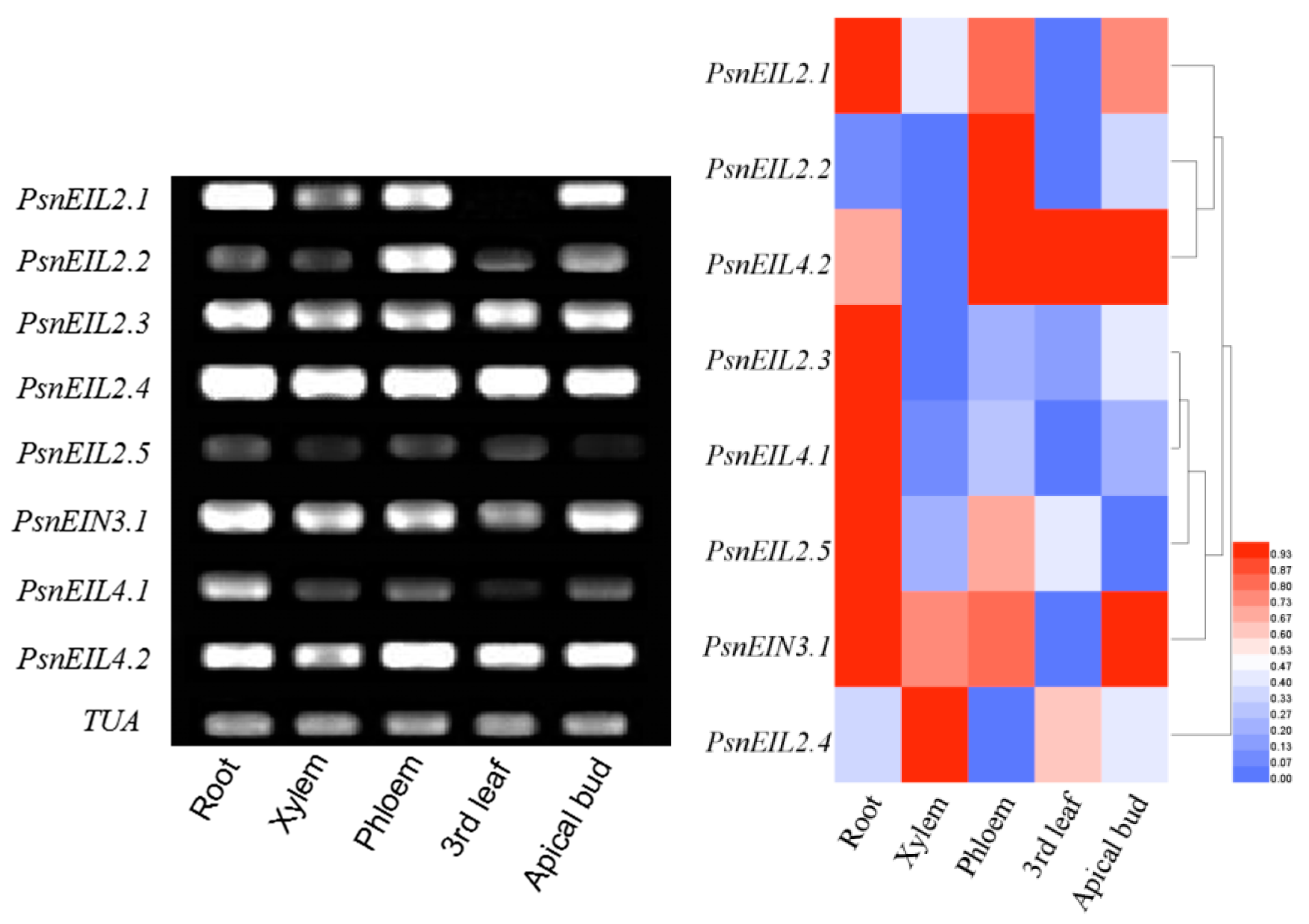
3.3.1. Characterization of Tissue Expression of PsnEIN3/EIL in Populus × xiaohei
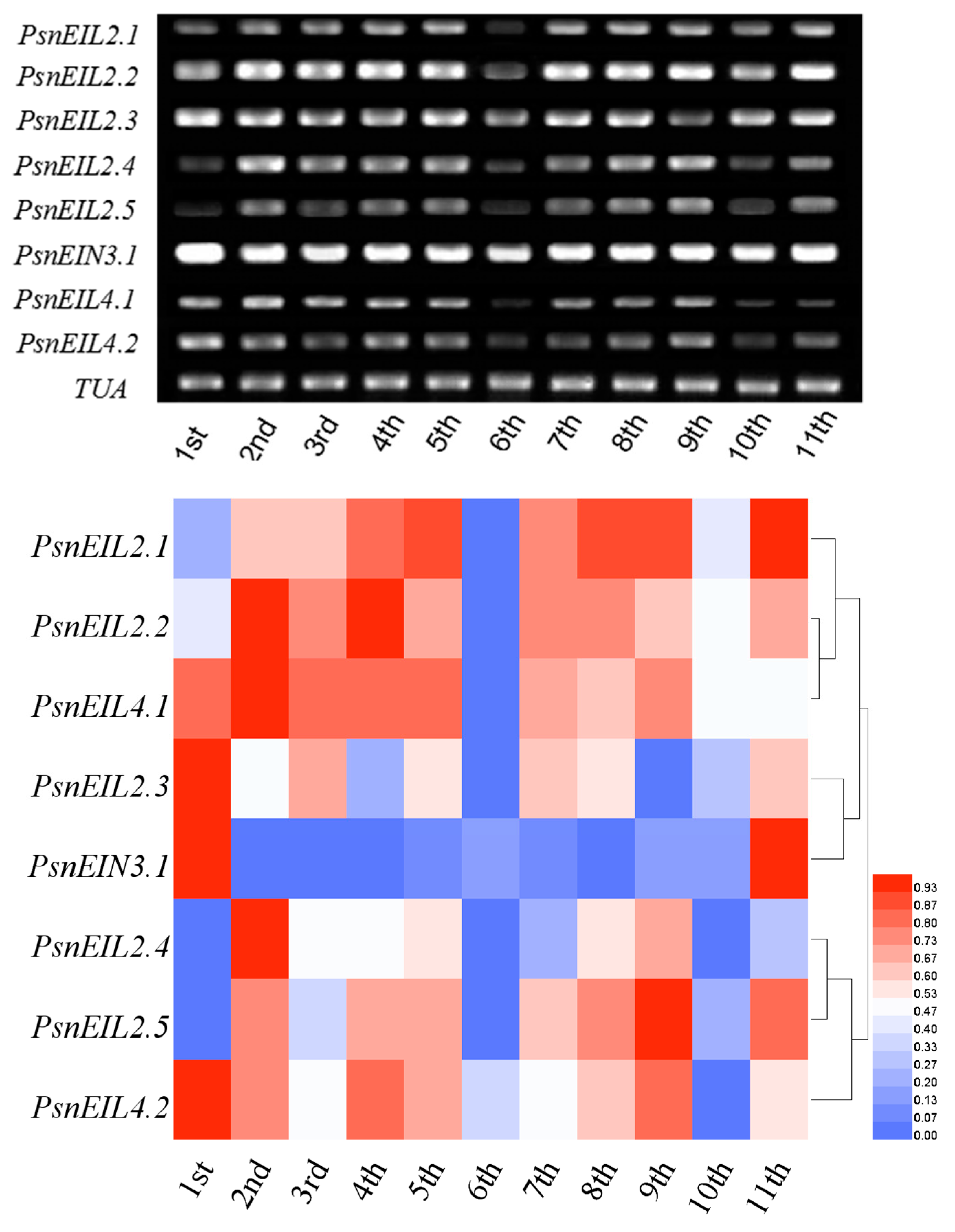
3.3.2. PsnEIN3/EIL Genes Showed Different Expression Patterns during the Development of Stems of Populus × xiaohei
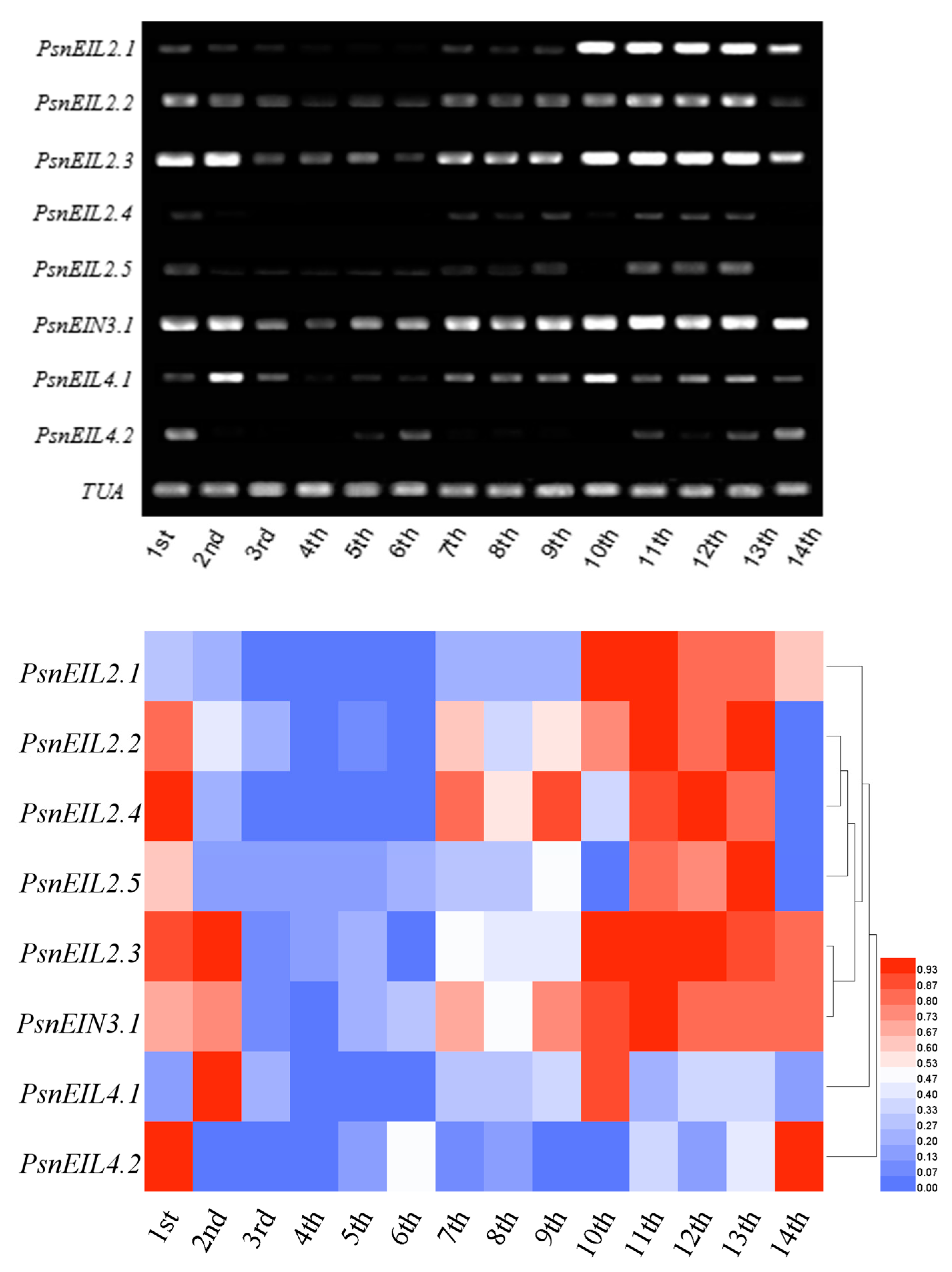
3.3.3. Most PsnEIN3/EIL Genes Are Involved in the Maturation and Senescence Processes of Leaves
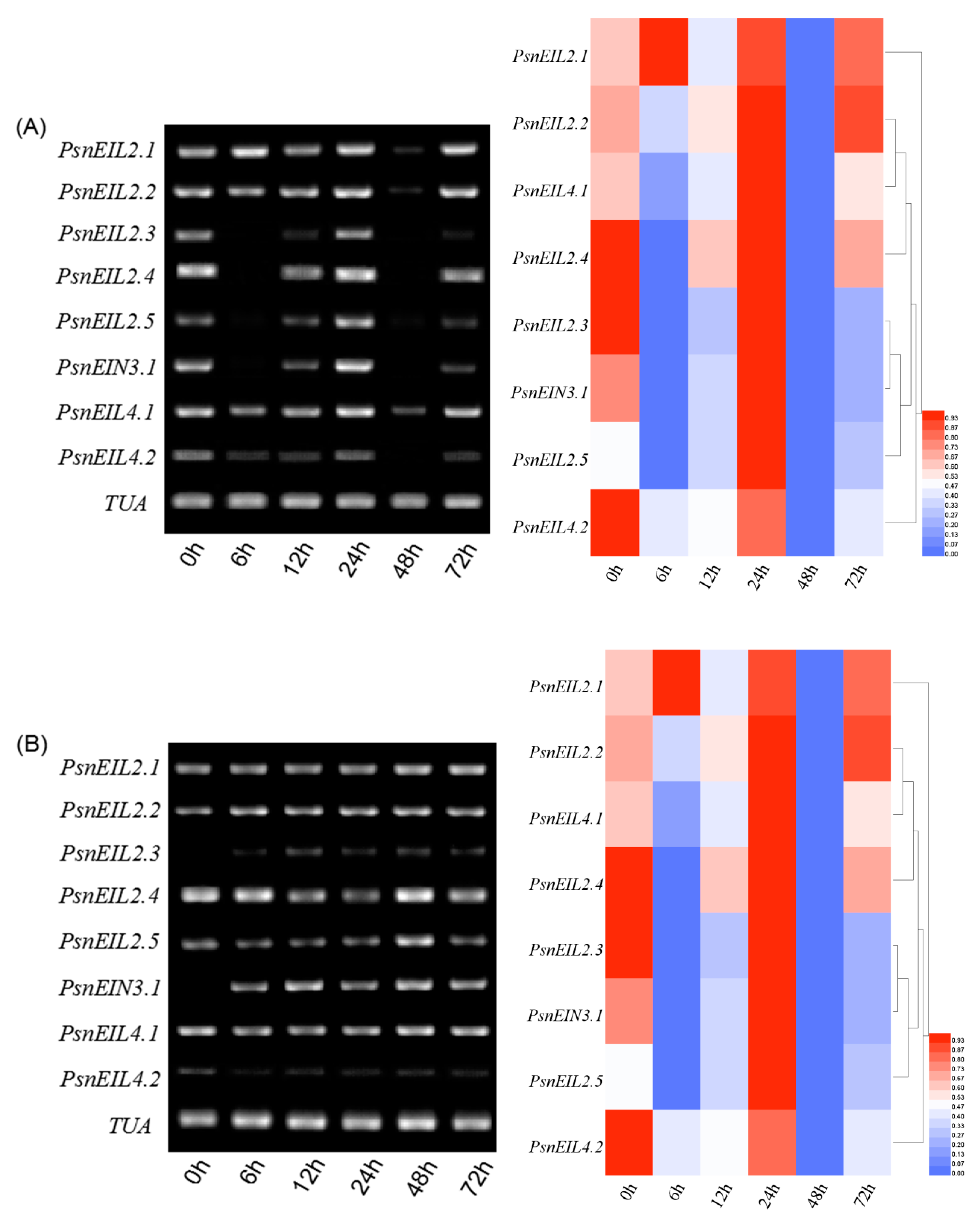
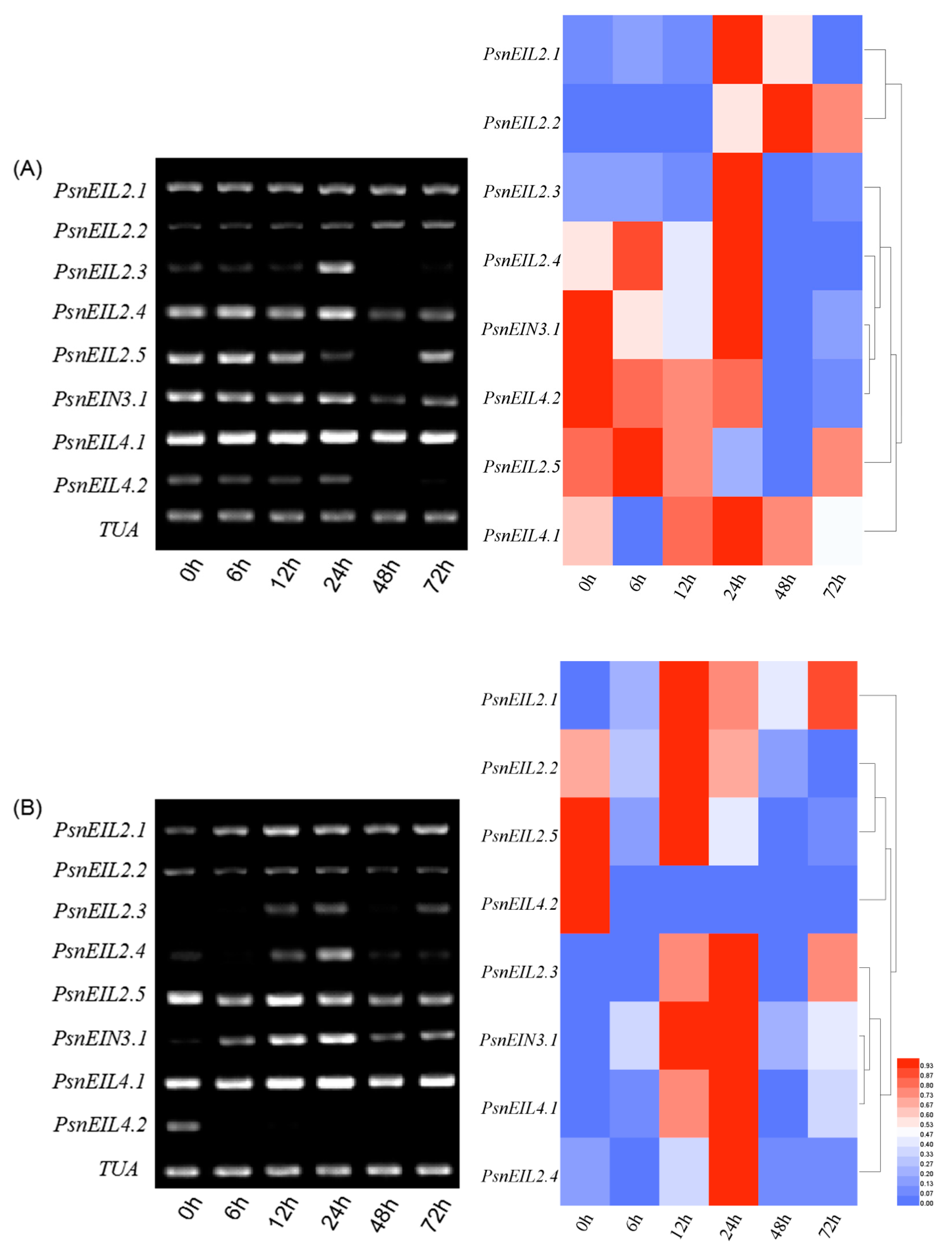
3.3.4. The PsnEIN3/EIL Gene of Populus × xiaohei Could Be Induced by High Salt Concentrations
3.3.5. Involvement of PsnEIN3/EIL in the Response to Heavy Metal Stress in Populus × xiaohei
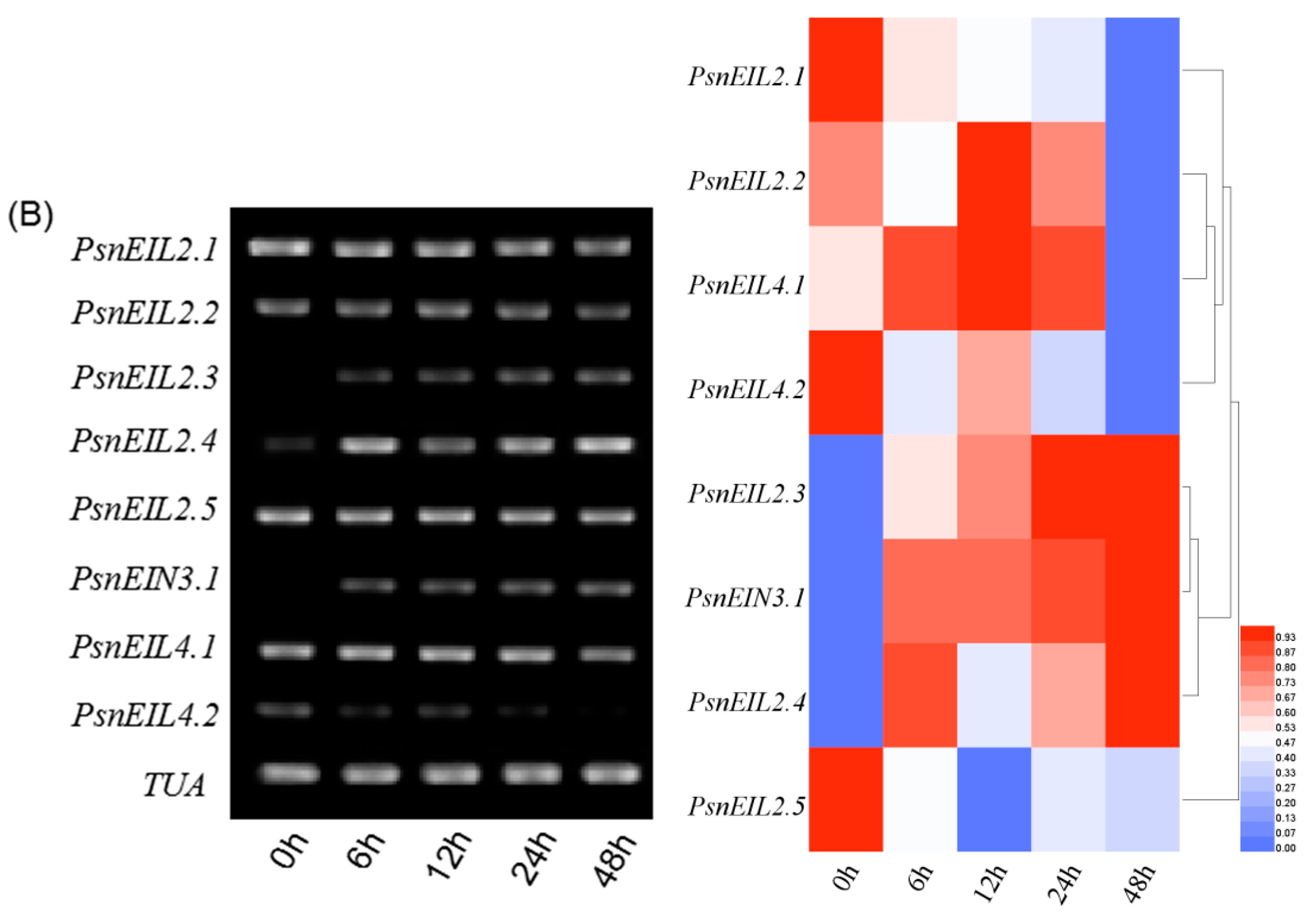
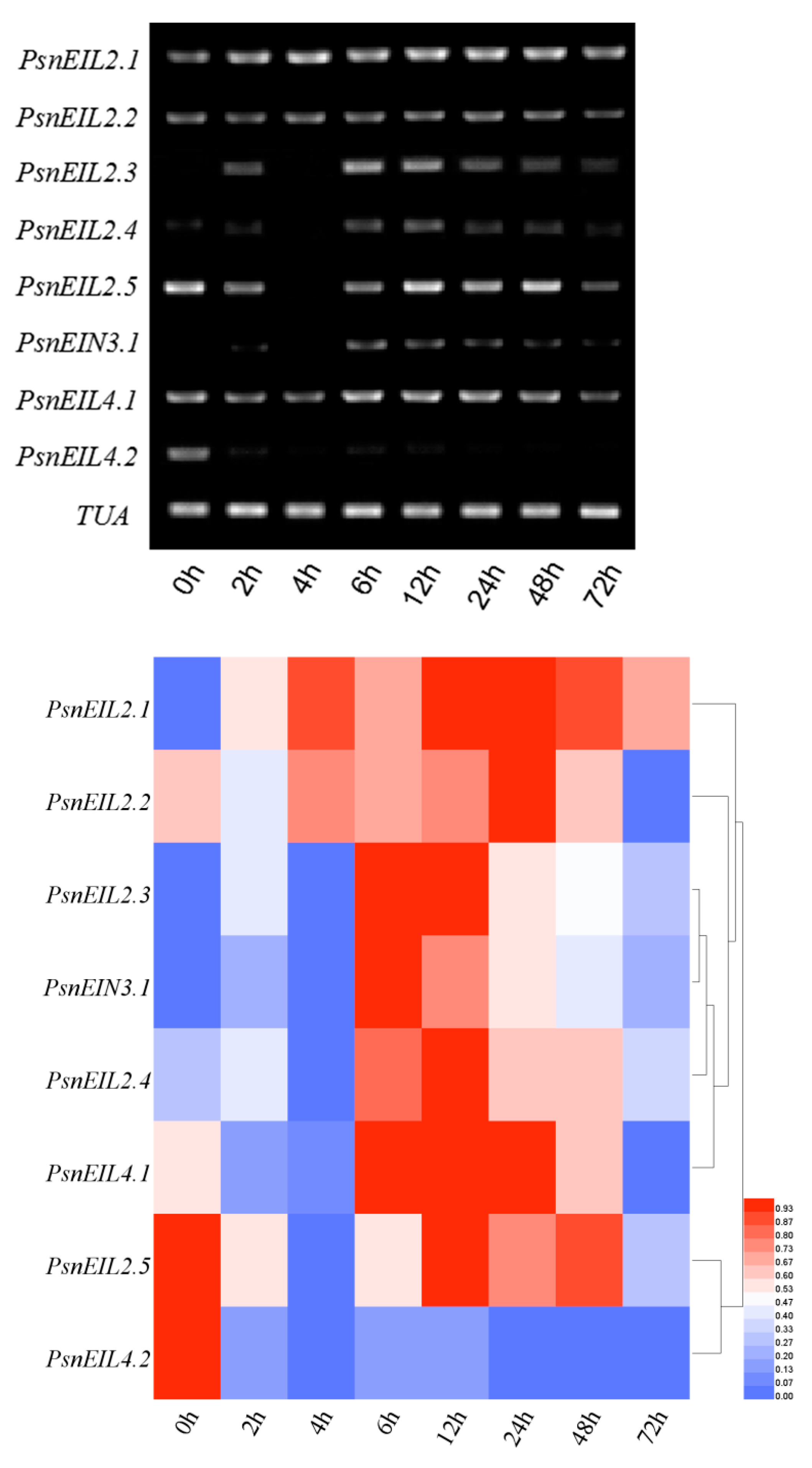
3.3.6. Response of PsnEIN3/EIL Genes to NaHCO3 in Populus × xiaohei
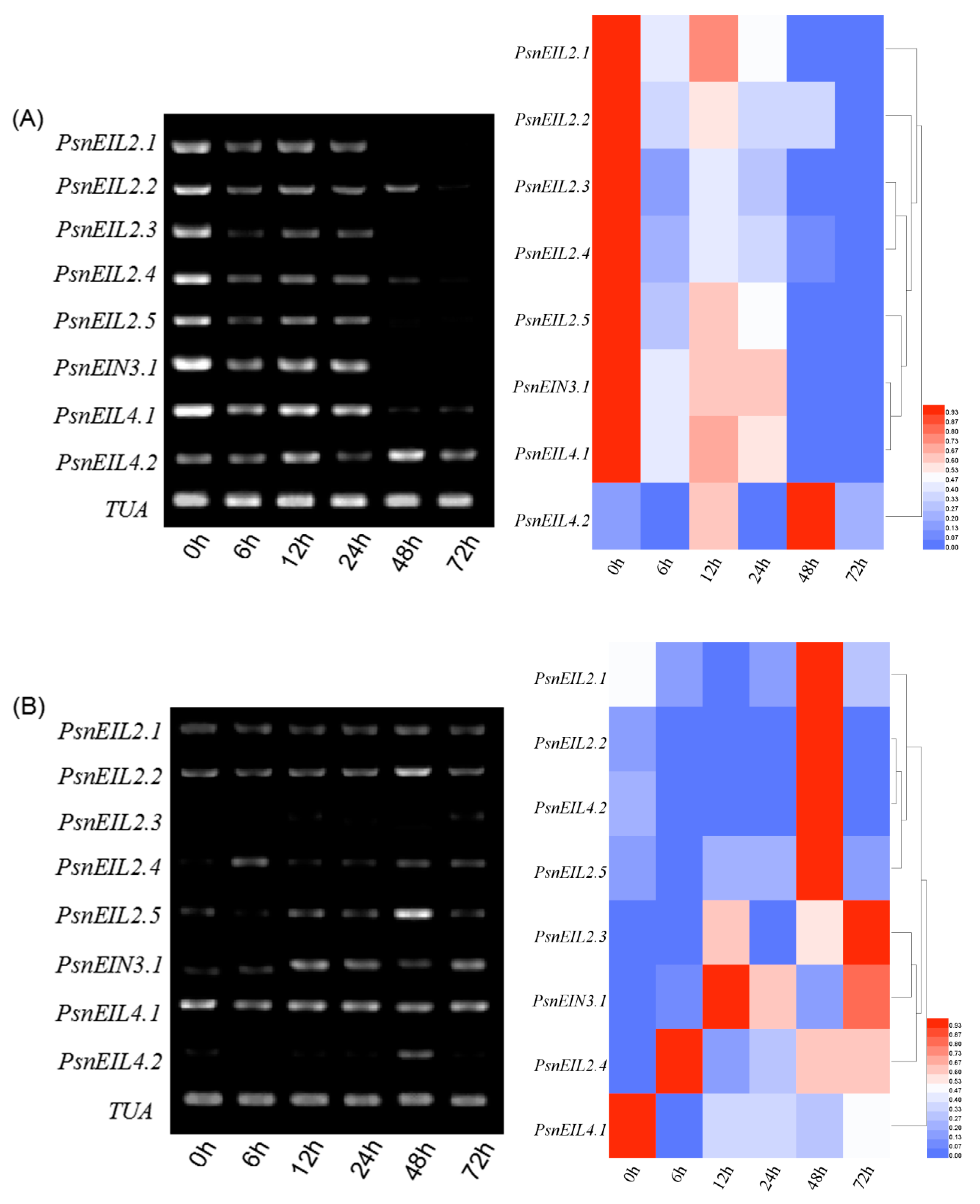
3.3.7. Response of PsnEIN3/EIL Genes to PEG in Populus × xiaohei
3.3.8. The PsnEIN3/EIL Genes were Involved in the Abscisic Acid Response of Populus × xiaohei
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Suttle, J.C.; Hultstrand, J.F. Ethylene-induced leaf abscission in cotton seedlings: The physiological bases for age-dependent differences in sensitivity. Plant Physiol. 1991, 95, 29–33. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Johnson, P.R.; Ecker, J.R. The ethylene gas signal transduction pathway: A molecular perspective. Annu. Rev. Genet. 1998, 32, 227–254. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.M.; Hu, C.Y.; Pang, Y.; Kastaniotis, A.J.; Hiltunen, J.K.; Zhu, Y.X. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell 2007, 19, 3692–3704. [Google Scholar] [CrossRef]
- Wang, D.H.; Li, F.; Duan, Q.H.; Han, T.; Xu, Z.H.; Bai, S.N. Ethylene perception is involved in female cucumber flower development. Plant J. 2010, 61, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Wuriyanghan, H.; Zhang, B.; Cao, W.H.; Ma, B.; Lei, G.; Liu, Y.F.; Wei, W.; Wu, H.J.; Chen, L.J.; Chen, H.W.; et al. The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell 2009, 21, 1473–1494. [Google Scholar] [CrossRef] [PubMed]
- Etchells, J.P.; Provost, C.M.; Turner, S.R. Plant vascular cell division is maintained by an interaction between PXY and ethylene signalling. PLoS Genet. 2012, 8, e1002997. [Google Scholar] [CrossRef]
- Corbineau, F.; Xia, Q.; Bailly, C.; El-Maarouf-Bouteau, H. Ethylene, a key factor in the regulation of seed dormancy. Front. Plant Sci. 2014, 5, 539. [Google Scholar] [CrossRef]
- Dubois, M.; Van den Broeck, L.; Inze, D. The Pivotal Role of Ethylene in Plant Growth. Trends Plant Sci. 2018, 23, 311–323. [Google Scholar] [CrossRef]
- Feng, G.; Sanderson, B.J.; Keefover-Ring, K.; Liu, J.; Ma, T.; Yin, T.; Smart, L.B.; DiFazio, S.P.; Olson, M.S. Pathways to sex determination in plants: How many roads lead to Rome? Curr. Opin. Plant Biol. 2020, 54, 61–68. [Google Scholar] [CrossRef]
- van Loon, L.C.; Geraats, B.P.; Linthorst, H.J. Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 2006, 11, 184–191. [Google Scholar] [CrossRef]
- Clarke, S.M.; Cristescu, S.M.; Miersch, O.; Harren, F.J.M.; Wasternack, C.; Mur, L.A.J. Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol. 2009, 182, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiol. 2010, 154, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Wu, N.; Cui, F.; You, L.; Li, X.; Xiong, L. A homolog of Ethylene Overproducer, OSETOL1, differentially modulates drought and submergence tolerance in rice. Plant J. 2014, 78, 834–849. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Su, J.; Meng, X.; Li, S.; Liu, Y.; Xu, J.; Zhang, S. Multilayered Regulation of Ethylene Induction Plays a Positive Role in Arabidopsis Resistance against Pseudomonas syringae. Plant Physiol. 2015, 169, 299–312. [Google Scholar] [CrossRef]
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef] [PubMed]
- Dolgikh, V.A.; Pukhovaya, E.M.; Zemlyanskaya, E.V. Shaping Ethylene Response: The Role of EIN3/EIL1 Transcription Factors. Front. Plant Sci. 2019, 10, 1030. [Google Scholar] [CrossRef] [PubMed]
- Chao, Q.; Rothenberg, M.; Solano, R.; Roman, G.; Terzaghi, W.; Ecker, J.R. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 1997, 89, 1133–1144. [Google Scholar] [CrossRef]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic. Acids Res. 2000, 28, 960–967. [Google Scholar] [CrossRef]
- Tieman, D.M.; Ciardi, J.A.; Taylor, M.G.; Klee, H.J. Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J. 2001, 26, 47–58. [Google Scholar] [CrossRef]
- Rieu, I.; Mariani, C.; Weterings, K. Expression analysis of five tobacco EIN3 family members in relation to tissue-specific ethylene responses. J. Exp. Bot. 2003, 54, 2239–2244. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Tamura, S.; Nakano, R.; Inaba, A.; Kubo, Y. Characterization of a novel tomato EIN3-like gene (LeEIL4). J. Exp. Bot. 2003, 54, 2775–2776. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ecker, J.R. The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 2004, 7, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Wang, S.; Jia, Q.; Wu, P. OsEIL1, a rice homolog of the Arabidopsis EIN3 regulates the ethylene response as a positive component. Plant Mol. Biol. 2006, 61, 141–152. [Google Scholar] [CrossRef]
- Hiraga, S.; Sasaki, K.; Hibi, T.; Yoshida, H.; Uchida, E.; Kosugi, S.; Kato, T.; Mie, T.; Ito, H.; Katou, S.; et al. Involvement of two rice ETHYLENE INSENSITIVE3-LIKE genes in wound signaling. Mol. Genet. Genom. 2009, 282, 517–529. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, F.; Shi, X.; Liu, X.; Yang, H.; Zhang, Z. Identification and Characterization of the Cyclin-Dependent Kinases Gene Family in Silkworm, Bombyx mori. DNA Cell Biol. 2016, 35, 13–23. [Google Scholar] [CrossRef]
- Li, Z.; Peng, J.; Wen, X.; Guo, H. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 2013, 25, 3311–3328. [Google Scholar] [CrossRef]
- Hua, Q.; Zhijin, Z.; Juan, W.; Xinbing, C.; Pengcheng, W.; Rongfeng, H.; Hao, Y. The activation of OsEIL1 on YUC8 transcription and auxin biosynthesis is required for ethylene-inhibited root elongation in rice early seedling development. PLoS Genet. 2017, 13, e1006955. [Google Scholar]
- Hibi, T.; Kosugi, S.; Iwai, T.; Kawata, M.; Seo, S.; Mitsuhara, I.; Ohashi, Y. Involvement of EIN3 homologues in basic PR gene expression and flower development in tobacco plants. J. Exp. Bot. 2007, 58, 3671–3678. [Google Scholar] [CrossRef]
- Zhong, S.; Zhao, M.; Shi, T.; Shi, H.; An, F.; Zhao, Q.; Guo, H. EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 2009, 106, 21431–21436. [Google Scholar] [CrossRef]
- Liu, X.; Liu, R.; Li, Y.; Shen, X.; Zhong, S.; Shi, H. EIN3 and PIF3 Form an Interdependent Module That Represses Chloroplast Development in Buried Seedlings. Plant Cell 2017, 29, 3051–3067. [Google Scholar] [CrossRef] [PubMed]
- Sirko, A.; Wawrzynska, A.; Rodriguez, M.C.; Sektas, P. The family of LSU-like proteins. Front. Plant Sci. 2014, 5, 774. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Z.; Quan, R.; Li, G.; Wang, R.; Huang, R. An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol. 2011, 157, 854–865. [Google Scholar] [CrossRef]
- Liu, C.; Li, J.; Zhu, P.; Yu, J.; Hou, J.; Wang, C.; Long, D.; Yu, M.; Zhao, A. Mulberry EIL3 confers salt and drought tolerances and modulates ethylene biosynthetic gene expression. PeerJ 2019, 7, e6391. [Google Scholar] [CrossRef] [PubMed]
- Yang, L. Ethylene Signal is Involved in Regulating Arabidopsis Response to Drought Stress Stimulated by PEG. Plant Divers. 2010, 32, 89–96. [Google Scholar]
- Kong, X.; Li, C.; Zhang, F.; Yu, Q.; Gao, S.; Zhang, M.; Tian, H.; Zhang, J.; Yuan, X.; Ding, Z. Ethylene promotes cadmium-induced root growth inhibition through EIN3 controlled XTH33 and LSU1 expression in Arabidopsis. Plant Cell Environ. 2018, 41, 2449–2462. [Google Scholar] [CrossRef]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Mu, A.; Jiang, Z.; Kim, J.M.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef]
- Shi, H.; Zhang, Y.; Chen, L. Expression and Regulation of PpEIN3b during Fruit Ripening and Senescence via Integrating SA, Glucose, and ACC Signaling in Pear (Pyrus pyrifolia Nakai. Whangkeumbae). Genes 2019, 10, 476. [Google Scholar] [CrossRef]
- Li, Y.; Jin, C.; Liu, Y.; Wang, L.; Li, F.; Wang, B.; Liu, G.; Jiang, J.; Li, H. Global Analysis of the WOX Transcription Factor Gene Family in Populus × xiaohei T. S. Hwang et Liang Reveals Their Stress−Responsive Patterns. Forests 2022, 13, 122. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Bolser, D.M.; Staines, D.M.; Perry, E.; Kersey, P.J. Ensembl Plants: Integrating Tools for Visualizing, Mining, and Analyzing Plant Genomic Data. Methods Mol. Biol. 2017, 1533, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Rozas, R. DnaSP version 2.0: A novel software package for extensive molecular population genetics analysis. Comput. Appl. Biosci. 1997, 13, 307–311. [Google Scholar] [PubMed]
- Wang, F. Semi-Quantitative RT-PCR: An Effective Method to Explore the Regulation of Gene Transcription Level Affected by Environmental Pollutants. Methods Mol. Biol. 2021, 2326, 95–103. [Google Scholar] [CrossRef]
- Abdelmigid, H.M. Expression analysis of Type 1 and 2 Metallothionein genes in Rape (Brassica napus L.) during short-term stress using sqRT-PCR analysis. Indian J. Exp. Biol. 2016, 54, 212–218. [Google Scholar]
- Aguilar-Hernandez, H.S.; Santos, L.; Leon-Galvan, F.; Barrera-Pacheco, A.; Espitia-Rangel, E.; De Leon-Rodriguez, A.; Guevara-Gonzalez, R.G.; Barba de la Rosa, A.P. Identification of calcium stress induced genes in amaranth leaves through suppression subtractive hybridization. J. Plant Physiol. 2011, 168, 2102–2109. [Google Scholar] [CrossRef]
- Mukhopadhyay, P.; Tyagi, A.K. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 2015, 5, 9998. [Google Scholar] [CrossRef]
- Zhou, M.; Li, D.; Li, Z.; Hu, Q.; Yang, C.; Zhu, L.; Luo, H. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 2013, 161, 1375–1391. [Google Scholar] [CrossRef]
- Ji, R.; Ge, W.; Wang, H.; Zhao, Y.; Feng, H. BrSKS13, a multiple-allele-inherited male sterility-related gene in Chinese cabbage (Brassica rapa L. ssp. pekinensis), affects pollen development and pollination/fertilization process. Gene 2019, 696, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Samanta, S.K.; Ghosh, S.K. The chitin biosynthesis pathway in Entamoeba and the role of glucosamine-6-P isomerase by RNA interference. Mol. Biochem. Parasitol. 2012, 186, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.B.; Ji, Z.R.; Chi, F.M.; Qiao, Z.; Xu, C.N.; Zhang, J.X.; Zhou, Z.S.; Dong, Q.L. Genome-wide identification and expression analysis of the WRKY gene family in peach. Yi Chuan 2016, 38, 254–270. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, R.; Liang, Z.; Wu, X.; Wang, J. Genome-wide identification and analysis of the EIN3/EIL gene family in allotetraploid Brassica napus reveal its potential advantages during polyploidization. BMC Plant Biol. 2019, 19, 110. [Google Scholar] [CrossRef]
- Liu, L.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Smith, C.; Bevan, M.W. The activities of acidic and glutamine-rich transcriptional activation domains in plant cells: Design of modular transcription factors for high-level expression. Plant Mol. Biol. 1998, 36, 195–204. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Xue, Y.; Zhu, B.; Xue, P.; Tian, H.; Luo, Y. Construction of Plant Antisense Gene Expression Vector pBI121-LeEIL2 and Transformation in Tomato. J. Henan Agric. Sci. 2003, 47–50. [Google Scholar]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Tomo, Y.; Terada, T.; et al. Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3. J. Mol. Biol. 2005, 348, 253–264. [Google Scholar] [CrossRef]
- Solano, R.; Stepanova, A.; Chao, Q.; Ecker, J.R. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998, 12, 3703–3714. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Li, Z.; Wen, X.; Li, W.; Shi, H.; Yang, L.; Zhu, H.; Guo, H. Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 2014, 10, e1004664. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, X.; Guo, W.; Zhang, P.; Zhang, W. Sequence Analysis of EIN3/EIL Gene Family and Cloning of GhEIL3 in Cotton. Acta Bot. Boreal.-Occident. Sin. 2017, 37, 445–452. [Google Scholar]
- Filiz, E.; Vatansever, R.; Ozyigit, I.I.; Uras, M.E.; Sen, U.; Anjum, N.A.; Pereira, E. Genome-wide identification and expression profiling of EIL gene family in woody plant representative poplar (Populus trichocarpa). Arch. Biochem. Biophys. 2017, 627, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Tu, Y.; Chen, X.L.; Liu, X.G. Genome-wide Identification and Phylogenetic Analysis of the EIN3/EILs Gene Family in Melons. Genom. Appl. Biol. 2014, 33, 105–112. [Google Scholar]
- Wagner, A. The fate of duplicated genes: Loss or new function? Bioessays 1998, 20, 785–788. [Google Scholar] [CrossRef]
- Li, G.; Zhang, L.; Wu, J.; Yue, X.; Wang, M.; Sun, L.; Di, D.; Kronzucker, H.J.; Shi, W. OsEIL1 protects rice growth under NH4+ nutrition by regulating OsVTC1-3-dependent N-glycosylation and root NH4+ efflux. Plant Cell Environ. 2022, 1–17. [Google Scholar] [CrossRef]
- Feng, Y.; Xu, P.; Li, B.; Li, P.; Wen, X.; An, F.; Gong, Y.; Xin, Y.; Zhu, Z.; Wang, Y.; et al. Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 13834–13839. [Google Scholar] [CrossRef]
- Qiu, K.; Li, Z.; Yang, Z.; Chen, J.; Wu, S.; Zhu, X.; Gao, S.; Gao, J.; Ren, G.; Kuai, B.; et al. EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399. [Google Scholar] [CrossRef]
- Wang, S.; Li, L.X.; Zhang, Z.; Fang, Y.; Li, D.; Chen, X.S.; Feng, S.Q. Ethylene precisely regulates anthocyanin synthesis in apple via a module comprising MdEIL1, MdMYB1, and MdMYB17. Hortic. Res. 2022, uhac034. [Google Scholar] [CrossRef]
- Xiao, F.; Gong, Q.; Zhao, S.; Lin, H.; Zhou, H. MYB30 and ETHYLENE INSENSITIVE3 Antagonistically Modulate Root Hair Growth in Arabidopsis. Plant J. 2021, 106, 480–492. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ko, E.E.; Tran, J.; Qiao, H. TREE1-EIN3–mediated transcriptional repression inhibits shoot growth in response to ethylene. Proc. Natl. Acad. Sci. USA 2020, 117, 29178–29189. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pan, Y.; Chang, B.; Wang, Y.; Tang, Z. NO Promotes Seed Germination and Seedling Growth Under High Salt May Depend on EIN3 Protein in Arabidopsis. Front. Plant Sci. 2015, 6, 1203. [Google Scholar] [CrossRef] [PubMed]


| Gene | Chromosome | ORF Length | Protein Length /Amino Acids | PI | Molecular Weight (Da) | Subcellular Localization |
|---|---|---|---|---|---|---|
| PsnEIN3.1 | Chr04 | 1854 | 617 | 5.68 | 70,025.84 | Nucleus |
| PsnEIL2.1 | Chr10 | 1539 | 556 | 5.97 | 62,939.99 | Nucleus |
| PsnEIL2.2 | Chr10 | 1980 | 659 | 5.49 | 74,463.65 | Nucleus |
| PsnEIL2.3 | Chr08 | 1989 | 662 | 5.28 | 74,991.26 | Nucleus |
| PsnEIL2.4 | Chr08 | 1989 | 662 | 5.29 | 74,934.21 | Nucleus |
| PsnEIL2.5 | Chr08 | 1962 | 653 | 5.38 | 73,785.89 | Nucleus |
| PsnEIL4.1 | Chr01 | 1197 | 398 | 8.24 | 44,013.25 | Nucleus |
| PsnEIL4.2 | Chr03 | 1788 | 595 | 5.60 | 66,742.84 | Nucleus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Jin, C.; Li, Y.; Wang, L.; Li, F.; Wang, B.; Jiang, J.; Zheng, Z.; Li, H. Identification and Analysis of the EIN3/EIL Gene Family in Populus × xiaohei T. S. Hwang et Liang: Expression Profiling during Stress. Forests 2022, 13, 382. https://doi.org/10.3390/f13030382
Liu Y, Jin C, Li Y, Wang L, Li F, Wang B, Jiang J, Zheng Z, Li H. Identification and Analysis of the EIN3/EIL Gene Family in Populus × xiaohei T. S. Hwang et Liang: Expression Profiling during Stress. Forests. 2022; 13(3):382. https://doi.org/10.3390/f13030382
Chicago/Turabian StyleLiu, Yuting, Chunhui Jin, Yue Li, Lili Wang, Fangrui Li, Bo Wang, Jing Jiang, Zhimin Zheng, and Huiyu Li. 2022. "Identification and Analysis of the EIN3/EIL Gene Family in Populus × xiaohei T. S. Hwang et Liang: Expression Profiling during Stress" Forests 13, no. 3: 382. https://doi.org/10.3390/f13030382
APA StyleLiu, Y., Jin, C., Li, Y., Wang, L., Li, F., Wang, B., Jiang, J., Zheng, Z., & Li, H. (2022). Identification and Analysis of the EIN3/EIL Gene Family in Populus × xiaohei T. S. Hwang et Liang: Expression Profiling during Stress. Forests, 13(3), 382. https://doi.org/10.3390/f13030382





