Analyzing TLS Scan Distribution and Point Density for the Estimation of Forest Stand Structural Parameters
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
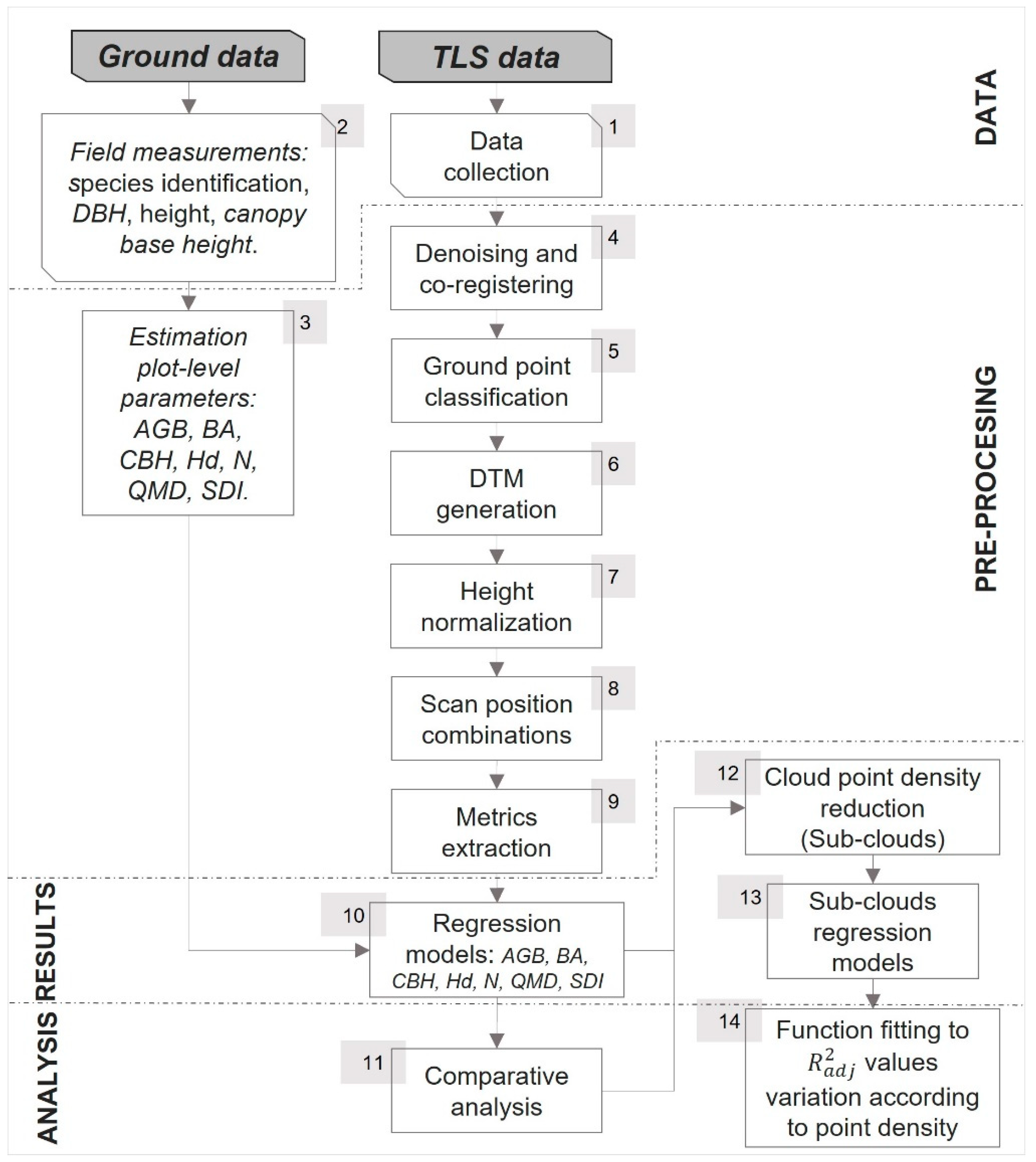
2.2. Methodology Overview
2.3. Field Data Collection
2.3.1. Reference Field Data
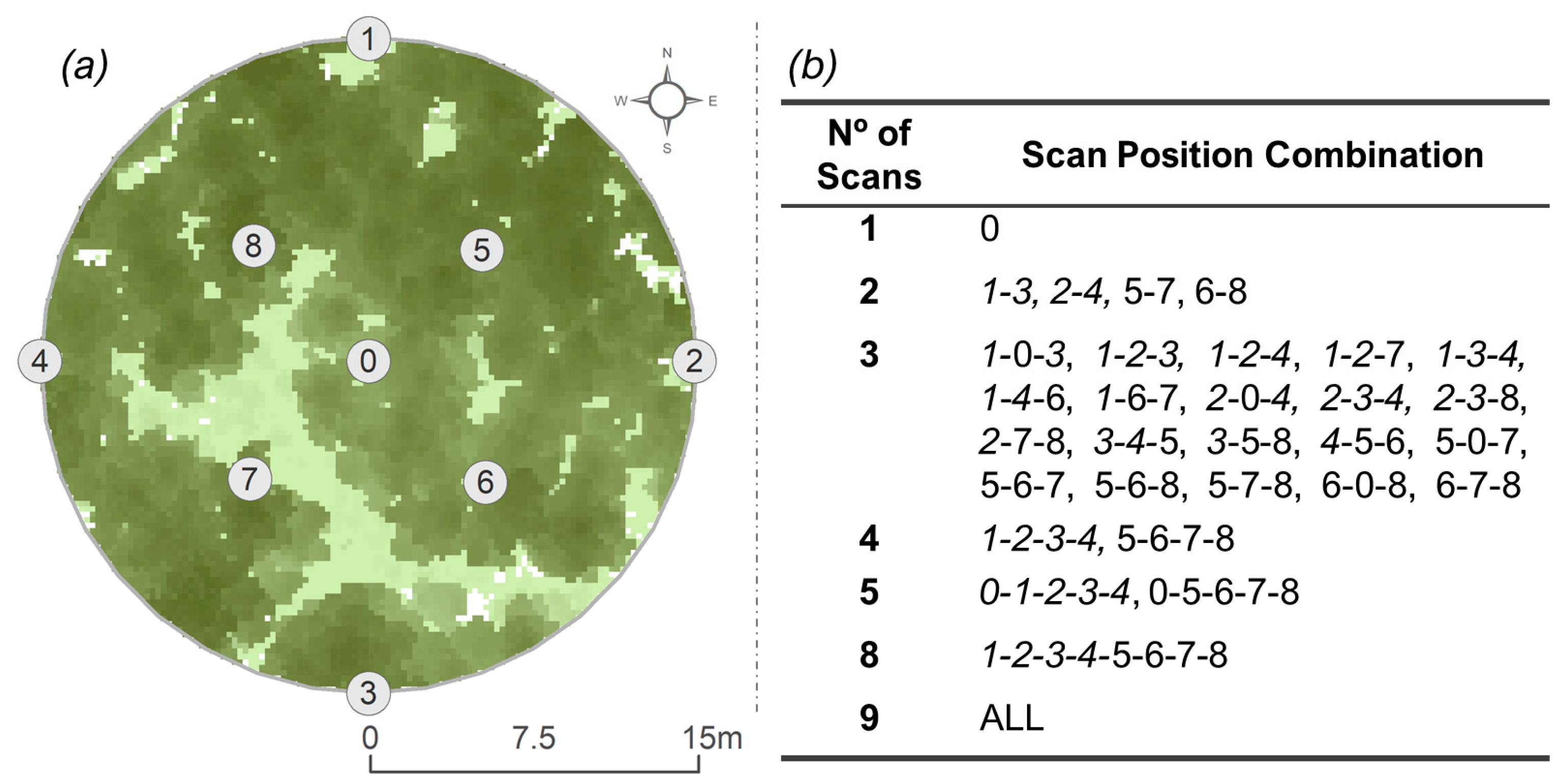
2.3.2. TLS Data Collection
2.4. Data Pre-Processing
2.5. Regression Models
2.6. TLS Point Cloud Density Reduction
3. Results
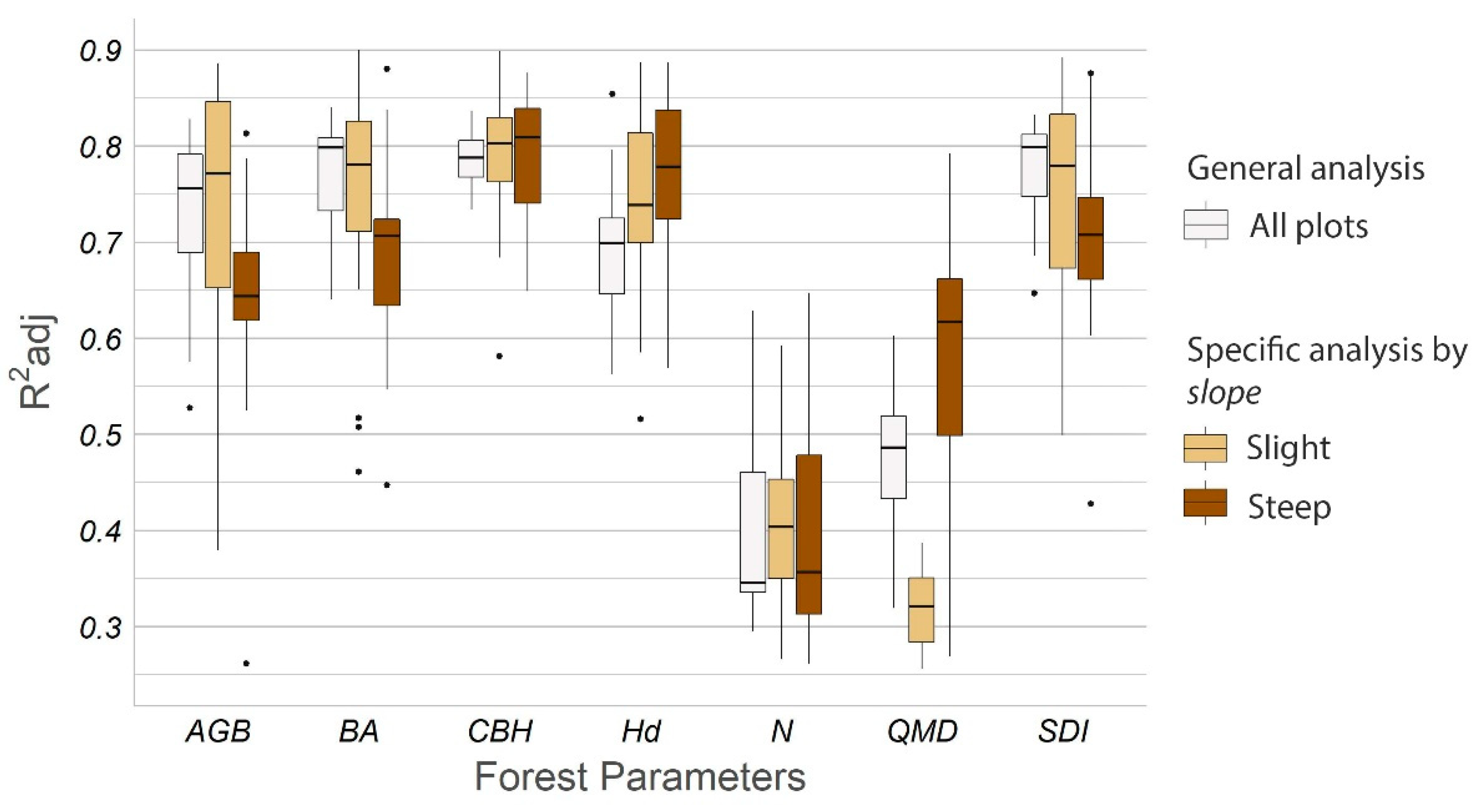
3.1. Estimate of Forest Parameters Varying the Number and Distribution of TLS Scans
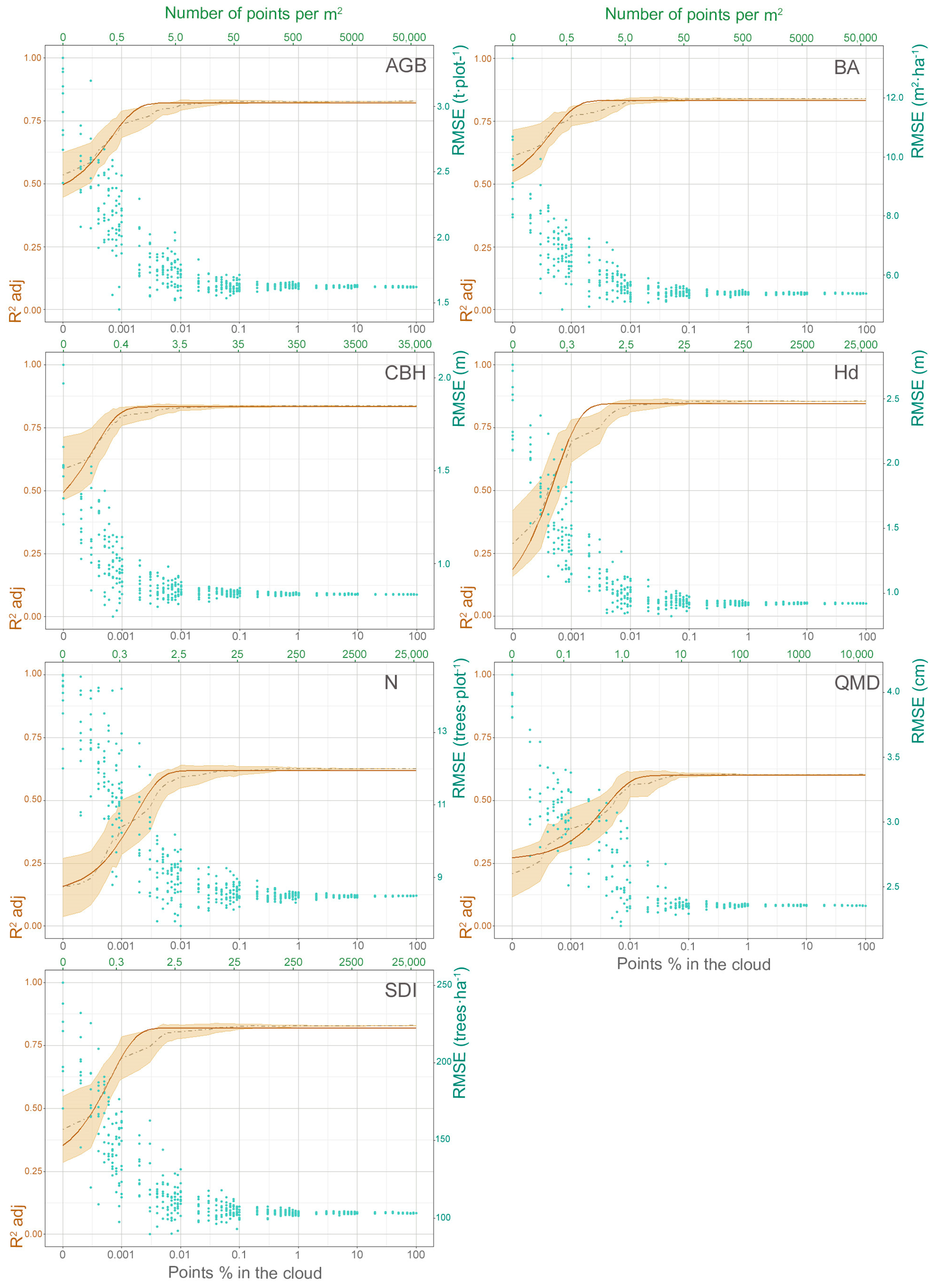
3.2. Analysis of the TLS Point Cloud Density Reduction
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tinkham, W.T.; Mahoney, P.R.; Hudak, A.T.; Domke, G.M.; Falkowski, M.J.; Woodall, C.W.; Smith, A.M.S. Applications of the United States Forest Inventory and Analysis Dataset: A Review and Future Directions. Can. J. For. Res. 2018, 48, 1251–1268. [Google Scholar] [CrossRef]
- Lister, A.J.; Andersen, H.; Frescino, T.; Gatziolis, D.; Healey, S.; Heath, L.S.; Liknes, G.C.; McRoberts, R.; Moisen, G.G.; Nelson, M.; et al. Use of Remote Sensing Data to Improve the Efficiency of National Forest Inventories: A Case Study from the United States National Forest Inventory. Forests 2020, 11, 1364. [Google Scholar] [CrossRef]
- Newnham, G.J.; Armston, J.D.; Calders, K.; Disney, M.I.; Lovell, J.L.; Schaaf, C.B.; Strahler, A.H.; Mark Danson, F. Terrestrial Laser Scanning for Plot-Scale Forest Measurement. Curr. For. Rep. 2015, 1, 239–251. [Google Scholar] [CrossRef]
- Wilkes, P.; Lau, A.; Disney, M.; Calders, K.; Burt, A.; Gonzalez de Tanago, J.; Bartholomeus, H.; Brede, B.; Herold, M. Data Acquisition Considerations for Terrestrial Laser Scanning of Forest Plots. Remote Sens. Environ. 2017, 196, 140–153. [Google Scholar] [CrossRef]
- LaRue, E.A.; Wagner, F.W.; Fei, S.; Atkins, J.W.; Fahey, R.T.; Gough, C.M.; Hardiman, B.S. Compatibility of Aerial and Terrestrial LiDAR for Quantifying Forest Structural Diversity. Remote Sens. 2020, 12, 1407. [Google Scholar] [CrossRef]
- Liang, X.; Kankare, V.; Hyyppä, J.; Wang, Y.; Kukko, A.; Haggrén, H.; Yu, X.; Kaartinen, H.; Jaakkola, A.; Guan, F.; et al. Terrestrial Laser Scanning in Forest Inventories. ISPRS J. Photogramm. Remote Sens. 2016, 115, 63–77. [Google Scholar] [CrossRef]
- Liang, X.; Hyyppä, J.; Kaartinen, H.; Lehtomäki, M.; Pyörälä, J.; Pfeifer, N.; Holopainen, M.; Brolly, G.; Francesco, P.; Hackenberg, J.; et al. International Benchmarking of Terrestrial Laser Scanning Approaches for Forest Inventories. ISPRS J. Photogramm. Remote Sens. 2018, 144, 137–179. [Google Scholar] [CrossRef]
- Bauwens, S.; Bartholomeus, H.; Calders, K.; Lejeune, P. Forest Inventory with Terrestrial LiDAR: A Comparison of Static and Hand-Held Mobile Laser Scanning. Forests 2016, 7, 127. [Google Scholar] [CrossRef]
- Liang, X.; Kukko, A.; Kaartinen, H.; Hyyppä, J.; Yu, X.; Jaakkola, A.; Wang, Y. Possibilities of a Personal Laser Scanning System for Forest Mapping and Ecosystem Services. Sensors 2014, 14, 1228–1248. [Google Scholar] [CrossRef]
- Zimbres, B.; Shimbo, J.; Bustamante, M.; Levick, S.; Miranda, S.; Roitman, I.; Silvério, D.; Gomes, L.; Fagg, C.; Alencar, A. Savanna Vegetation Structure in the Brazilian Cerrado Allows for the Accurate Estimation of Aboveground Biomass Using Terrestrial Laser Scanning. For. Ecol. Manage. 2020, 458, 117798. [Google Scholar] [CrossRef]
- Crespo-Peremarch, P.; Fournier, R.A.; Nguyen, V.; van Lier, O.R.; Ruiz, L.Á. A Comparative Assessment of the Vertical Distribution of Forest Components Using Full-Waveform Airborne, Discrete Airborne and Discrete Terrestrial Laser Scanning Data. For. Ecol. Manage. 2020, 473, 118268. [Google Scholar] [CrossRef]
- Danson, F.M.; Hetherington, D.; Morsdorf, F.; Koetz, B.; Allgower, B. Forest Canopy Gap Fraction From Terrestrial Laser Scanning. IEEE Geosci. Remote Sens. Lett. 2007, 4, 157–160. [Google Scholar] [CrossRef]
- Lovell, J.L.; Jupp, D.L.B.; Culvenor, D.S.; Coops, N.C. Using Airborne and Ground-Based Ranging Lidar to Measure Canopy Structure in Australian Forests. Can. J. Remote Sens. 2003, 29, 607–622. [Google Scholar] [CrossRef]
- Calders, K.; Adams, J.; Armston, J.; Bartholomeus, H.; Bauwens, S.; Bentley, L.P.; Chave, J.; Danson, F.M.; Demol, M.; Disney, M.; et al. Terrestrial Laser Scanning in Forest Ecology: Expanding the Horizon. Remote Sens. Environ. 2020, 251, 112102. [Google Scholar] [CrossRef]
- Domingo, D.; Lamelas, M.; Montealegre, A.; García-Martín, A.; de la Riva, J. Estimation of Total Biomass in Aleppo Pine Forest Stands Applying Parametric and Nonparametric Methods to Low-Density Airborne Laser Scanning Data. Forests 2018, 9, 158. [Google Scholar] [CrossRef]
- van Leeuwen, M.; Nieuwenhuis, M. Retrieval of Forest Structural Parameters Using LiDAR Remote Sensing. Eur. J. For. Res. 2010, 129, 749–770. [Google Scholar] [CrossRef]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote Sensing Technologies for Enhancing Forest Inventories: A Review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef]
- Srinivasan, S.; Popescu, S.C.; Eriksson, M.; Sheridan, R.D.; Ku, N.W. Multi-Temporal Terrestrial Laser Scanning for Modeling Tree Biomass Change. For. Ecol. Manage. 2014, 318, 304–317. [Google Scholar] [CrossRef]
- Torralba, J.; Crespo-Peremarch, P.; Ruiz, L.Á. Assessing the Use of Discrete, Full-Waveform LiDAR and TLS to Classify Mediterranean Forest Species Composition. Rev. Teledetección 2018, 52, 27. [Google Scholar] [CrossRef]
- Gollob, C.; Ritter, T.; Wassermann, C.; Nothdurft, A. Influence of Scanner Position and Plot Size on the Accuracy of Tree Detection and Diameter Estimation Using Terrestrial Laser Scanning on Forest Inventory Plots. Remote Sens. 2019, 11, 1602. [Google Scholar] [CrossRef]
- Molina-Valero, J.A.; Martínez-Calvo, A.; Ginzo Villamayor, M.J.; Novo Pérez, M.A.; Álvarez-González, J.G.; Montes, F.; Pérez-Cruzado, C. Operationalizing the Use of TLS in Forest Inventories: The R Package FORTLS. Environ. Model. Softw. 2022, 150, 105337. [Google Scholar] [CrossRef]
- Torralba, J.; Ruiz, L.Á.; Carbonell-Rivera, J.P.; Crespo-Peremarch, P. Análisis de Posiciones y Densidades TLS (Terrestrial Laser Scanning) Para Optimizar La Estimación de Parámetros Forestales. Ed. Univ. De Valladolid 2019, 443–446. [Google Scholar]
- Astrup, R.; Ducey, M.J.; Granhus, A.; Ritter, T.; von Lüpke, N. Approaches for Estimating Stand-Level Volume Using Terrestrial Laser Scanning in a Single-Scan Mode. Can. J. For. Res. 2014, 44, 666–676. [Google Scholar] [CrossRef]
- Lovell, J.L.; Jupp, D.L.B.; Newnham, G.J.; Culvenor, D.S. Measuring Tree Stem Diameters Using Intensity Profiles from Ground-Based Scanning Lidar from a Fixed Viewpoint. ISPRS J. Photogramm. Remote Sens. 2011, 66, 46–55. [Google Scholar] [CrossRef]
- Liang, X.; Litkey, P.; Hyyppa, J.; Kaartinen, H.; Vastaranta, M.; Holopainen, M. Automatic Stem Mapping Using Single-Scan Terrestrial Laser Scanning. IEEE Trans. Geosci. Remote Sens. 2012, 50, 661–670. [Google Scholar] [CrossRef]
- Li, L.; Mu, X.; Soma, M.; Wan, P.; Qi, J.; Hu, R.; Zhang, W.; Tong, Y.; Yan, G. An Iterative-Mode Scan Design of Terrestrial Laser Scanning in Forests for Minimizing Occlusion Effects. IEEE Trans. Geosci. Remote Sens. 2021, 59, 3547–3566. [Google Scholar] [CrossRef]
- Kankare, V.; Liang, X.; Vastaranta, M.; Yu, X.; Holopainen, M.; Hyyppä, J. Diameter Distribution Estimation with Laser Scanning Based Multisource Single Tree Inventory. ISPRS J. Photogramm. Remote Sens. 2015, 108, 161–171. [Google Scholar] [CrossRef]
- Pueschel, P.; Newnham, G.; Rock, G.; Udelhoven, T.; Werner, W.; Hill, J. The Influence of Scan Mode and Circle Fitting on Tree Stem Detection, Stem Diameter and Volume Extraction from Terrestrial Laser Scans. ISPRS J. Photogramm. Remote Sens. 2013, 77, 44–56. [Google Scholar] [CrossRef]
- Donager, J.J.; Sankey, T.T.; Sankey, J.B.; Sanchez Meador, A.J.; Springer, A.E.; Bailey, J.D. Examining Forest Structure With Terrestrial Lidar: Suggestions and Novel Techniques Based on Comparisons Between Scanners and Forest Treatments. Earth Sp. Sci. 2018, 5, 753–776. [Google Scholar] [CrossRef]
- Bastrup-Birk, A.; Reker, J.; Zal, N. European Forest Ecosystems: State and Trends; European Environment Agency: Copenhagen, Denmark, 2016; ISBN 978-92-9213-728-1. [Google Scholar]
- Assmann, E. The Principles of Forest Yield Studies: Studies in the Organic Production, Structure, Increment and Yield of Forest Stands; Davis, P.W., Ed.; Pergamon Press Ltd.: Oxford, UK, 1970; ISBN 978-0-08-006658-5. [Google Scholar]
- Montero, G.; Ruiz-Peinado, R.; Muñoz, M. Produccion de Biomasa y Fijación de CO2 Por Los Bosques Españoles; INIA: Madrid, Spain, 2005; ISBN 8474985129. [Google Scholar]
- West, P.W. Tree and Forest Measurement, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2009; ISBN 978-3-540-95965-6. [Google Scholar]
- Reineke, L.H. Perfecting a Stand-Density Index for Even-Aged Forests. J. Agric. Res. 1933, 46, 627–638. [Google Scholar]
- Axelsson, P. DEM Generation from Laser Scanner Data Using Adaptive TIN Models. Int. Arch. Photogramm. Remote Sens. 2000, 33, 110–117. [Google Scholar] [CrossRef]
- Isenburg, M. LAStools-Efficient Tools for LiDAR Processing. (Version 180409). 2018. Available online: http://rapidlasso.com/lastools (accessed on 6 January 2018).
- Crespo-Peremarch, P.; Torralba, J.; Carbonell-Rivera, J.P.; Ruiz, L.Á. Comparing the Generation of DTM in a Forest Ecosystem Using TLS, ALS and UAV-DAP, and Different Software Tools. ISPRS-Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, XLIII-B3-2, 575–582. [Google Scholar] [CrossRef]
- Ritter, T.; Gollob, C.; Nothdurft, A. Towards an Optimization of Sample Plot Size and Scanner Position Layout for Terrestrial Laser Scanning in Multi-Scan Mode. Forests 2020, 11, 1099. [Google Scholar] [CrossRef]
- McGaughey, R. FUSION/LDV: Software for LIDAR Data Analysis and Visualization. United States Dep. Agric. For. Serv. Pacific Northwest Res. Stn. 2016, 211. [Google Scholar]
- Akaike, H. Information Theory and an Extension of the Maximum Likelihood Principle. In Proceedings of the 2nd International Symposium on Information, Budapest, Hungary, 1973; 1973; pp. 267–281. [Google Scholar]
- Girardeu-Montatut, D. CloudCompare-Open Source Project. Version 2.10.2 (Zephyrus) Stereo. 2019. Available online: http://www.cloudcompare.org/ (accessed on 28 November 2022).
- David, M. Geostatistical Ore Reserve Estimation; Developments in Geomathematics; Elsevier: Amsterdam, The Netherlands, 1977; Volume 2, ISBN 9780444415325. [Google Scholar]
- Crespo-Peremarch, P.; Ruiz, L.Á.; Balaguer-Beser, Á.; Estornell, J. Analyzing the Role of Pulse Density and Voxelization Parameters on Full-Waveform LiDAR-Derived Metrics. ISPRS J. Photogramm. Remote Sens. 2018, 146, 453–464. [Google Scholar] [CrossRef]
- Boucher, P.B.; Paynter, I.; Orwig, D.A.; Valencius, I.; Schaaf, C. Sampling Forests with Terrestrial Laser Scanning. Ann. Bot. 2021, 128, 689–708. [Google Scholar] [CrossRef] [PubMed]
- Hilker, T.; van Leeuwen, M.; Coops, N.C.; Wulder, M.A.; Newnham, G.J.; Jupp, D.L.B.; Culvenor, D.S. Comparing Canopy Metrics Derived from Terrestrial and Airborne Laser Scanning in a Douglas-Fir Dominated Forest Stand. Trees 2010, 24, 819–832. [Google Scholar] [CrossRef]
- Martins Neto, R.P.; Buck, A.L.B.; Silva, M.N.; Lingnau, C.; Machado, Á.M.L.; Pesck, V.A. Avaliação Da Varredura Laser Terrestre Em Diferentes Distâncias Da Árvore Para Mensurar Variáveis Dendrométricas. Bol. Ciências Geodésicas 2013, 19, 420–433. [Google Scholar] [CrossRef][Green Version]
- Molina-Valero, J.A.; Ginzo Villamayor, M.J.; Novo Pérez, M.A.; Álvarez-Gónzalez, J.G.; Pérez-Cruzado, C. Estimación Del Área Basimétrica En Masas Maduras de Pinus Sylvestris En Base a Una Única Using Single-Scan Terrestrial Laser Scanner (TLS). Cuad. la Soc. Española Ciencias For. 2019, 45, 97–116. [Google Scholar] [CrossRef]
- Giannetti, F.; Puletti, N.; Quatrini, V.; Travaglini, D.; Bottalico, F.; Corona, P.; Chirici, G. Integrating Terrestrial and Airborne Laser Scanning for the Assessment of Single-Tree Attributes in Mediterranean Forest Stands. Eur. J. Remote Sens. 2018, 51, 795–807. [Google Scholar] [CrossRef]
- Fleck, S.; Mölder, I.; Jacob, M.; Gebauer, T.; Jungkunst, H.F.; Leuschner, C. Comparison of Conventional Eight-Point Crown Projections with LIDAR-Based Virtual Crown Projections in a Temperate Old-Growth Forest. Ann. For. Sci. 2011, 68, 1173–1185. [Google Scholar] [CrossRef]
- Liang, X.; Hyyppä, J. Automatic Stem Mapping by Merging Several Terrestrial Laser Scans at the Feature and Decision Levels. Sensors 2013, 13, 1614–1634. [Google Scholar] [CrossRef] [PubMed]
- Maas, H.G.; Bienert, A.; Scheller, S.; Keane, E. Automatic Forest Inventory Parameter Determination from Terrestrial Laser Scanner Data. Int. J. Remote Sens. 2008, 29, 1579–1593. [Google Scholar] [CrossRef]
- Saarinen, N.; Kankare, V.; Vastaranta, M.; Luoma, V.; Pyörälä, J.; Tanhuanpää, T.; Liang, X.; Kaartinen, H.; Kukko, A.; Jaakkola, A.; et al. Feasibility of Terrestrial Laser Scanning for Collecting Stem Volume Information from Single Trees. ISPRS J. Photogramm. Remote Sens. 2017, 123, 140–158. [Google Scholar] [CrossRef]
- Bravo, F.; Montero, G.; Del Río, M. Indices de Densidad de Las Masas Forestales. Ecología 1997, 11, 177–187. [Google Scholar]
- Abegg, M.; Kükenbrink, D.; Zell, J.; Schaepman, M.; Morsdorf, F. Terrestrial Laser Scanning for Forest Inventories—Tree Diameter Distribution and Scanner Location Impact on Occlusion. Forests 2017, 8, 184. [Google Scholar] [CrossRef]
- Zong, X.; Wang, T.; Skidmore, A.K.; Heurich, M. The Impact of Voxel Size, Forest Type, and Understory Cover on Visibility Estimation in Forests Using Terrestrial Laser Scanning. GIScience Remote Sens. 2021, 58, 323–339. [Google Scholar] [CrossRef]
- Ruiz, L.Á.; Hermosilla, T.; Mauro, F.; Godino, M. Analysis of the Influence of Plot Size and LiDAR Density on Forest Structure Attribute Estimates. Forests 2014, 5, 936–951. [Google Scholar] [CrossRef]
- Kankare, V.; Puttonen, E.; Holopainen, M.; Hyyppä, J. The Effect of TLS Point Cloud Sampling on Tree Detection and Diameter Measurement Accuracy. Remote Sens. Lett. 2016, 7, 495–502. [Google Scholar] [CrossRef]
- Litkey, P.; Puttonen, E.; Liang, X. Comparison of Point Cloud Data Reduction Methods in Single-Scan TLS for Finding Tree Stems in Forest. In Proceedings of the SilviLaser 2011, 11th International Conference on LiDAR Applications for Assessing Forest Ecosystems, Hobart, Australia, 16–20 October 2011; pp. 626–635. [Google Scholar]




| Number of sample plots | 28 | |||
| Main species | P. halepensis, P. pinaster and Q. suber | |||
| DBH range (cm) | 5.0–82.0 | |||
| Mean | SD | Min | Max | |
| Slope (%) | 21.5 | 11.6 | 0.0 | 45.6 |
| Herbaceous cover (%) | 40 | 30 | 0 | 90 |
| Shrub cover (%) | 40 | 20 | 10 | 90 |
| AGB (T·plot−1) | 7.90 | 4.11 | 1.67 | 19.40 |
| BA/ha (m2·ha−1) | 31.99 | 14.16 | 9.03 | 73.50 |
| CBH (m) | 6.6 | 2.2 | 1.1 | 10.2 |
| Hd (m) | 14.4 | 2.6 | 7.6 | 18.9 |
| N/plot (trees·plot−1) | 53 | 16 | 25 | 81 |
| N/ha (trees·ha−1) | 731 | 209 | 354 | 1103 |
| QMD (cm) | 23.4 | 4.0 | 12.7 | 29.6 |
| SDI (trees·ha−1) | 661 | 264 | 240 | 1406 |
| Sensor | Faro Focus 3D 120 |
| Accuracy | ±2 mm at 25 m |
| Range | 0.6–120 m |
| Pulse frequency | 97 Hz |
| Scan angle | H: 360°/V: 305° |
| Wavelength | 905 nm |
| Beam divergence | 0.19 mrad |
| Measurement speed | 122.000–976.000 points/sec |
| Size | 24.1 × 20.3 × 10.2 cm |
| Weight | 5.2 Kg |
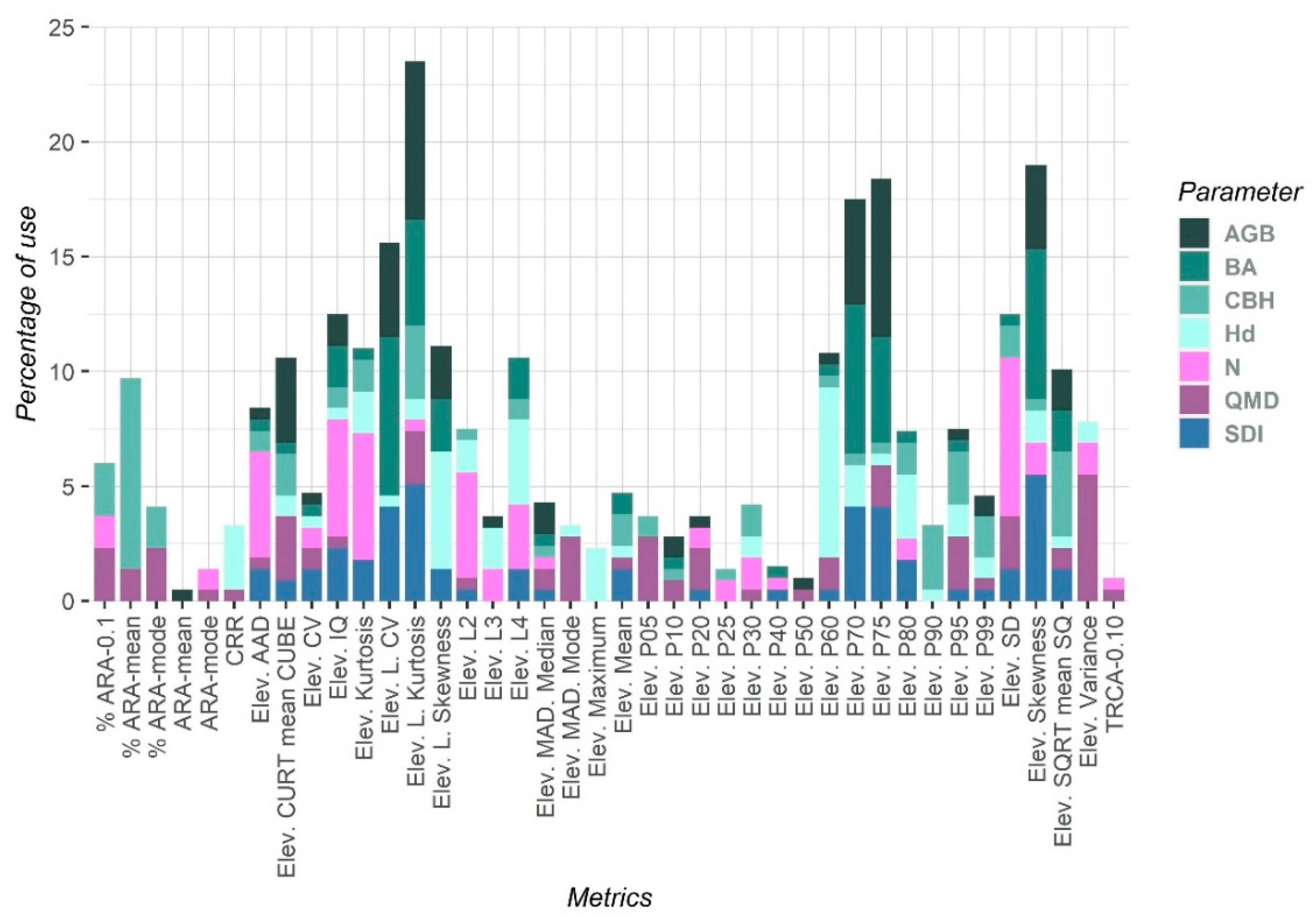
| Type | Metrics Name | Abbreviations |
|---|---|---|
| Forest heigh metrics | Mean elevation | Elev. Mean |
| Maximum elevation | Elev. Maximum | |
| L moment 2–4 elevation | Elev. L2–L4 | |
| 05th to 99th percentile of the return heights | Elev. P05–P99 | |
| Elevation quadratic mean | Elev. SQRT mean SQ | |
| Elevation cubic mean | Elev. CURT mean CUBE | |
| Forest height variability metrics | Standard deviation for the distribution of point heights | Elev. SD |
| Coefficient of variation for the distribution of point heights | Elev. CV | |
| Variance for the distribution of point heights | Elev. Variance | |
| Skewness for the distribution of point heights | Elev. Skewness | |
| Kurtosis for the distribution of point heights | Elev. Kurtosis | |
| L moment coefficient of variation for the distribution of point heights | Elev. L. CV | |
| L moment skewness for the distribution of point heights | Elev. L. Skewness | |
| L moment kurtosis for the distribution of point heights | Elev. L. Kurtosis | |
| Interquartile distance for the distribution of point heights | Elev. IQ | |
| Average Absolute Deviation for the distribution of point heights | Elev. AAD | |
| Median of the absolute deviations from the overall median for the distribution of point heights | Elev. MAD. Median | |
| Median of the absolute deviations from the overall mode for the distribution of point heights | Elev. MAD. Mode | |
| Forest density metrics | Canopy relief ratio | CRR |
| Percentage of all returns above the 0.1 m | % ARA-0.1 | |
| Percentage of all returns above the mean | % ARA-mean | |
| Percentage of all returns above the mode | % ARA-mode | |
| All returns above mean divided by the total first returns x 100 | ARA-mean | |
| All returns above mode divided by the total first returns x 100 | ARA-mode | |
| Total return count above 0.10 | TRCA-0.10 |
| Forest Parameter | Scan Combination | R2adj | RMSE | nRMSE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Min. | Mean | Max. | Mean | Max. | |||||
| AGB (t·plot−1) | 5-6-7-8-0 | 0.828 | 0.730 | 0.528 | 1.61 | 2.00 | 2.68 | 0.09 | 0.11 | 0.15 |
| 0 | 0.700 | 2.13 | 0.12 | |||||||
| ALL | 0.808 | 1.70 | 0.10 | |||||||
| BA (m2·ha−1) | 5-6-7-8-0 | 0.840 | 0.773 | 0.64 | 5.35 | 6.34 | 8.02 | 0.08 | 0.10 | 0.12 |
| 0 | 0.728 | 7.00 | 0.11 | |||||||
| ALL | 0.798 | 6.01 | 0.09 | |||||||
| CBH (m) | 5-0-7 | 0.837 | 0.787 | 0.734 | 0.83 | 0.95 | 1.06 | 0.09 | 0.10 | 0.12 |
| 0 | 0.778 | 0.97 | 0.11 | |||||||
| ALL | 0.825 | 0.86 | 0.09 | |||||||
| Hd (m) | 1-4-6 | 0.854 | 0.694 | 0.563 | 0.92 | 1.33 | 1.60 | 0.08 | 0.12 | 0.14 |
| 0 | 0.736 | 1.23 | 0.11 | |||||||
| ALL | 0.711 | 1.29 | 0.11 | |||||||
| N (trees·plot−1) | 1-3-4 | 0.628 | 0.43 | 0.321 | 8.45 | 10.51 | 11.72 | 0.16 | 0.20 | 0.22 |
| 0 | 0.363 | 11.33 | 0.21 | |||||||
| ALL | 0.343 | 11.38 | 0.21 | |||||||
| QMD (cm) | 0 | 0.603 | 0.472 | 0.319 | 2.35 | 2.72 | 3.09 | 0.14 | 0.16 | 0.18 |
| ALL | 0.520 | 2.58 | 0.15 | |||||||
| SDI (trees·ha−1) | 3-4-5 | 0.832 | 0.780 | 0.647 | 101.94 | 116.5 | 147.99 | 0.09 | 0.10 | 0.13 |
| 0 | 0.756 | 123.01 | 0.11 | |||||||
| ALL | 0.799 | 111.79 | 0.10 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torralba, J.; Carbonell-Rivera, J.P.; Ruiz, L.Á.; Crespo-Peremarch, P. Analyzing TLS Scan Distribution and Point Density for the Estimation of Forest Stand Structural Parameters. Forests 2022, 13, 2115. https://doi.org/10.3390/f13122115
Torralba J, Carbonell-Rivera JP, Ruiz LÁ, Crespo-Peremarch P. Analyzing TLS Scan Distribution and Point Density for the Estimation of Forest Stand Structural Parameters. Forests. 2022; 13(12):2115. https://doi.org/10.3390/f13122115
Chicago/Turabian StyleTorralba, Jesús, Juan Pedro Carbonell-Rivera, Luis Ángel Ruiz, and Pablo Crespo-Peremarch. 2022. "Analyzing TLS Scan Distribution and Point Density for the Estimation of Forest Stand Structural Parameters" Forests 13, no. 12: 2115. https://doi.org/10.3390/f13122115
APA StyleTorralba, J., Carbonell-Rivera, J. P., Ruiz, L. Á., & Crespo-Peremarch, P. (2022). Analyzing TLS Scan Distribution and Point Density for the Estimation of Forest Stand Structural Parameters. Forests, 13(12), 2115. https://doi.org/10.3390/f13122115










