Abstract
Wood identification is an important tool in many areas, from biology to cultural heritage. In the fight against illegal logging, it has a more necessary and impactful application. Identifying a wood sample to genus or species level is difficult, expensive and time-consuming, even when using the most recent methods, resulting in a growing need for a readily accessible and field-applicable method for scientific wood identification. Providing fast results and ease of use, computer vision-based technology is an economically accessible option currently applied to meet the demand for automated wood identification. However, despite the promising characteristics and accurate results of this method, it remains a niche research area in wood sciences and is little known in other fields of application such as cultural heritage. To share the results and applicability of computer vision-based wood identification, this paper reviews the most frequently cited and relevant published research based on computer vision and machine learning techniques, aiming to facilitate and promote the use of this technology in research and encourage its application among end-users who need quick and reliable results.
1. Introduction
Illegal logging is one of the most pressing environmental issues, particularly in tropical countries with large forest areas and botanical groups that are highly valued in international markets. Illegal logging is currently the most profitable ecological crime worldwide, accounting for 10 to 30% of the global timber trade [1,2].
Although Amazonian forests are traditionally seen as the hotspot of illegal logging, areas such as Southeast Asia, Central Africa and Russia, home to roughly 60% of the world’s forests, are unfortunately experiencing a surge in this crime [1,3]. The financial impact of illegal logging is estimated at 52 to 157 billion dollars a year [1], but, more importantly, the environmental damage, in many cases irreversible, can also have a global ecological impact [4].
Several institutional and international legal measures have been put in place to prevent overexploitation and irreversible loss of species and habitats [5,6,7]. Innovative programmes are emerging [8,9], solid research is under way [10,11,12,13,14], and research with significant impact and news items are being shared worldwide [15].
The impact of wood identification extends beyond illegal trading and ecological issues. Wood identification is paramount for the timber industry, civil and structural engineering, criminology, archaeology, art history, ethnography, and conservation and restoration, and many other disciplines.
Despite the multiple wood identification methods now available, the varied results, costs, accessibility, deployment time and limiting factors hinder their applicability to real-world identification. This paper presents an overview of the changes that have occurred in wood identification methods and a review of computer vision-based wood identification, which is currently one of the fastest developing research areas in artificial intelligence (AI) with very promising results and high identification accuracy. In this technique, visual data are processed from any given image to extract the relevant features in order to make a decision.
Analogic and Digital Systems
Historically, wood identification methods mainly comprised the study of chemical and physical and anatomical features aspects of wood. Methods such as macroscopy, which uses the physical characteristics of wood observable to the naked eye or with a 10× hand lens, and microscopy, which resorts to light compound microscopes to the observation of multiple cell typologies that constitute the wood, were the first to be used. The main limitation of these methods is that wood cannot always be identified at the species level. As a result, there has been an emergence of multiple techniques such as near-infrared spectroscopy [16,17,18], DNA barcoding [19,20,21], mass spectrometry [22,23,24], and X-ray tomography [25,26,27], with optical microscopy still used as a confirmation method for the results of these techniques.
However, an important contribution was made with the advent of computer-based technologies, which rapidly became a preferred option for constructing species databases and hosting identification tools. Several wood identification databases and software based on digital technology have been made available, including GUESS [28,29], CSIROID [30] and the DELTA system [31], three of the most significant early programmes in achieving the goal of wood identification. As a proof of concept, the importance of these systems was fundamental for the development of what is today defined as computer-assisted wood identification. The results they obtained made considerable progress compared to earlier methods, especially with regard to the time required and identification accuracy. The DELTA-Intkey for commercial timbers is the only one of these three systems still in use.
2. Online Reference Databases for Wood Identification
This section briefly describes all the digital reference databases available online, to the best of our knowledge. The common objective of online identification keys is to enable and facilitate analysis of wood anatomical features and, ultimately, identification of the wood. Table 1 summarises the computer-assisted wood identification systems mentioned above.

Table 1.
Summary of computer-assisted wood identification systems.
2.1. Commercial Timbers: Descriptions, Illustrations, Identification and Information Retrieval
Among the several DELTA-INTKEY online identification keys that have been developed, including CITESwoodID and Softwoods, this interactive identification key developed in 2000 and updated in 2018–2019 is an integrated database of microscopic descriptions and illustrations of 409 internationally traded hardwood taxa [32]. It covers major forest regions of the world and is freely available online.
2.2. Anatomy of European and North American Woods
Created in 2000, this system includes 426 wood taxa [33] and provides an interactive identification key for the common non-commercial wood species of Europe and North America. It includes 325 hardwood species and 101 softwood species (native and introduced) with 145 features and 15 sets. It has two extra sets that isolate the features applicable to identification of modified and carbonised wood from palaeobotanical contexts. A useful feature, which is missing from the commercial timbers key, is a set that isolates the features extracted from the IAWA standards.
2.3. Wood Database of the Forestry and Forest Products Research Institute
This online database created in 2003 focuses on the identification of 781 Japanese tree species, substantiating the descriptions on the IAWA list of hardwood features [52]. It includes a multiple-entry key and an image database and is freely available online [34].
2.4. InsideWood
Developed in 2004, InsideWood is by far the largest and best-known online identification key [35]. It is a multiple access key based on the IAWA hardwood list [52] and is freely available online. It includes keys for hardwoods, softwoods and fossil hardwoods, and more than 10,030 microscopic anatomic descriptions covering all regions of the world, with more than 63,435 searchable images. It is a centralised database that integrates all the anatomical data available for modern wood.
2.5. Wood Anatomy of Central European Species
This is a completely revised and updated version of Schweingruber’s work [53], created in 2004 [36] and last updated in 2007. It is a web-based identification key with 133 species, accompanied by macroscopic and microscopic descriptions. It also provides information such as sample preparation, staining and other procedures, and is freely available online.
2.6. CITESwoodID
CITES (Convention on International Trade in Endangered Species of Wild Fauna and Flora) [54] is an agreement that was drawn up in 1963 after a meeting of members of IUCN (The World Conservation Union). One of many initiatives intended to contribute to the goals of the agreement, CITESwoodID was developed in 2005 [37] and last updated in 2017. As the name indicates, the platform focuses on CITES species and is an interactive identification key of macroscopic descriptions with an integrated database. It includes illustrations of 44 CITES protected woods and 31 look-a-like trade species. It provides comprehensive, detailed information about each species, with advice on how to avoid misinterpretations, and numerous explanatory notes of the relevant features and procedures for description and identification. It is freely available online.
2.7. Key to a Selection of Arid Australian Hardwoods and Softwoods
Stemming from doctoral research [38], this interactive key focuses on Australian woods. It is hosted on the Lucid website and includes 58 wood-producing species of arid Australia, particularly non-commercial species. It is mostly based on specimens from northeast South Australia, southwest Queensland and far western New South Wales and is freely available online.
2.8. Brazilian Commercial Timbers—Interactive Wood Identification Key
As the name suggests, this is an interactive identification system focusing on Brazilian species. Made available in 2010 [39], it was developed in collaboration with the Forest Products Laboratory (LPF) and the Brazilian Forest Service (SFB). It is hosted on the Lucid website and includes 275 species, among them Brazilian CITES-listed timber species. All the nomenclature was revised in 2020 according to the Brazilian Flora Species List. The key works by analysing macroscopic features and chemical and physical tests on the woods. It is freely available online.
2.9. Pl@ntwood
Pl@ntwood [40] was developed in 2011 and is described by the authors as an interactive graphical identification tool based on the IDAO system, specifically designed to be user friendly. It comprises 110 Amazonian tree species belonging to 34 angiosperm families and includes microscopic morphological features.
2.10. The Forest Species Database—Microscopy (FSDM)
Created in 2013, this online database [41,42] comprises 2240 microscopic images of 112 species, 85 genera and 30 families of both hardwoods and softwoods. It is freely available online.
2.11. The Forest Species Database—Macroscopy (FSDM)
This online database for forest species identification [43,44] was made available in 2014. It includes 2942 macroscopic images of 41 Brazilian forest species and is freely available online.
2.12. MacroHOLZdata
MacroHOLZdata [45], created in 2002 and made available for the first time in 2016, is another interactive identification key with an integrated database for macroscopic wood descriptions. Completely redesigned in 2022, it is available in German, English and Spanish, and includes 150 common hardwood and softwood commercial timbers. The database is free of charge.
2.13. Forest Species Classifier
Made available in 2018, Forest Species Classifier is the result of a master’s degree [46]. It is a user-friendly online database focusing on Brazilian forest species. It uses macroscopic [43] and microscopic [41] databases and includes microscopic images of 112 species and macroscopic images of 41 species, with a total of 5182 images. It is freely available online.
2.14. UTForest—UTFPR Classificador
This new version of the Forest Species Classifier platform [50] has been available since 2021. It allows macroscopic identification of 44 native species of Brazil and includes 1318 images.
2.15. Charcoal
Developed in 2018, this database comprises charcoal samples of 44 Brazilian hardwood forest species, using 528 images [47,55]. It is available for research purposes only.
2.16. CharKey
This 2019 electronic identification key is described by the authors [48] as the first computer-aided identification key designed for charcoals from French Guiana. It uses SEM photographs to illustrate the anatomical features of 507 species belonging to 274 genera and 71 families. Most of the descriptions were taken from Détienne et al. [56], and follow the IAWA list of microscopic features for hardwood identification [52]. The key contains 289 “items”, and its main aim is to identify specimens to the genus level. It is freely available online.
2.17. Softwood Retrieval System (SRS) for Coniferous Wood
The Softwood Retrieval System (SRS) for Coniferous Wood [49] is an online identification key with descriptions and micrographs of 180 Chinese coniferous wood species (155 species with descriptions and microphotographs and 25 species with only microphotographs) from nine families and more than 1000 images showing anatomical details. The system is searchable by an interactive multiple-entry key. The microphotographs were collected from slices of 115 coniferous species provided by the Wood Collection of the Chinese Academy of Forestry (Beijing, CAFw) and 40 coniferous species from the Herbarium of Southwest Forestry University (Kunming, SWFUw). The descriptions use features from the IAWA List of Microscopic Features for Softwood Identification [57]. The system supports three retrieval methods for coniferous wood retrieval: species name, anatomical characteristics, and microscopic anatomical images (in test).
2.18. Mader App
This mobile app is under development. Its goal is to contribute to the global wood identification effort and the fight against illegal logging using AI [51]. The project comprises 26 species, with a vast image database of 1000 images per species, aiming to obtain maximum intraspecific variability for each species. The images were taken using a portable microscope and the app aims to obtain real-time recognition of samples. Preliminary data from the authors indicate that accuracy is 95%. The authors intend to make the app available soon on the Play Store, and the database used will be freely available for neural network training [51].
3. Computer Vision-Based Wood Identification
The digital systems described above are the foundation of the systems which, despite their limitations, are currently used to identify wood, mostly based on computer vision technology. They are applicable to several fields of research and industry, including neurobiology, autonomous vehicles, and facial recognition. Computer vision systems process visual data from any given image or video to extract the required and relevant features to make a decision [58].
This image recognition ability, also known as image classification, is one of the most important research areas in AI and is most frequently based on supervised learning. In this case, the network is required to create a model that learns from labelled images to determine classification rules, then it classifies the input data based on these same rules (generally used for image classification). In the case of unsupervised learning, it is the model that obtains unknown information through unlabelled data (generally used for image clustering) [59].
Machine learning can also decide what to do without human assistance from the data recognised by computer vision (input data), using predesigned algorithms [60,61]. This removes the need to teach the model the necessary features or procedures for wood identification [62].
Computer vision technology is very appealing to many researchers because of its verifiable potential for field application [63] and proven ability to recognise and quantify wood structure variations that are not easily discernible using strictly “human” analysis. It is also an affordable resource [64] and, therefore, scalable. However, for the software to correctly interpret the specific architecture structure of the samples analysed to such a high level of precision, reference material must be constantly entered into the image database so that it can recognise natural variations in wood structure [12].
Computer vision-based wood identification is the real-world application of combining two types of software with different approaches within AI [65,66].
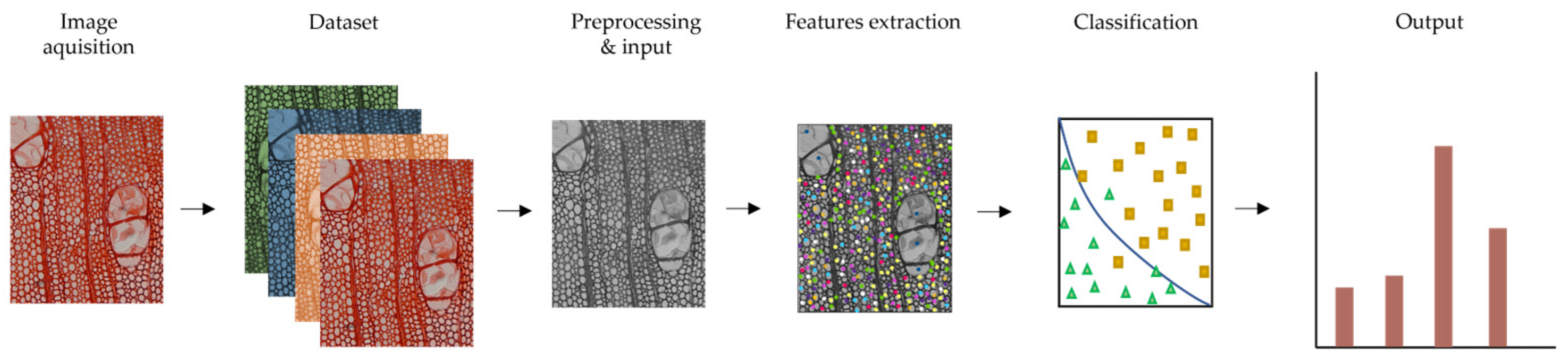
Figure 1 shows a pipeline of this method.
Figure 1.
General scheme of machine learning method for image classification (based on [62,67]).
3.1. Machine Learning
Machine learning operates primarily as software that recognises patterns from input images that are processed to define a descriptive structure to which the unknown image will be referenced [68]. This involves various stages, as follows.
3.2. Image Acquisition
The most frequently used types of image are macroscopic images (obtained without magnification using a normal digital camera) [69,70,71,72], stereograms (stereoscopic images obtained with hand lens magnification, ca. 10×) [65,66,73,74,75], micrographs (optical microscopic images) [76,77,78], SEM images (up to 10,000×) [79], and X-ray computed tomography (CT) images [26,80].
Light control and uniformity are significant issues in image processing [66,81,82]. They include techniques that are used to filter and normalise image brightness [83,84,85].
3.3. Image Datasets
Image dataset construction or availability is one of the most significant factors among the multiple issues that can affect the performance of computer vision-based wood identification systems.
The more extensive the dataset is, the more naturally occurring biological variations within a species will be accessed and learned by the model. However, because constructing a dataset of wood samples is such a difficult and time-consuming task, most studies use wood collections for references [41,69,74,81,86,87,88,89].
This limitation is countered to some extent by initiatives such as ImageNet [90]. Aiming to advance computer vision and deep learning research, the ImageNet dataset was made freely available to researchers worldwide. It contains 14.2 million images across more than 20,000 classes. A similar process is under way with herbaria digitalisation [91,92,93]. However, despite the efforts made [42,78,87,94,95], the lack of free access to worldwide wood image datasets continues to be the main constraint for computer vision-based wood identification [62].
Table 2 shows the main currently available datasets that have useful data for computer vision-based wood identification research.

Table 2.
Wood image datasets available for computer vision-based wood identification research, adapted from [62].
3.4. Image Processing
Machine learning comprises two independent procedures: feature processing, also known as extraction (extraction of relevant features from input images), and classification (learning extracted features and querying image classification). There is, however, a previous step to image processing.
Pre-processing aims to convert the image into data that a specific algorithm can use to extract the required features, thus reducing computational complexity and facilitating subsequent processing [99]. The techniques used for this include greyscale conversion and image cropping [71,74,78,88,100,101], filtering [83,85], image sharpening [74,102] and denoising [79,103,104].
Another important pre-processing procedure is data splitting, where the dataset is split into subsets, most commonly training, validation, and test sets. Data splitting is ultimately used to create a training set, a validation set and a test set in order to later evaluate the model performance. To understand the reasons for these sets, one should think that machine learning systems mimic the human learning process based on examples. From this training set, the system will learn to generalise in order to correctly classify the images. The validation set is used to avoid the system learning the images “from memory” during this generalization process. Finally, the test set is used to check the reliability of the learning process. The use of these distinct data samples is one of the earliest pre-processing steps needed to evaluate any model’s performance.
More specifically, the system will interpret the images extracted from the designated training set as nothing more than a combination of pixels. Each pixel will have a specific intensity represented by a number, and in this way a matrix of numbers is formed. Image processing is based on extracting elements such as points, blobs, angles, corners and edges, and the patterns they form. Variability in the anatomy of each wood species is represented as patterns of distinct pixel intensities, arrangement, distribution, and aggregation. The variations detected by computer vision will be learned by machine learning. This process is the fundamental operating system of computer vision for all applications, including wood identification [62].
After the extracted features have been learned, a classification model is established by a classifier and a test set is formed to evaluate the system’s learning. The images are then input, allowing the classification model to complete the identification through feedback of the predicted classes of each image [62].
Computer vision detects and “sees” the input image using multiple feature extraction algorithms, while machine learning selects the types of features to be extracted, in most cases texture and local features.
Texture features work with the combination and arrangement of image elements (pixel intensities and resulting patterns) [65,74,96,102,105]. The most frequently used techniques are grey level co-occurrence matrix (GLCM), grey level aura matrix (GLAM), local binary pattern (LBP), higher local order autocorrelation (HLAC), and Gabor filter-based features (GFBF). Despite the individual capabilities of each technique, texture fusion of different types of texture features has shown superior classification accuracy [101,106,107].
Local features differ from texture features by not describing an image as a unit, but by describing significant and important specific features (keypoints) such as edges, corners or points. The most frequently used algorithms are scale-invariant feature transform (SIFT), speeded up robust features (SURF), oriented features-from-accelerated-segment-test (FAST) and rotated binary-robust-independent-elementary-feature (BRIEF) (ORB), and Accelerated-KAZE (AKAZE).
Beyond features typology, factors such as dimensionality reduction and feature selection are also important, as a large number of features extracted from an image can substantially reduce the computational efficiency of classification models. To achieve this balance, methods such as R AutoEncoder [108], principal component analysis (PCA), linear discriminant analysis (LDA), and genetic algorithms (GA) are used for dimension reduction of data sets [109].
Another important element is the classification models created to learn the extracted features and establish classification rules. The most frequently used classifiers are k-nearest neighbours (k-NN), support vector machines (SVM), and artificial neural networks (ANN) [110,111]. These classification procedures can be executed either in on-site hardware [112] or on a cloud-based interface [89].
Machines can be easily misled by factors such as the source of images, which can be acquired in the field using mobile phones [89] or in a laboratory-controlled environment [70,113], and variables including different thicknesses, orientation, staining, digital artefacts and other variations, which is why many thin sections from historical wood collections are useful only to the trained human eye [10].
4. Deep Learning
Deep learning is among the most notable and promising of the many branches of machine learning research.
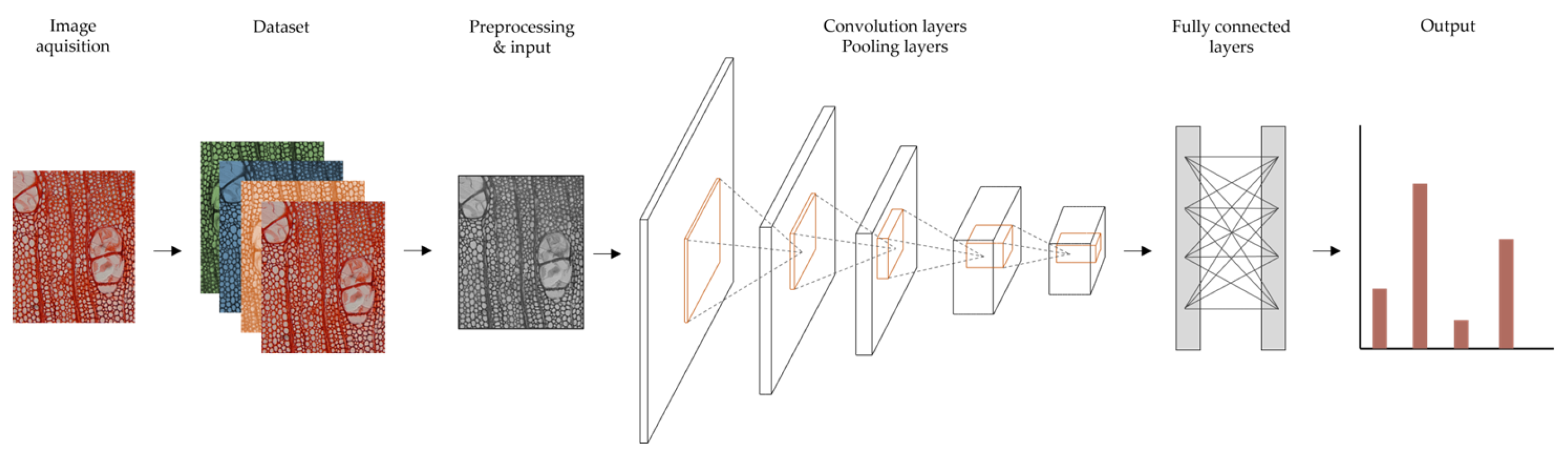
As a neural network that attempts to simulate the function, structure and behaviour of the human brain (Figure 2), it has the capacity to process and “learn” large amounts of data [114,115].
Figure 2.
General pipeline of deep learning models for image classification (based on [62,67]).
Its multiple different architectures include ANN [116], deep neural networks (DNN) [117], recurrent neural networks (RNN) [118], deep reinforcement learning (DRL) [119], and convolutional neural networks (CNN) [120]. The fields to which these have been applied are so vast that they are very difficult to summarise, but they include computer vision [121], forensic research [122], climate science [123], machine translation [124], classic literature [125] and bioinformatics [126], to name just a few.
Among these multiple architectures, it is mostly ANNs and CNNs that are applied to wood characterisation and identification.
Table 3 summarises this research and the applications of deep learning technologies.

Table 3.
Research and applications of deep learning technologies.
4.1. Artificial Neural Networks (ANN)
Artificial neural networks are not only one of the main investigation methods, but also constitute the foundation of deep learning [62]. These mathematical structures inspired by biological neural networks are a form of supervised or unsupervised learning that show high ability to learn from examples given to them and extrapolate the information when applied to future non-identified samples. This ability to reproduce, model and “learn” nonlinear processes has given ANNs widespread applications in multiple disciplines [78,116].
In the field of wood differentiation and identification, examples of research applying ANNs include:
- -
- Esteban et al. [127] used a feedforward multilayer perceptron (MLP) network, which uses a similar structure to ANN to distinguish between Juniperus cedrus and J. phoenicea var. canariensis, obtaining a 92% probability of correctly differentiating the species;
- -
- Mallik et al. [79] applied SEM to wood cross sections with 1500× magnification to obtain species-level identification through the shape, number, area and distribution of earlywood tracheids, processed by image segmentation, object recognition and statistical methods. Their results showed that when distinguishing between hardwoods and softwoods, a 0.89 accuracy was obtained using leave-one-out cross-validation and 0.93 using an external validation test (EVT), and when differentiating seven wood species, they obtained a 0.81 accuracy using one-leave-out cross-validation and 0.80 using an EVT;
- -
- The same microscopic features analysis was applied by Martins et al. [77], who used microscopic transverse sections applying local phase quantisation (LPQ), local binary patterns (LBP) and grey-level co-occurrence matrix (GLOM) to identify Brazilian species. The process was applied to 112 species, 85 genera and 30 families, obtaining a recognition rate of 98.6% for differentiation of hardwoods and softwoods and 86% for discrimination of the 112 species;
- -
- Turhan [128] used the SVM as a machine learning algorithm to differentiate Salix alba, S. caprea and S. eleagnos, obtaining a 95.2% success rate;
- -
- Filho et al. [71] used a two-level divide-and-conquer classification strategy to differentiate 41 species of Brazilian flora, obtaining the highest accuracy level, of 97.77%;
- -
- Esteban et al. [130] used a multilayer perceptron (MP) to differentiate Pinus sylvestris L. and P. nigra Arn subsp. salzmannii (Dunal) Franco, obtaining 81.2% accuracy in the testing set;
- -
- Silva et al. [78] used microscopic images of cross sections of 77 commercial wood species from the Democratic Republic of the Congo for surface texture analysis, reporting 88% successful identifications at species level, 89% at genus level and 90% at family level.
- -
- He et al. [135] applied machine learning classifiers SVM, Naive Bayes (NB), Decision Tree C5.0 and ANN) to discriminate between Swietenia macrophylla King, S. mahagoni (L.) Jacq and S. humilis Zucc. The best results were obtained with SVM, with an overall accuracy of 91.4%;
- -
- Deklerck et al. [136] used machine learning not for image-based data processing, but for metabolome profile obtained through direct analysis in real-time (DART™) ionisation combined with time-of-flight mass spectrometry (TOFMS) to study the heartwood of 175 samples of 10 species of the Meliaceae family. Combining these techniques resulted in accuracy levels of 82.2%;
- -
- de Andrade et al. [73] generated 2000 macroscopic images of 21 species using a smartphone and samples manually polished with a knife to replicate field conditions. A grey level co-occurrence matrix for the development of classifiers based on SVM was used, resulting in accuracies of 97.7%;
- -
- Silva et al. [140] used 77 Congolese wood species as a reference base for applying a multi-view random forest (MVRF) model for species-level identification. To ensure information was not missed, the authors used images of the three anatomical planes. The results showed that the concatenation of features from the transverse and tangential planes clearly outperforms transverse-only analysis, while adding the radial plane minimally improves the results obtained. The use of the MVRF model outperformed concatenation of LPQ features. The results showed that the supplementary information added using three planes analysis and the model type considerably improve the final results. Moreover, when evaluating the performance of the systems developed, using the k-fold cross-validation scheme could have led to overestimation of the results, so the authors applied a leave-k-tree-out approach during cross-validation. The results showed that implementing this approach dramatically decreased accuracy compared with traditional cross-validation schemes.
4.2. Convolutional Neural Networks (CNN)
Convolutional neural networks are one of the most significant applications of ANNs. In the AI context, a CNN is a class of feedforward ANN that has been successfully applied to digital image processing analysis.
A CNN processes images more effectively by applying filtering techniques to ANNs [115]. This is a powerful and accurate way of solving classification problems, and CNNs are mainly credited for their role in image analysis, recognition, and classification. The architecture of a CNN typically has multiple layers between input and output: three convolutional layers, a pooling layer and a fully connected layer. These layers process different tasks during the image’s course. As the images progress through the distinct layers, features such as edges, colours and shapes are extracted and interpreted. These features are then learned and classified by the deep neural network, resulting ultimately in the network’s ability to identify a specific object [62,115,141]. Other advantages are the capacity of automatically recognise important features without human supervision.
CNNs have difficulty dealing with variance in the data presented, as tilted or rotated images. This results in a limitation to encode an object’s orientation and position or process spatially invariant data.
Research examples applied to wood identification include:
- -
- Hafemann et al. [129] applied the CNN model 3-ConvNeta to identify macro images of 41 species and micro images of 112 species. The results obtained 95.77% accuracy for macroscopic images and 97.32% accuracy for microscopic images;
- -
- Kwon et al. [131] applied six LeNet and MiniVGGNet CNN models to identify five Korean softwood species (Cryptomeria japonica, Chamaecyparis obtuse, Pinus koraiensis, P. densiflora, Larix kaempferi), using an iPhone 7 camera to obtain macroscopic images of rough sawn surfaces from cross sections. Of all the CNN models tested, LeNet3 achieved the highest results and stability, with two extra layers added to the original LeNet architecture. The identification accuracy obtained was 99.3%. The authors reported that the software weight of the CNN created is small enough for installation on a mobile device such as a smartphone;
- -
- Maintaining the objective of ensuring field applicability, Kwon et al. [133] acknowledged the real-world limitations of not including longitudinal wood surfaces. Using mobile device cameras to obtain macroscopic images, they applied a combination of models, obtaining the best results with LeNet2, LeNet3 and MiniVGGNet4. Their results showed an overall accuracy of 98% and an improvement on their earlier study, particularly in the case of P. koraiensis and P. densiflora;
- -
- Figueroa-Mata et al. [86] applied deep convolutional networks for identification of 41 Brazilian forest species from xylotheque samples at species level, achieving an accuracy of 98.3%;
- -
- Ravindran et al. [112] used CNNs to identify 10 neotropical species in the Meliaceae family (Cabralea canjerana, Carapa guianensis, Guarea glabra, G. grandifolia, Khaya ivorensis, K. senegalensis, and the CITES-listed Swietenia macrophylla, S. mahagoni, Cedrela fissilis, and C. odorata), using only the transverse surface. The results showed an accuracy of 87.4 to 97.5%;
- -
- To develop an automatic classification system for charcoal, Maruyama et al. [47] applied two LBP configurations of as texture descriptors. As state-of-the-art machine learning classifiers, SVM and random forests (RF) have shown the best results. Inception_v3 CNN was applied for representation learning evaluation. The database comprised 44 charcoal samples from Brazilian native species from natural forests. The authors reported that both handcrafted features and RL achieved results of around 95% recognition rate;
- -
- Oliveira et al. [132] used databases developed by Filho et al. [71] and Martins et al. [77] to access cross sections of 2942 wood macroscopic images of 41 species and 2240 microscopic images of 112 species, applying CNNs to create three models. Based on the results, the authors reported 100% recognition accuracy for the scale model, 98.73% for the macroscopic model, and 99.11% for the microscopic model;
- -
- Kanayama et al. [134] applied a deep CNN approach to near-infrared hyperspectral imaging (NIR-HSI) using a principal component (PC) algorithm to identify 120 samples of 38 hardwood species. The results obtained showed 90.5% accuracy;
- -
- A CNN was also used by Ravindran and Wiedenhoeft [66] to compare the macroscopic field identification programme XyloTron, using an ImageNet pre-trained ResNet34 CNN, with mass spectrometry to differentiate 10 Meliaceae species used by Deklerck et al. [136]. The results showed identification accuracy of 81.9% at the species level and 96.1% at the genus level compared to 74.9% and 91.4%, respectively, in the work by Deklerck et al. [136];
- -
- Lopes et al. [81] applied the InceptionV4_ResNetV2 CNN to analyse macroscopic images of the end-grain of 10 xylarium North American hardwood species, producing 1869 images using a smartphone fitted with a 14× macro lens. Their results showed an accuracy of 92.6%;
- -
- de Geus et al. [137] applied the DenseNet CNN to recognise 281 species, using the largest dataset of microscopic transverse, radial and tangential images available at the time. Rotation invariant LPQ (RiLPQ) showed the best results of the feature descriptors used. The authors reported an identification accuracy of 98.8%;
- -
- Olschofsky and Köhl [69] applied Inception-v3, an image classification model using a CNN for feature recognition and classification, pre-trained with 1.2 million images. The CITES-protected species Cedrella odorata was chosen and compared with 13 other tropical tree species for recognition. The results with the pre-trained CNNs had 98% accuracy, but when other tree species not used for training were added, the classification accuracy fell to 87%;
- -
- The ResNet101 CNN, associated with an SVM as classifier, was applied by Lens et al. [76] to species-level identification of 112 mainly neotropical tree species, using only transverse sections but focusing on microscopic rather than macroscopic analysis. The results showed successful identification in 95.6% of cases;
- -
- Wu et al. [138] applied deep convolutional neural networks (CNNs) for the identification of 11 rough saw hardwood North American species based on tangential plane images only. CNNs ResNet-50, DenseNet-121, as well as MobileNet-V2 were tested, resulting in an overall accuracy of 98.2%.
- -
- Shugar et al. [139] combined X-ray fluorescence spectrometry (XRF) and a CNN to identify 48 wood specimens of both hardwoods and softwoods, mostly from heartwood and using either tangential or radial sections. They reported 99% identification accuracy from the 66 datasets;
- -
- In the study by Fabijańska et al. [72], a CNN with residual connections was tested to identify 312 wood core scanned images of 14 European softwood and hardwood tree species, developing a wood patch classification and a wood core classification. The results showed that the proposed model correctly recognised patch images in 93% of cases and wood core images in 98.7%. Comparison of the results also showed that this model outperformed the state-of-the-art convolutional neural network-based model.
4.3. Generative Adversarial Networks (GANs)
Within deep learning, GANs [142] are described as neural networks that can learn to generate realistic samples from the data on which they were trained.
They use a neural network as a generator that takes a random distribution of data as input and learns to map that information to output the desired distribution of data. A second neural network, known as a discriminator (a binary classifier), will use the input and output images to determine the probability of the image originating as a training image (real) or on the generator (fake), thus assessing the most likely class to which the output image belongs [143].
Generative adversarial networks can produce highly realistic images using CNNs in an unsupervised manner [144]. Their application extends to multiple fields of scientific research, but they remain poorly explored in wood sciences [145,146,147].
- -
- Addressing the possibility of eliminating economic and processing burdens in acquiring images of worldwide wood species for machine-learning training purposes, Lopes et al. [144] accessed 119 hardwood species references on the publicly available Xylarium Digital Database [87]. Applying a style-based GAN, they successfully generated highly realistic and anatomically meaningful synthetic microscopic cross-sectional images of hardwood species which they reported as virtually indistinguishable from real cross-sectional images.
- -
- To evaluate the resemblance, quality and pattern evaluation between the synthetic and real cross sections, a structural similarity index measure (SSIM) and Fréchet inception distance (FID) were applied and a visual Turing test (VTT) was performed by wood anatomists to confirm the usefulness and realism of the GAN-generated images. The results showed that the artificially generated images were indistinguishable from real microscopic cross-sectional images.
- -
- The authors [144] reported that it is even feasible to generate synthetic hybrids based on microscopic cross-sectional images from two parental species. This would have considerable implications on wood science and technology, especially for estimating the wood permeability, strength, density, or hydraulic potential, for example, of a species that has not even been planted.
5. Field Applicable Wood Identification Systems
One of the most interesting features of computer vision-based wood identification systems is their field application capability. Despite the consensus that it will be a long time before this technology becomes readily available not only to researchers and law enforcement bodies, but also the general public, it is evident that this goal is reachable. Programmes already developed or under development to respond to field application needs include:
5.1. MyWood-ID
Described as an automated wood identification mobile app [89], MyWood-ID uses a smartphone with a retrofitted macro lens and machine vision for macroscopic wood identification. The system uses a database of 20 species of timber native to Malaysia and provides a simple and effective way to acquire macroscopic wood digital images. The images are then uploaded to a cloud server via an internet connection for immediate identification results. It is intended to be cost-effective, easily accessible and intuitive, and to provide fast results.
These characteristics are evident when compared with other field deployable systems [64,74]. As differentiating features of their wood identification system, the authors cite its portability, lower initial cost, faster field deployment time, intuitive use, and continual online database update. However, it requires a constant internet connection for results and is operating-system-dependent (running only on iPhone 6 and 7). The main limitation of this system is the lack of consistent light control for wood image acquisition, although the authors indicate that this can be mitigated using the learning capability of a deep learning algorithm. The results achieved by this system have an accuracy of 96% to 98%. It is a paid app.
5.2. MyWood-Premium
This is an update of the previous app, developed by FRIM (Wood Anatomy Lab of Forest Research Institute Malaysia) and UTAR (Universiti Tunku Abdul Rahman) [148]. The updated version comprises a database of 100 wood species native to Malaysia. It is available only on iPhone, iPod touch and Mac, and requires iOS 8.0 or later. It recommends the Ollo-clip™ Macro Lens with 21× magnification for optimum performance. It is a free app.
5.3. Xylorix
Another recent approach to rapid field wood identification is Xylorix [149,150], a platform that combines a suite of apps, tools and services. Xylorix Inspector is a wood identification mobile app that uses macroscopic features for automated identification. It is based on trained AI models to automatically identify the wood genus or species. However, a Xylorix WIDK-24X01 illuminated macro lens must be attached to the mobile phone camera for correct performance. It is available on either Apple iOS or Android operating systems and is supported by most mobile phones. Of the 24 species in the system database, 11 are free and the other 13 are paid.
5.4. XyloTron
XyloTron is a paid, open-source, image-based macroscopic field identification programme designed for wood and charcoal identification [64,151]. It features adjustable and controlled visible light, UV illumination capacity, and all the necessary software to control the device, capture images, and deploy the trained classification models.
It works by capturing high-quality images of wood or charcoal samples with visible or UV light. The identification accuracy for wood is described as 97.7%, increasing with the use of UV light to 99.1% (e.g., identification confusion between Albizia sp., fluorescent, and Inga sp., not fluorescent) and 98.7% for charcoal.
One limitation is that it is not a simple or easily deployable on-site system to use, because it requires a permanent connection to a laptop computer.
Ravindran and Wiedenhoeft [66] compared the performance of XyloTron and MS for species- and genus-level identification of 10 species of Meliaceae. The results showed a similar species-level accuracy of the XyloTron and MS models, but higher genus-level accuracy with XyloTron [66].
5.5. XyloPhone
To overcome visual aberrations (field distortion and spherical aberration), uncontrolled light sources, high prices, and a lack of real field applicability, Wiedenhoeft [82] proposed the XyloPhone. Described as an open-source, 3D-printed imaging attachment adaptable to virtually any smartphone for macroscopic image capture, it is a small, closed plastic box that provides a fixed focal distance, exclusion of ambient light, and a choice of visible or UV illumination. It is powered by a rechargeable external battery and a commercially available lens, making it affordable and, according to the author, providing comparable image quality to XyloTron.
To document features such as evenness of illumination, distortion, maximum resolution, and spherical aberration, the author compared the Xylophone + iPhone (XPi), the XyloPhone + Samsung (XPs), the Ollo Clip 14× + iPhone (OCi), and the Xylorix + iPhone, with two distinct configurations. He reported that XyloPhone’s optical performance, especially when used with more recent smartphones, is clearly superior to the lenses/lighting arrays of other systems [82].
5.6. WIDER
WIDER is a battery-charged portable system that uses spectroscopy measurement and machine-learning-based identification software. It comprises a database of 15 species and the authors [152] reported accuracy results of 95%. It is part of a larger project that was completed in 2021 and brought into use by USAID PEER Cycle 8 (Development of Wood Identification System and Timber Tracking Database to Support Legal Trade). The same project developed the ECVT 4D Dynamic [152] technology for monitoring tree physiological processes.
5.7. IMAIapp
IMAIapp [153] is a wood identification mobile app that uses a lens attached to a smartphone. Its purpose is to use convolutional neural networks (CNN) capable of carrying out timber identification through machine learning of macroscopic elements observed in photo enlargements. The difficulty of the problem lies in the number of classes that the method is required to recognise automatically, from a total of 400 wood species and a high number of macroscopic images. EfficientNet architecture is enhanced by a novel approach for pre-processing that combines computer vision and data augmentation techniques applied to the original dataset. The use of classification models based on deep learning is the leading technique with the best performance at present, and the innovative approach to increase the quality of the training data makes the model integrated in IMAIapp robust to rotation, illumination and zoom invariants. This means the app can be used in the field. Using TensorFlow Lite libraries for Apple and Google platforms, the application works standalone, is 100% executable from the mobile device and does not require a connection to the Internet. IMAIapp is therefore a design using an edge-computing method that is intended to avoid computing constraints on the mobile device on which it is installed. The project is under development and the app will be free for Android and iOS.
6. Discussion
In the last 100 years, what we now call the traditional wood identification method based on anatomical descriptions has followed well defined, standardised features to successfully distinguish and identify the multiple families, genera and species of angiosperm and gymnosperm trees. However, despite the many positive aspects of this method, it is now evident that it is reaching its limit.
The main limitations are the identification uncertainty at species level, the time-consuming methodology, the lack of anatomists with the necessary training for the task, and the associated costs of these professionals, mainly when on-site identifications are required. These limitations have a notable impact on the type of monitoring that can be carried out, e.g., in the fight against illegal logging. Faster, more accurate and economically scalable methods are urgently needed.
As a valuable response to this need, the results obtained so far by computer vision-based identification (CVBI) of wood and, in particular, deep learning approaches, clearly demonstrate that this method has enormous potential for wood identification and quantitative wood anatomy.
Despite the many obstacles remaining, this method is steadily adapting to overcome limitations such as inter- and intra-anatomical variability, high anatomical resemblance, non-homogeneous illumination, staining or deformed samples, and limited image databases, among many other issues.
Of all the resources discussed, deep learning appears to be the most significant and promising solution in AI developments, and CNN models applied to wood sciences are one of the leading and most rapidly evolving systems. CNNs have exhibited a notably more efficient capacity and accuracy for quantitative wood anatomy and feature recognition, alongside computer cost reduction.
Field-deployable identification systems appear to be the most important and impactful option in computer vision-based wood identification. This resource, based on advances in communication technologies, will enable more prolific, increasingly accurate and faster screening by authorities without human prejudice, particularly with regard to illegal timber and charcoal trading.
Computer vision-based identification technology could become one of the most effective and unavoidable weapons in the fight against the illegal timber and charcoal trade, as it enables individuals who are untrained in traditional identification to obtain highly accurate and legally binding identifications on the spot.
The multiple future contributions of the technologies underpinning CVBI for wood sciences are difficult to fully envision at present. However, it is of utmost importance to overcome or at least mitigate the limitations that are severely hampering the development and implementation of these systems.
Two of the most pressing issues are the limited number of digital databases, which are specific to geographically restricted areas/species or inaccessible to the global research community, and the lack of extensive field testing and verification hindering the accuracy quantification of the systems.
The most urgent actions required are the construction of a freely accessible global digital wood image database, the availability of this tool in a cloud-based system for access everywhere, by everyone, and priority inclusion of CITES-listed species and their look-a-likes.
Author Contributions
Conceptualisation, writing and original draft preparation J.L.S.; review and editing R.B., J.P., P.d.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by national funds through Fundação para a Ciência e Tecnologia (FCT), within the scope of the Project UID/EAT/0622/2016 of CITAR—Centro de Investigação da Universidade Católica Escola das Artes, Porto, Portugal. J.L.S. is recipient of a PhD fellowship (UI/BD/151009/2021) with financial support from FCT.
Data Availability Statement
Data are contained within the article and are also available from the corresponding author.
Acknowledgments
I would like to acknowledge Yong Haur Tay for his kind clarifications about Xylorix, Pedro Luis Paula Filho for his kind clarifications about The Forest Species Database—Macroscopic, Forest Species Classifier and UTForest—UTFPR Classificador, Hisashi Abe for his kind clarifications about the Wood database of the Forestry and Forest Products Research Institute, and Dra Cassiana Ferreira for her kind clarifications about the Mader app.
Conflicts of Interest
The authors declare no conflict of interest.
References
- May, C. Transnational Crime and the Developing World; Global Financial Integrity: Washington, DC, USA, 2017. [Google Scholar]
- Nellemann, C. Green Carbon, Black Trade: A Rapid Response Assessment on Illegal Logging, Tax Fraud and Laundering in the World’s Tropical Forests. 2012. Available online: https://wedocs.unep.org/20.500.11822/8030 (accessed on 27 April 2022).
- EU; EC. Forests. Available online: https://ec.europa.eu/environment/forests/illegal_logging.htm (accessed on 27 April 2022).
- Bösch, M. Institutional quality, economic development and illegal logging: A quantitative cross-national analysis. Eur. J. For. Res. 2021, 140, 1049–1064. [Google Scholar] [CrossRef]
- UN. Convention on International Trade in Endangered Species of Wild Fauna and Flora. Available online: https://cites.org/sites/default/files/eng/disc/CITES-Convention-EN.pdf (accessed on 27 April 2022).
- EU. European Union Timber Regulation. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32010R0995 (accessed on 27 April 2022).
- European Union; Austrian Development Cooperation; The World Bank; IUCN. WWF ENPI EAST FLEG—European Neighborhood and Paternaship Instrument East Countries Forest Law Enforcement and Governance Program. Available online: https://www.enpi-fleg.org/ (accessed on 6 June 2022).
- FAOUN; UNDP. UNEP UN-REDD Programme. Available online: https://www.un-redd.org/ (accessed on 27 April 2022).
- UNDER; UNEP. FAOUN Preventing, Halting and Reversing The Degradation Of Ecosystems Worldwide. Available online: https://www.decadeonrestoration.org/ (accessed on 27 April 2022).
- Schmitz, N.; Beeckman, H.; Blanc-Jolivet, C.; Boeschoten, L.E.; Braga, J.J.W.B.; Cabezas, J.A.; Chaix, G.; Crameri, S.; Degen, B.; Deklerck, V.; et al. Overview of Current Practices in Data Analysis for Wood Identification. A Guide for the Different Timber Tracking Methods; GTTN-European Forest Institute: Joensuu, Finland, 2020. [Google Scholar]
- Schmitz, N.; Beeckman, H.; Cabezas, J.A.; Cervera, M.T.; Espinoza, E.; Fernandez-Golfin, J.; Gasson, P.; Hermanson, J.; Jaime Arteaga, M.; Koch, G.; et al. The Timber Tracking Tool Infogram. Overview of Wood Identification Methods’ Capacity; Global Timber Tracking Network, GTTN Secretariat, European Forest Institute and Thünen Institute: Joensuu, Finland, 2019. [Google Scholar]
- Dormontt, E.E.; Boner, M.; Braun, B.; Breulmann, G.; Degen, B.; Espinoza, E.; Gardner, S.; Guillery, P.; Hermanson, J.C.; Koch, G.; et al. Forensic timber identification: It’s time to integrate disciplines to combat illegal logging. Biol. Conserv. 2015, 191, 790–798. [Google Scholar] [CrossRef]
- United Nations. International Consortium on Combating Wildlife Crime. In Best Practice Guide for Forensic Timber Identification; United Nations Office on Drugs and Crime: New York, NY, USA, 2016. [Google Scholar]
- ITTO. Biennial Review and Assessment of the World Timber Situation. Available online: https://www.itto.int/direct/topics/topics_pdf_download/topics_id=6783&no=1 (accessed on 27 April 2022).
- Interpol Illegal Logging in Latin America and Caribbean Inflicting Irreversible Damage-INTERPOL. Available online: https://www.interpol.int/News-and-Events/News/2022/Illegal-logging-in-Latin-America-and-Caribbean-inflicting-irreversible-damage-INTERPOL (accessed on 27 April 2022).
- Abe, H.; Watanabe, K.; Ishikawa, A.; Noshiro, S.; Fujii, T.; Iwasa, M.; Kaneko, H.; Wada, H. Simple separation of torreya nucifera and chamaecyparis obtusa wood using portable visible and near-infrared spectrophotometry: Differences in light-conducting properties. J. Wood Sci. 2016, 62, 210–212. [Google Scholar] [CrossRef]
- Pace, J.H.C.; Latorraca, J.-V.D.F.; Hein, P.R.G.; de Carvalho, A.M.; Castro, J.P.; da Silva, C.-E.S. Wood species identification from Atlantic forest by near infrared spectroscopy. For. Syst. 2019, 28, e015. [Google Scholar] [CrossRef]
- Snel, F.A.; Braga, J.W.B.; da Silva, D.; Wiedenhoeft, A.C.; Costa, A.; Soares, R.; Coradin, V.T.R.; Pastore, T.C.M. Potential field-deployable NIRS identification of seven dalbergia species listed by CITES. Wood Sci. Technol. 2018, 52, 1411–1427. [Google Scholar] [CrossRef]
- Akhmetzyanov, L.; Copini, P.; Sass-Klaassen, U.; Schroeder, H.; de Groot, G.A.; Laros, I.; Daly, A. DNA of centuries-old timber can reveal its origin. Sci. Rep. 2020, 10, 20316. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Liu, X.; Jiang, X.; Yin, Y. Extraction and amplification of DNA from aged and archaeological populus euphratica wood for species identification. Holzforschung 2015, 69, 925–931. [Google Scholar] [CrossRef]
- Wagner, S.; Lagane, F.; Seguin-Orlando, A.; Schubert, M.; Leroy, T.; Guichoux, E.; Chancerel, E.; Bech-Hebelstrup, I.; Bernard, V.; Billard, C.; et al. High-throughput DNA sequencing of ancient wood. Mol. Ecol. 2018, 27, 1138–1154. [Google Scholar] [CrossRef]
- Carmona, R.J.; Wiemann, M.C.; Baas, P.; Barros, C.; Chavarria, G.D.; McClure, P.J.; Espinoza, E.O. Forensic identification of CITES appendix I cupressaceae using anatomy and mass spectrometry. IAWA J. 2020, 41, 720–739. [Google Scholar] [CrossRef]
- Espinoza, E.O.; Wiemann, M.C.; Barajas-Morales, J.; Chavarria, G.D.; McClure, P.J. Forensic analysis of cites-protected dalbergia timber from the americas. IAWA J. 2015, 36, 311–325. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, G.J.; Liu, B.; He, T.; Guo, J.; Jiang, X.; Yin, Y. Wood discrimination analyses of pterocarpus tinctorius and endangered pterocarpus santalinus using DART-FTICR-MS coupled with multivariate statistics. IAWA J. 2019, 40, 58–74. [Google Scholar] [CrossRef]
- Ge, Z.; Chen, L.; Luo, R.; Wang, Y.; Zhou, Y. The detection of structure in wood by X-ray CT imaging technique. BioResources 2018, 13, 3674–3685. [Google Scholar] [CrossRef]
- Kobayashi, K.; Hwang, S.-W.; Okochi, T.; Lee, W.-H.; Sugiyama, J. Non-destructive method for wood identification using conventional X-ray computed tomography data. J. Cult. Herit. 2019, 38, 88–93. [Google Scholar] [CrossRef]
- Tazuru, S.; Sugiyama, J. Wood identification of japanese shinto deity statues in matsunoo-taisha shrine in kyoto by synchrotron X-ray microtomography and conventional microscopy methods. J. Wood Sci. 2019, 65, 60. [Google Scholar] [CrossRef]
- Wheeler, E.A.; LaPasha, C.A. A microcomputer based system for computer-aided wood identification. IAWA Bull. 1987, 8, 347–354. [Google Scholar]
- LaPasha, C.A. General unknown entry and search system. A program package for computer-assisted identification. Suppl. N. C. Agric. Resour. Serv. 1986, 474, 18. [Google Scholar]
- Ilic, J. Computer aided wood identification using csiroid. IAWA J. 1993, 14, 333–340. [Google Scholar] [CrossRef]
- Dallwitz, M.J. A general system for coding taxonomic descriptions. Taxon 1980, 29, 41–46. [Google Scholar] [CrossRef]
- Richter, H.G.; Dallwitz, M.J. Commercial Timbers: Descriptions, Illustrations, Identification, and Information Retrieval. Available online: https://www.delta-intkey.com/wood/en/index.htm (accessed on 10 October 2021).
- Heiss, A.G. Anatomy of European and North American Woods—An Interactive Identification Key. Available online: http://www.holzanatomie.at/ (accessed on 9 February 2022).
- Forestry & Forest Products Research Institute. Wood Database of the Forestry & Forest Products Research Institute. Available online: https://db.ffpri.go.jp/WoodDB/index-E.html (accessed on 2 May 2022).
- Wheeler, E.A. InsideWood. Available online: https://insidewood.lib.ncsu.edu/search;jsessionid=hYhHqrsAfkTKM8JGVm0e3WjZLOdRCfo3_1Y5k6Zq?0 (accessed on 9 January 2022).
- Schoch, W.; Heller-Kellenberger, I.; Schweingruber, F.; Kienast, F.; Schmatz, D. Wood Anatomy of Central European Species. Available online: http://www.woodanatomy.ch/authors.html (accessed on 3 May 2022).
- Richter, H.G.; Gembruch, K.; Koch, G. CITESwoodID: Descriptions, Illustrations, Identification, and Information Retrieval. Available online: https://www.delta-intkey.com/citeswood/index.htm (accessed on 9 February 2022).
- Barker, J.A.; Flinders, B.A.H. Key to a Selection of Arid Australian Hardwoods & Softwoods. Available online: https://keys.lucidcentral.org/keys/v3/arid/default_wip.htm (accessed on 4 January 2022).
- Coradin, V.T.R.; Camargos, J.A.A.; Pastore, T.C.M.; Christo, A.G. Brazilian Commercial Timbers: Interactive Identification Key Based on General and Macroscopic Features Madeiras Comerciais Do Brasil: Chave Interativa de Identificação Baseada em Caracteres Gerais e Macroscópicos. Available online: https://keys.lucidcentral.org/keys/v4/madeiras_comerciais_do_brasil/index_en.html (accessed on 2 May 2022).
- Sarmiento, C.; Détienne, P.; Heinz, C.; Molino, J.-F.; Grard, P.; Bonnet, P. Pl@ntwood: A computer-assisted identification tool for 110 species of amazon trees based on wood anatomical features. IAWA J. 2011, 32, 221–232. [Google Scholar] [CrossRef]
- Martins, J.; Oliveira, L.S.; Nisgoski, S.; Sabourin, R. The Forest Species Database—Microscopy. Available online: https://web.inf.ufpr.br/vri/databases/forest-species-database-microscopic/ (accessed on 3 May 2022).
- UFPR Forest Species Database—Microscopic. Available online: https://web.inf.ufpr.br/vri/databases/forest-species-database-microscopic/ (accessed on 21 April 2022).
- Filho, P.L.P.; Oliveira, L.S.; Nisgoski, S.; Britto, A.S. The Forest Species Database—Macroscopic. Available online: https://web.inf.ufpr.br/vri/databases/forest-species-database-macroscopic/ (accessed on 3 May 2022).
- UFPR Forest Species Database—Macroscopic. Available online: https://web.inf.ufpr.br/vri/databases/forest-species-database-macroscopic/ (accessed on 23 April 2021).
- Richter, H.G.; Oelker, M.; Koch, G. MacroHOLZdata—Computer Aided Macroscopic Wood Identification and Information on Properties and Utilization of Trade Timbers. CD-ROM. Available online: http://macroholzdata.appstor.io/ (accessed on 23 April 2021).
- De Oliveira, W. Forest Species Classifier. Available online: http://reconhecimentoflorestal.md.utfpr.edu.br./#/pt/classificador (accessed on 3 May 2022).
- Maruyama, T.M.; Oliveira, L.S.; Britto, A.S.; Nisgoski, S. Automatic classification of native wood charcoal. Ecol. Inform. 2018, 46, 1–7. [Google Scholar] [CrossRef]
- Bodin, S.C.; Scheel-Ybert, R.; Beauchêne, J.; Molino, J.-F.; Bremond, L. CharKey: An electronic identification key for wood charcoals of French Guiana. IAWA J. 2019, 40, 75-S20. [Google Scholar] [CrossRef]
- EyeWood, S.F.U. Softwood Retrieval System for Coniferous Wood. Available online: http://woodlab.swfu.edu.cn/#/ (accessed on 23 April 2021).
- Filho, P.L.d.P. UTForest—UTFPR Classificador. Available online: https://clb.lamia.sh.utfpr.edu.br/classification (accessed on 8 June 2022).
- Ferreira, C.A.; Inga, J.G.; Vidal, O.D.; Goytendia, W.E.; Moya, S.M.; Centeno, T.B.; Vélez, A.; Gamarra, D.; Tomazello-Filho, M. Identification of tree species from the peruvian tropical amazon “selva central” forests according to wood anatomy. BioResources 2021, 16, 7161–7179. [Google Scholar] [CrossRef]
- Wheeler, E.A.; Baas, P.; Gasson, P.E. IAWA List of microscopic features for hardwood identification: With an appendix on non-anatomical information. IAWA Bull. 1989, 10, 219–332. [Google Scholar] [CrossRef]
- Schweingruber, F.H. Microscopic Wood Anatomy: Structural Variability of Stems and Twigs in Recent and Subfossil Woods from Central Europe; Swiss Federal Institute for Forest: Birmensdorf, Swiss, 1990; ISBN 3905620022. [Google Scholar]
- CITES. Available online: https://cites.org/eng (accessed on 2 May 2022).
- Menon, L.T.; Laurensi, I.A.; Penna, M.C.; Oliveira, L.E.S.; Britto, A.S. Data augmentation and transfer learning applied to charcoal image classification. In Proceedings of the 2019 International Conference on Systems, Signals and Image Processing (IWSSIP), Osijek, Croatia, 5–7 June 2019; pp. 69–74. [Google Scholar]
- Détienne, P.; Jacquet, P.; Mariaux, A. Manuel d’identification des bois tropicaux. tome 3: Guyane Française. In Manuel D’identification des Bois Tropicaux; CIRAD: Montpellier, France, 1982; Volume 3, p. 315. ISBN 139782876145962. [Google Scholar]
- Richter, H.G.; Grosser, D.; Heinz, I.; Gasson, P.E. IAWA list of microscopic features for softwood identification. IAWA J. 2004, 25, 1–70. [Google Scholar] [CrossRef]
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep learning for computer vision: A brief review. Comput. Intell. Neurosci. 2018, 2018, 7068349. [Google Scholar] [CrossRef]
- Li, N.; Shepperd, M.; Guo, Y. A Systematic review of unsupervised learning techniques for software defect prediction. Inf. Softw. Technol. 2020, 122, 106287. [Google Scholar] [CrossRef]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef]
- Khan, A.I.; Al-Habsi, S. Machine learning in computer vision. Procedia Comput. Sci. 2020, 167, 1444–1451. [Google Scholar] [CrossRef]
- Hwang, S.-W.; Sugiyama, J. Computer vision-based wood identification and its expansion and contribution potentials in wood science: A review. Plant Methods 2021, 17, 47. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, P.; Ebanyenle, E.; Ebeheakey, A.A.; Abban, K.B.; Lambog, O.; Soares, R.; Costa, A.; Wiedenhoeft, A.C. Image based identifcation of ghanaian timbers using the xylotron: Opportunities, risks and challenges. In Proceedings of the 33rd Conference on Neural Information Processing Systems (NeurIPS 2019), Vancouver, BC, Canada, 9–14 December 2019. [Google Scholar]
- Ravindran, P.; Thompson, B.J.; Soares, R.K.; Wiedenhoeft, A.C. The xylotron: Flexible, open-source, image-based macroscopic field identification of wood products. Front. Plant Sci. 2020, 11, 1015. [Google Scholar] [CrossRef] [PubMed]
- Tou, J.Y.; Tou, P.; Lau, P.Y.; Tay, Y.H. Computer vision-based wood recognition system. In Proceedings of the International Workshop on Advanced Image Technology; 2007. Available online: https://scholar.google.com/scholar?hl=en&as_sdt=0%2C5&q=Computer+vision-based+wood+recognition+system.+In+Proceedings+of+International+Workshop+on+Advanced+Image+Technology%2C+2007&btnG= (accessed on 27 April 2022).
- Ravindran, P.; Wiedenhoeft, A.C. Comparison of two forensic wood identification technologies for ten meliaceae woods: Computer vision versus mass spectrometry. Wood Sci. Technol. 2020, 54, 1139–1150. [Google Scholar] [CrossRef]
- Du, M.; Liu, N.; Hu, X. Techniques for interpretable machine learning. Commun. ACM 2019, 63, 68–77. [Google Scholar] [CrossRef]
- Martins, A.L.R.; Marcal, A.R.S.; Pissarra, J. Modified DBSCAN algorithm for microscopic image analysis of wood. In Iberian Conference on Pattern Recognition and Image Analysis; Springer: Cham, Switzerland, 2019; pp. 257–269. [Google Scholar]
- Olschofsky, K.; Köhl, M. Rapid field identification of cites timber species by deep learning. Trees For. People 2020, 2, 100016. [Google Scholar] [CrossRef]
- Barmpoutis, P.; Dimitropoulos, K.; Barboutis, I.; Grammalidis, N.; Lefakis, P. Wood species recognition through multidimensional texture analysis. Comput. Electron. Agric. 2018, 144, 241–248. [Google Scholar] [CrossRef]
- Filho, P.L.P.; Oliveira, L.S.; Nisgoski, S.; Britto, A.S. Forest species recognition using macroscopic images. Mach. Vis. Appl. 2014, 25, 1019–1031. [Google Scholar] [CrossRef]
- Fabijańska, A.; Danek, M.; Barniak, J. Wood species automatic identification from wood core images with a residual convolutional neural network. Comput. Electron. Agric. 2021, 181, 105941. [Google Scholar] [CrossRef]
- De Andrade, B.G.; Basso, V.M.; de Figueiredo Latorraca, J.V. Machine vision for field-level wood identification. IAWA J. 2020, 41, 681–698. [Google Scholar] [CrossRef]
- Khalid, M.; Lew, E.; Lee, Y.; Yusof, R.; Nadaraj, M. Design of an intelligent wood species recognition system. Int. J. Simul. Syst. Sci. Technol. 2008, 9, 9–19. [Google Scholar]
- Wang, H.; Zhang, G.; Qi, H. Wood recognition using image texture features. PLoS ONE 2013, 8, e76101. [Google Scholar] [CrossRef]
- Lens, F.; Liang, C.; Guo, Y.; Tang, X.; Jahanbanifard, M.; da Silva, F.S.C.; Ceccantini, G.; Verbeek, F.J. Computer-assisted timber identification based on features extracted from microscopic wood sections. IAWA J. 2020, 41, 660–680. [Google Scholar] [CrossRef]
- Martins, J.; Oliveira, L.S.; Nisgoski, S.; Sabourin, R. A database for automatic classification of forest species. Mach. Vis. Appl. 2013, 24, 567–578. [Google Scholar] [CrossRef]
- da Silva, N.R.; De Ridder, M.; Baetens, J.M.; Van den Bulcke, J.; Rousseau, M.; Bruno, O.M.; Beeckman, H.; Van Acker, J.; De Baets, B. Automated classification of wood transverse cross-section micro-imagery from 77 commercial central-African timber species. Ann. For. Sci. 2017, 74, 30. [Google Scholar] [CrossRef]
- Mallik, A.; Tarrío-Saavedra, J.; Francisco-Fernández, M.; Naya, S. Classification of wood micrographs by image segmentation. Chemom. Intell. Lab. Syst. 2011, 107, 351–362. [Google Scholar] [CrossRef]
- Kobayashi, K.; Akada, M.; Torigoe, T.; Imazu, S.; Sugiyama, J. Automated recognition of wood used in traditional japanese sculptures by texture analysis of their low-resolution computed tomography data. J. Wood Sci. 2015, 61, 630–640. [Google Scholar] [CrossRef]
- Lopes, D.J.V.; Burgreen, G.W.; Entsminger, E.D. North american hardwoods identification using machine-learning. Forests 2020, 11, 298. [Google Scholar] [CrossRef]
- Wiedenhoeft, A.C. The XyloPhone: Toward democratizing access to high-quality macroscopic imaging for wood and other substrates. IAWA J. 2020, 41, 699–719. [Google Scholar] [CrossRef]
- Yu, H.; Cao, J.; Luo, W.; Liu, Y. Image retrieval of wood species by color, texture, and spatial information. In Proceedings of the 2009 International Conference on Information and Automation, Zhuhai/Macau, China, 22–24 June 2009; pp. 1116–1119. [Google Scholar]
- Yusof, R.; Khalid, M.; Khairuddin, A.S.M. Application of kernel-genetic algorithm as nonlinear feature selection in tropical wood species recognition system. Comput. Electron. Agric. 2013, 93, 68–77. [Google Scholar] [CrossRef]
- Zamri, M.I.P.; Cordova, F.; Khairuddin, A.S.M.; Mokhtar, N.; Yusof, R. Tree species classification based on image analysis using improved-basic gray level aura matrix. Comput. Electron. Agric. 2016, 124, 227–233. [Google Scholar] [CrossRef]
- Figueroa-Mata, G.; Mata-Montero, E.; Valverde-Otarola, J.C.; Arias-Aguilar, D. Using deep convolutional networks for species identification of xylotheque samples. In Proceedings of the 2018 IEEE International Work Conference on Bioinspired Intelligence (IWOBI), San Carlos, Costa Rica, 18–20 July 2018; pp. 1–9. [Google Scholar]
- Kobayashi, K.; Kegasa, T.; Hwang, S.S.-W.; Sugiyama, J. Anatomical features of fagaceae wood statistically extracted by computer vision approaches: Some relationships with evolution. PLoS ONE 2019, 14, e0220762. [Google Scholar] [CrossRef]
- Souza, D.V.; Santos, J.X.; Vieira, H.C.; Naide, T.L.; Nisgoski, S.; Oliveira, L.E.S. An automatic recognition system of brazilian flora species based on textural features of macroscopic images of wood. Wood Sci. Technol. 2020, 54, 1065–1090. [Google Scholar] [CrossRef]
- Tang, X.J.; Tay, Y.H.; Siam, N.A.; Lim, S.C. MyWood-ID. Rapid and robust automated macroscopic wood identification system using smartphone with macro-lens. In Proceedings of the 2018 International Conference on Computational Intelligence and Intelligent Systems—CIIS 2018, Phuket, Thailand, 17–19 November 2018; ACM Press: New York, NY, USA, 2018; pp. 37–43. [Google Scholar]
- Stanford, V.L.; University, S.; University, P. ImageNet. Available online: https://www.image-net.org/ (accessed on 21 April 2021).
- Seregin, A.P. Moscow digital herbarium: A consortium since 2019. Taxon 2020, 69, 417–419. [Google Scholar] [CrossRef]
- New Your Botanical Garden, N. Index Herbariorum. Available online: http://sweetgum.nybg.org/science/ih/ (accessed on 21 April 2022).
- Soltis, P.S. Digitization of herbaria enables novel research. Am. J. Bot. 2017, 104, 1281–1284. [Google Scholar] [CrossRef]
- Sugiyama, J.; Hwang, S.W.; Kobayashi, K.; Zhai, S.; Kanai, I.; Kanai, K. Database of Cross Sectional Optical Micrograph from KYOw Lauraceae Wood. Available online: https://repository.kulib.kyoto-u.ac.jp/dspace/handle/2433/245888 (accessed on 21 April 2022).
- Wood-Auth; Barmpoutis, P. WOOD-AUTH Dataset A (Version 0.1). Available online: https://doi.org/10.2018/wood.auth (accessed on 21 April 2022).
- Nasirzadeh, M.; Khazael, A.A.; bin Khalid, M. Woods recognition system based on local binary pattern. In Proceedings of the Second International Conference on Computational Intelligence, Communication Systems and Networks, CICSyN 2010, Liverpool, UK, 28–30 July 2010; pp. 308–313. [Google Scholar]
- Khalid, M.; Yusof, R.; Khairuddin, A.S.M. Improved tropical wood species recognition system based on multi-feature extractor and classifier. Int. J. Electr. Comput. Eng. 2011, 5, 495–501. [Google Scholar]
- Damayanti, R.; Prakasa, E.; Krisdianto; Dewi, L.M.; Wardoyo, R.; Sugiarto, B.; Pardede, H.F.; Riyanto, Y.; Astutiputri, V.; Panjaitan, G.R.; et al. LignoIndo: Image database of indonesian commercial timber. IOP Conf. Ser. Earth Environ. Sci. 2019, 374, 12057. [Google Scholar] [CrossRef]
- Kour, A.; Yv, V.; Maheshwari, V.; Prashar, D. A review on image processing. Int. J. Electron. Commun. Comput. Eng. 2012, 4, 2278–4209. [Google Scholar]
- Martins, J.; Oliveira, L.S.; Britto, A.S.; Sabourin, R. Forest species recognition based on dynamic classifier selection and dissimilarity feature vector representation. Mach. Vis. Appl. 2015, 26, 279–293. [Google Scholar] [CrossRef]
- Yusof, R.; Khalid, M.; Khairuddin, A.S.M. Fuzzy logic-based pre-classifier for tropical wood species recognition system. Mach. Vis. Appl. 2013, 24, 1589–1604. [Google Scholar] [CrossRef]
- Yusof, R.; Rosli, N.R.; Khalid, M. Tropical wood species recognition based on gabor filter. In Proceedings of the 2009 2nd International Congress on Image and Signal Processing, Tianjin, China, 17–19 October 2009; pp. 1–5. [Google Scholar]
- Brunel, G.; Borianne, P.; Subsol, G.; Jaeger, M.; Caraglio, Y. Automatic identification and characterization of radial files in light microscopy images of wood. Ann. Bot. 2014, 114, 829–840. [Google Scholar] [CrossRef]
- Kobayashi, K.; Hwang, S.-W.; Lee, W.-H.; Sugiyama, J. Texture analysis of stereograms of diffuse-porous hardwood: Identification of wood species used in tripitaka koreana. J. Wood Sci. 2017, 63, 322–330. [Google Scholar] [CrossRef]
- Tou, J.Y.; Tay, Y.H.; Lau, P.Y. Rotational invariant wood species recognition through wood species verification. In Proceedings of the 2009 First Asian Conference on Intelligent Information and Database Systems, Dong hoi, Vietnam, 1–3 April 2009; pp. 115–120. [Google Scholar]
- Cavalin, P.R.; Kapp, M.N.; Martins, J.; Oliveira, L.E.S. A multiple feature vector framework for forest species recognition. In Proceedings of the 28th Annual ACM Symposium on Applied Computing—SAC ’13, Coimbra, Portugal, 18–22 March 2013; ACM Press: New York, NY, USA, 2013; p. 16. [Google Scholar]
- Yusof, R.; Khairuddin, U.; Rosli, N.R.; Ghafar, H.A.; Azmi, N.M.A.N.; Ahmad, A.; Khairuddin, A.S.M. A Study of feature extraction and classifier methods for tropical wood recognition system. In Proceedings of the TENCON 2018—2018 IEEE Region 10 Conference, Jeju, Republic of Korea, 28–31 October 2018; pp. 2034–2039. [Google Scholar]
- Lewis, N.D. Deep Learning Made Easy with R: A Gentle Introduction for Data Science; Platform, C.I.P., Ed.; CreateSpace Independent Publishing Platform: Scotts Valley, CA, USA, 2016; ISBN 1519514212. [Google Scholar]
- Zhuo, L.; Cheng, B.; Zhang, J. A Comparative study of dimensionality reduction methods for large-scale image retrieval. Neurocomputing 2014, 141, 202–210. [Google Scholar] [CrossRef]
- Fukushima, K. Neocognitron: A self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol. Cybern. 1980, 36, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Weng, Q. A Survey of image classification methods and techniques for improving classification performance. Int. J. Remote Sens. 2007, 28, 823–870. [Google Scholar] [CrossRef]
- Ravindran, P.; Costa, A.; Soares, R.; Wiedenhoeft, A.C. Classification of CITES-listed and other neotropical meliaceae wood images using convolutional neural networks. Plant Methods 2018, 14, 25. [Google Scholar] [CrossRef] [PubMed]
- Andrade, B.G.D.; Vital, B.R.; Carneiro, A.D.C.O.; Basso, V.M.; Pinto, F.D.A.D.C. Potential of texture analysis for charcoal classification. Floresta e Ambient. 2019, 26, e20171241. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, UK, 2016; ISBN 9780262035613. [Google Scholar]
- Abiodun, O.I.; Kiru, M.U.; Jantan, A.; Omolara, A.E.; Dada, K.V.; Umar, A.M.; Linus, O.U.; Arshad, H.; Kazaure, A.A.; Gana, U. Comprehensive review of artificial neural network applications to pattern recognition. IEEE Access 2019, 7, 158820–158846. [Google Scholar] [CrossRef]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Networks 2015, 61, 85–117. [Google Scholar] [CrossRef]
- Yu, Y.; Si, X.; Hu, C.; Zhang, J. A review of recurrent neural networks: LSTM cells and network architectures. Neural Comput. 2019, 31, 1235–1270. [Google Scholar] [CrossRef]
- Panzer, M.; Bender, B. Deep reinforcement learning in production systems: A systematic literature review. Int. J. Prod. Res. 2022, 60, 4316–4341. [Google Scholar] [CrossRef]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J.; et al. Recent advances in convolutional neural networks. Pattern Recognit. 2018, 77, 354–377. [Google Scholar] [CrossRef]
- Huang, T.S. Computer vision: Evolution and promise. Comput. Sci. 1996. [Google Scholar] [CrossRef]
- Carriquiry, A.; Hofmann, H.; Tai, X.H.; VanderPlas, S. Machine learning in forensic applications. Significance 2019, 16, 29–35. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Tuia, D.; Zhu, X.X.; Reichstein, M. Deep Learning for the Earth Sciences; Camps-Valls, G., Tuia, D., Zhu, X.X., Reichstein, M., Eds.; Wiley: Hoboken, NJ, USA, 2021; ISBN 9781119646143. [Google Scholar]
- Singh, S.P.; Kumar, A.; Darbari, H.; Singh, L.; Rastogi, A.; Jain, S. Machine translation using deep learning: An overview. In Proceedings of the 2017 International Conference on Computer, Communications and Electronics (Comptelix), Jaipur, India, 1–2 July 2017; pp. 162–167. [Google Scholar]
- Clanuwat, T.; Bober-Irizar, M.; Kitamoto, A.; Lamb, A.; Yamamoto, K.; Ha, D. Deep learning for classical japanese literature. In Proceedings of the Workshop on Machine Learning for Creativity and Design, Vancouver, BC, Canada, 9 December 2018. [Google Scholar]
- Li, Y.; Huang, C.; Ding, L.; Li, Z.; Pan, Y.; Gao, X. Deep learning in bioinformatics: Introduction, application, and perspective in the big data era. Methods 2019, 166, 4–21. [Google Scholar] [CrossRef]
- Esteban, L.G.; Fernández, F.G.; de Palacios, P.; Romero, R.M.; Cano, N.N. Artificial neural networks in wood identification: The case of two juniperus species from the canary islands. IAWA J. 2009, 30, 87–94. [Google Scholar] [CrossRef]
- Turhan, K.; Serdar, B. Support vector machines in wood identification: The case of three salix species from Turkey. Turk. J. Agric. For. 2013, 37, 249–256. [Google Scholar] [CrossRef]
- Hafemann, L.G.; Oliveira, L.S.; Cavalin, P. Forest species recognition using deep convolutional neural networks. In Proceedings of the 2014 22nd International Conference on Pattern Recognition, Stockholm, Sweden, 24–28 August 2014; pp. 1103–1107. [Google Scholar]
- Esteban, L.G.; de Palacios, P.; Conde, M.; Fernández, F.G.; García-Iruela, A.; González-Alonso, M. Application of artificial neural networks as a predictive method to differentiate the wood of Pinus sylvestris, L. and pinus nigra arn subsp. Salzmannii (dunal) franco. Wood Sci. Technol. 2017, 51, 1249–1258. [Google Scholar] [CrossRef]
- Kwon, O.; Lee, H.G.; Lee, M.-R.; Jang, S.; Yang, S.-Y.; Park, S.-Y.; Choi, I.-G.; Yeo, H. Automatic wood species identification of korean softwood based on convolutional neural networks. J. Korean Wood Sci. Technol. 2017, 45, 797–808. [Google Scholar] [CrossRef]
- De Oliveira, W.; Filho, P.L.d.P.; Martins, J.G. Software for forest species recognition based on digital images of wood. FLORESTA 2018, 49, 543–552. [Google Scholar] [CrossRef]
- Kwon, O.; Lee, H.G.; Yang, S.-Y.; Kim, H.; Park, S.-Y.; Choi, I.-G.; Yeo, H. Performance enhancement of automatic wood classification of korean softwood by ensembles of convolutional neural networks. J. Korean Wood Sci. Technol. 2019, 47, 265–276. [Google Scholar] [CrossRef]
- Kanayama, H.; Ma, T.; Tsuchikawa, S.; Inagaki, T. Cognitive spectroscopy for wood species identification: Near infrared hyperspectral imaging combined with convolutional neural networks. Analyst 2019, 144, 6438–6446. [Google Scholar] [CrossRef]
- He, T.; Marco, J.; Soares, R.; Yin, Y.; Wiedenhoeft, A. Machine learning models with quantitative wood anatomy data can discriminate between swietenia macrophylla and swietenia mahagoni. Forests 2019, 11, 36. [Google Scholar] [CrossRef]
- Deklerck, V.; Mortier, T.; Goeders, N.; Cody, R.B.; Waegeman, W.; Espinoza, E.; Van Acker, J.; Van den Bulcke, J.; Beeckman, H. A protocol for automated timber species identification using metabolome profiling. Wood Sci. Technol. 2019, 53, 953–965. [Google Scholar] [CrossRef]
- De Geus, A.R.; da Silva, S.F.; Gontijo, A.B.; Silva, F.O.; Batista, M.A.; Souza, J.R. An analysis of timber sections and deep learning for wood species classification. Multimed. Tools Appl. 2020, 79, 34513–34529. [Google Scholar] [CrossRef]
- Wu, F.; Gazo, R.; Haviarova, E.; Benes, B. Wood identification based on longitudinal section images by using deep learning. Wood Sci. Technol. 2021, 55, 553–563. [Google Scholar] [CrossRef]
- Shugar, A.N.; Drake, B.L.; Kelley, G. rapid identification of wood species using XRF and neural network machine learning. Sci. Rep. 2021, 11, 17533. [Google Scholar] [CrossRef]
- Da Silva, N.R.; Deklerck, V.; Baetens, J.; Van den Bulcke, J.; De Ridder, M.; Rousseau, M.; Bruno, O.M.; Beeckman, H.; Van den Acker, J.; De Baets, B.; et al. Improved wood species identification based on multi-view imagery of the three anatomical planes. Prepr. from Res. Sq. 2022, 18, 79. [Google Scholar] [CrossRef]
- Chai, J.; Zeng, H.; Li, A.; Ngai, E.W.T. Deep learning in computer vision: A critical review of emerging techniques and application scenarios. Mach. Learn. with Appl. 2021, 6, 100134. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014. [Google Scholar]
- Yi, X.; Walia, E.; Babyn, P. Generative adversarial network in medical imaging: A review. Med. Image Anal. 2019, 58, 101552. [Google Scholar] [CrossRef]
- Lopes, D.J.V.; Monti, G.F.; Burgreen, G.W.; Moulin, J.C.; dos, S.; Bobadilha, G.; Entsminger, E.D.; Oliveira, R.F. Creating high-resolution microscopic cross-section images of hardwood species using generative adversarial networks. Front. Plant Sci. 2021, 12, 760139. [Google Scholar] [CrossRef]
- Hu, K.; Wang, B.; Shen, Y.; Guan, J.; Cai, Y. Defect identification method for poplar veneer based on progressive growing generated adversarial network and MASK R-CNN model. BioResources 2020, 15, 3040–3052. [Google Scholar] [CrossRef]
- Habite, T.; Abdeljaber, O.; Olsson, A. Automatic detection of annual rings and pith location along norway spruce timber boards using conditional adversarial networks. Wood Sci. Technol. 2021, 55, 461–488. [Google Scholar] [CrossRef]
- Shu, D.; Park, S.W.; Kwon, J. 3D Point cloud generative adversarial network based on tree structured graph convolutions. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 3858–3867. [Google Scholar]
- UTAR; FRIM. MyWood-Premium. Available online: https://mywoodid.frim.gov.my/ (accessed on 20 July 2022).
- Tay, Y.H. XYLORIX: An AI-as-a-service platform for wood identification. In Proceedings of the IAWA-IUFRO International Symposium for Updating Wood Identification, Beijing, China, 20–22 May 2019. [Google Scholar]
- Agritix Xylorix. Available online: https://www.xylorix.com/products/ (accessed on 4 May 2022).
- XyloTron.org XyloTron. Available online: https://xylotron.org/ (accessed on 4 May 2022).
- The National Academies of Sciences, Engineering, and Medicine. Available online: https://sites.nationalacademies.org/PGA/PEER/PEERscience/PGA_195537 (accessed on 4 May 2022).
- Universidad Politécnica de Madrid, Universidad de Granada, Asociación Española del Comercio e Industria de la Madera. (AEIM) IMAI App.
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).