SLAF-Seq Technology-Based Genome-Wide Association and Population Structure Analyses of Ancient Camellia sinensis (L.) Kuntze in Sandu County, China
Abstract
1. Introduction
2. Materials and Methods
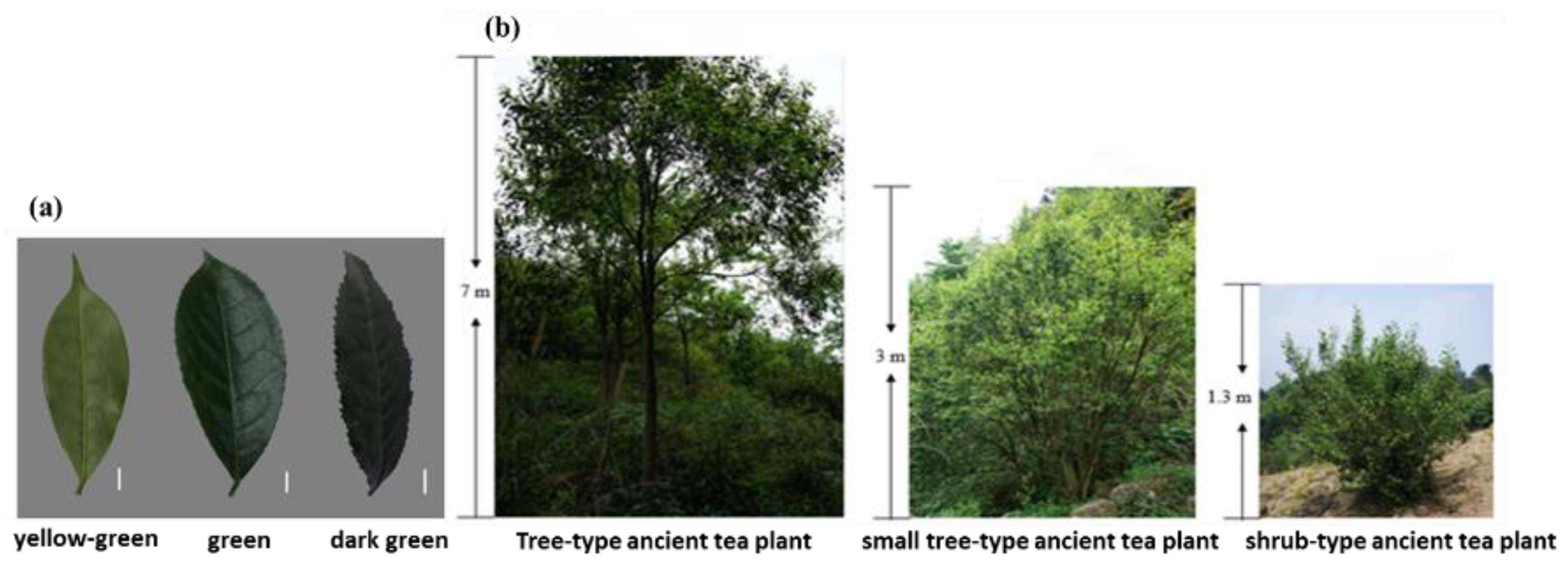
2.1. Plant Material and Phenotypic Statistics
2.2. DNA Isolation
2.3. SLAF-Seq Library Construction and High-Throughput Sequencing
2.4. SNPs Development
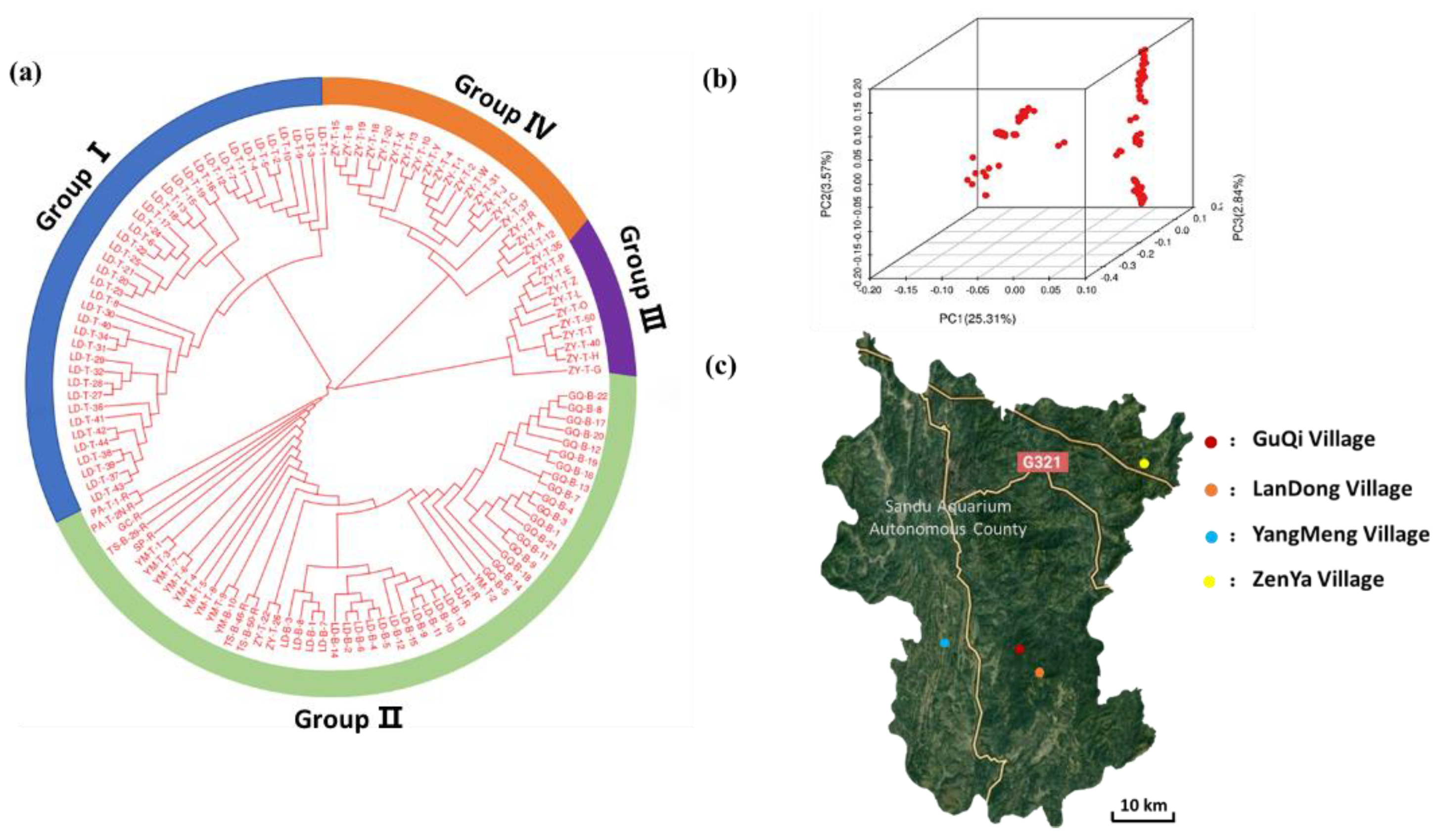
2.5. Phylogenetic and Population Structure Analyses
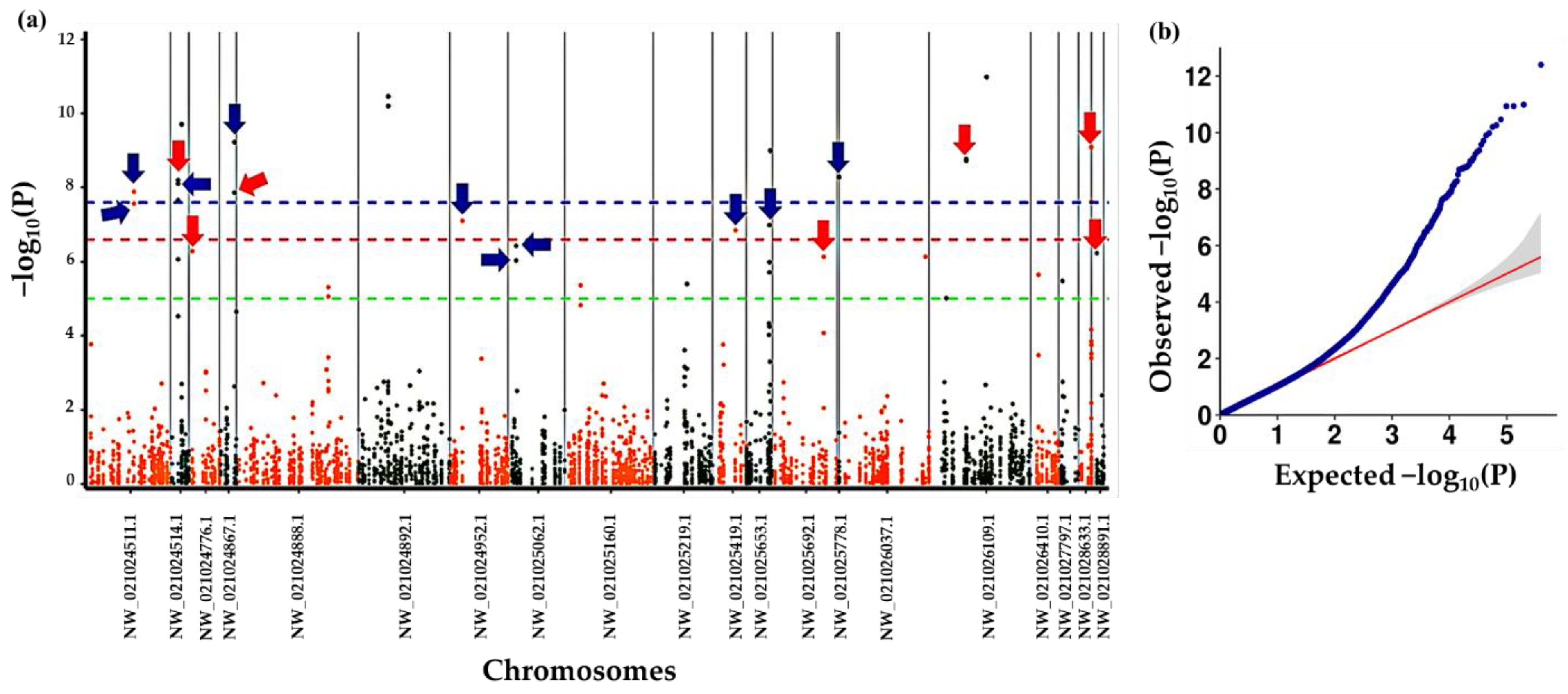
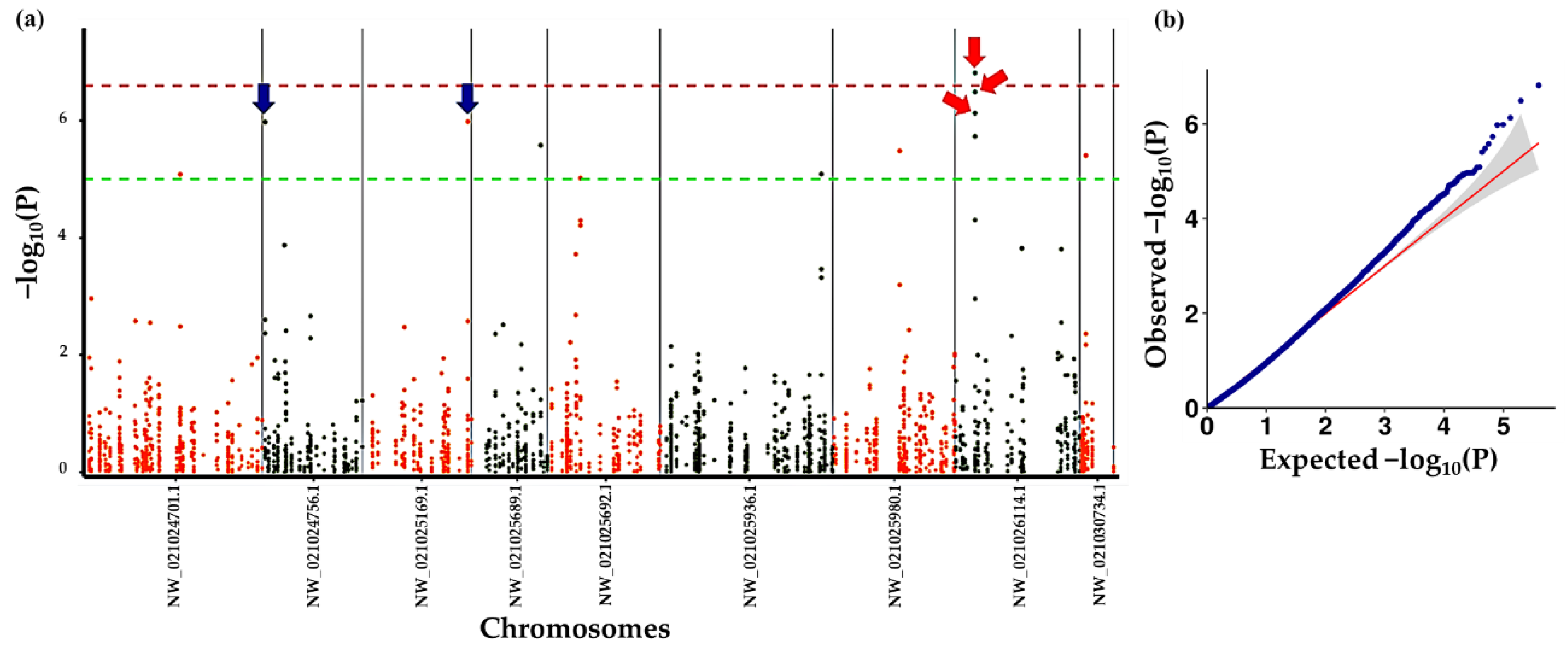
2.6. Genome-Wide Association Analysis
3. Results
3.1. Descriptive Statistics of Ancient Tea Plant Traits
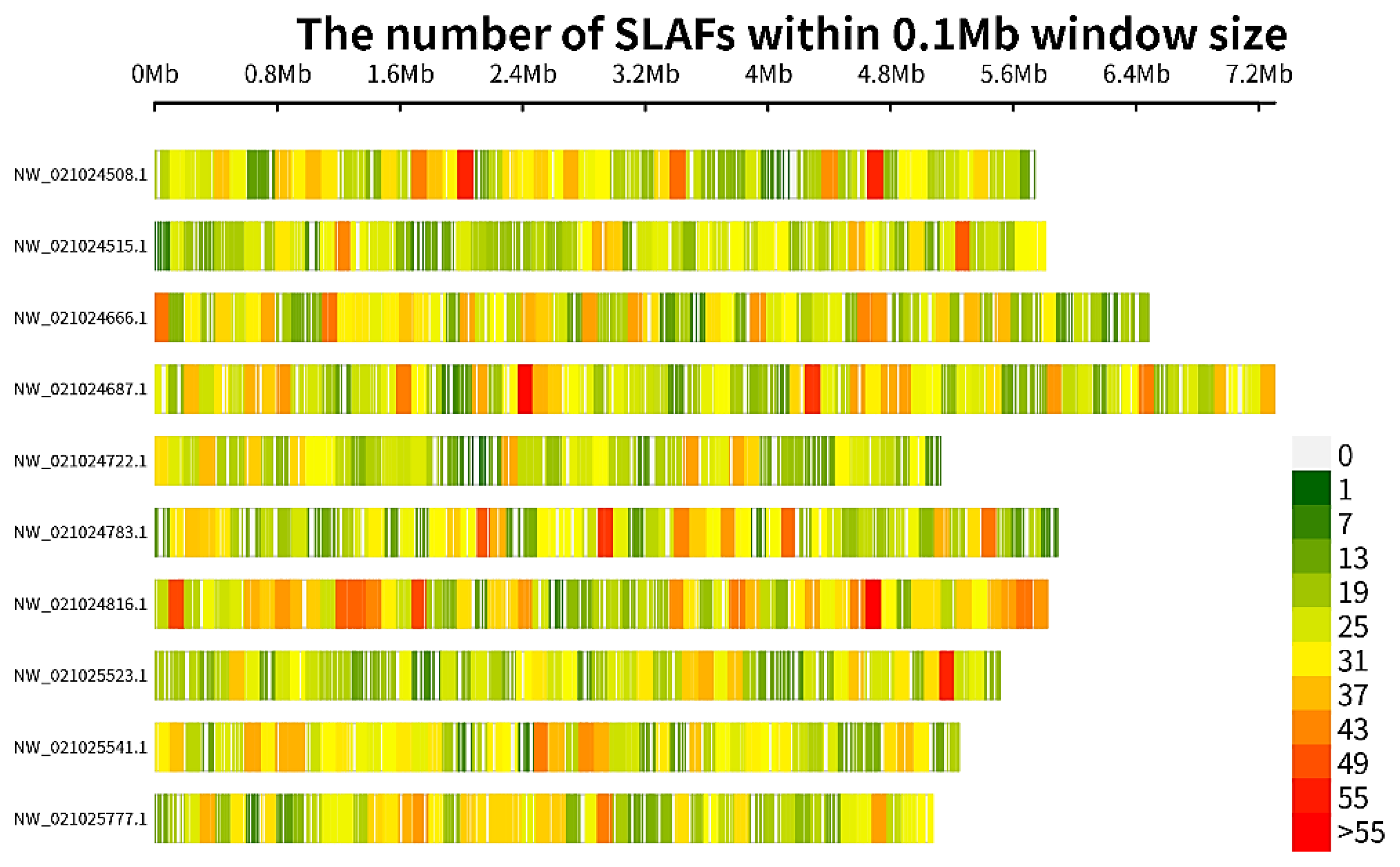
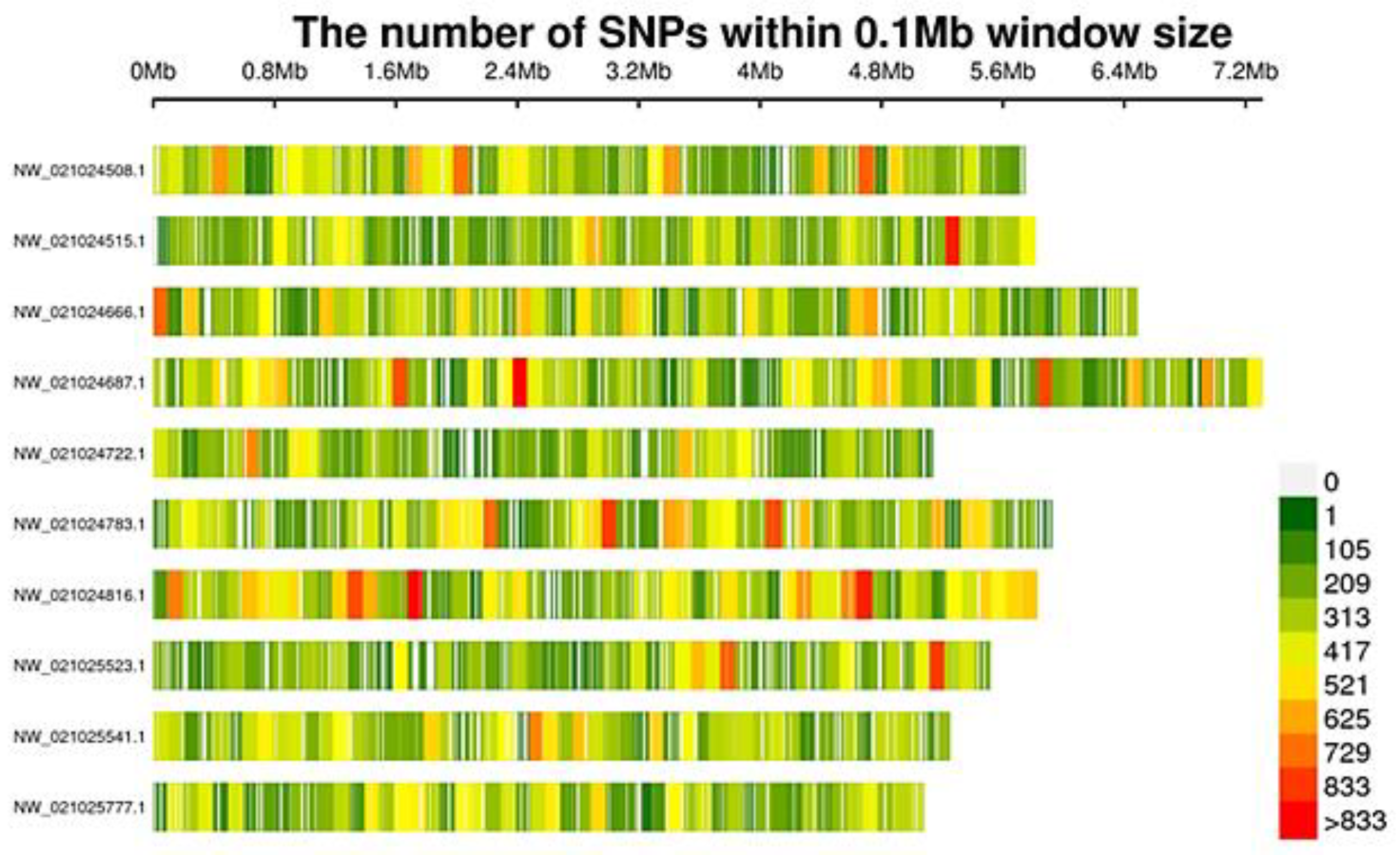
3.2. SLAF-Seq and SNPs
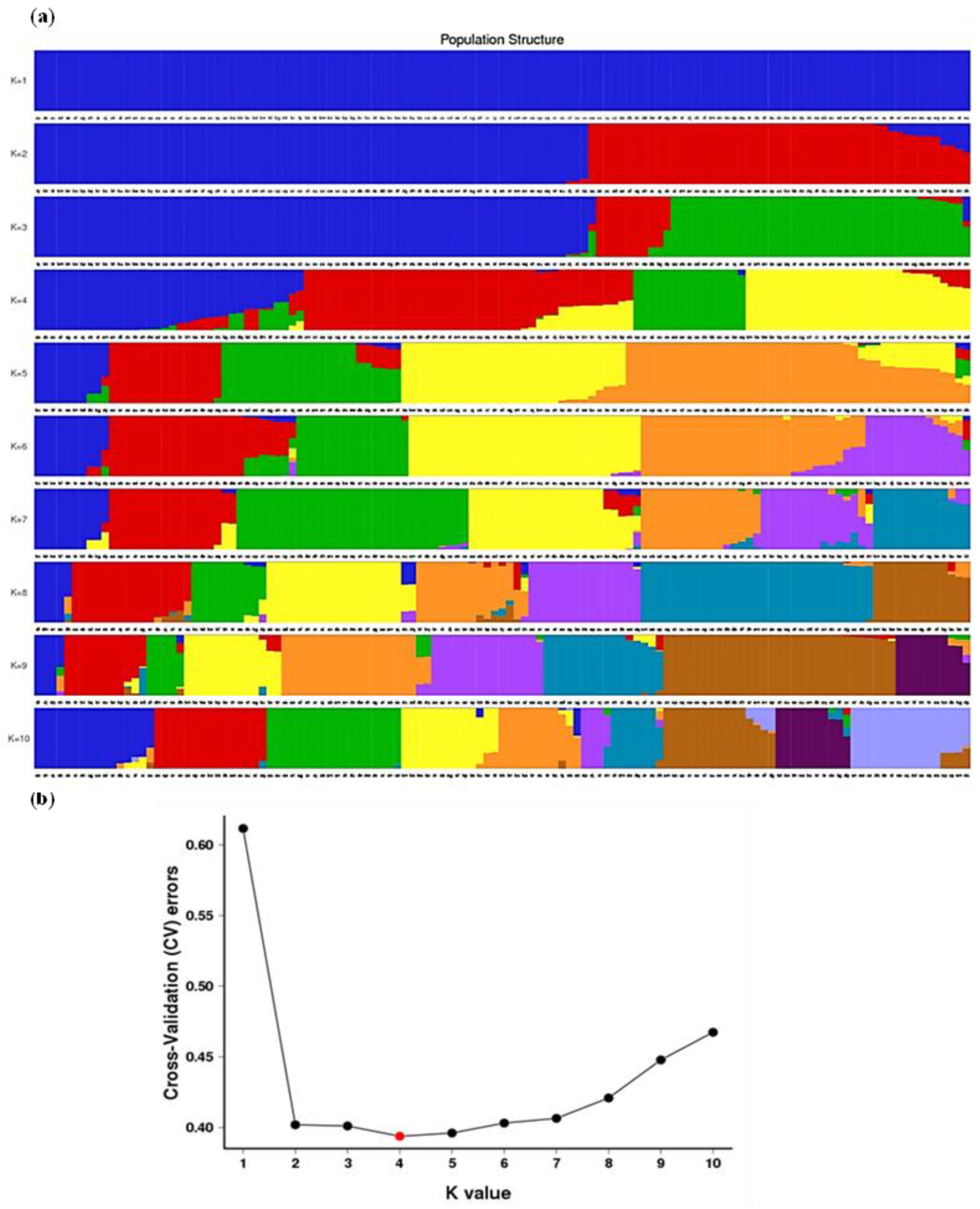
3.3. Population Structure Analyses
3.4. Association Analysis with Altitude at Origin of Plants
3.5. Association Analysis of Tree Shape
3.6. Association Analysis of Leaf Color
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Z.; Liu, H.; Cao, Y. Protection and utilization of variation type of wild tea resources and local tea varieties in Guizhou. Guizhou Agric. Sci. 2009, 21, 188–190. [Google Scholar]
- Zhao, Y.; Wang, R.; Liu, Q.; Dong, X.; Zhao, D. Genetic diversity of ancient Camellia sinensis (L.) O. Kuntze in Sandu County of Guizhou Province in China. Diversity 2021, 13, 276. [Google Scholar] [CrossRef]
- Lu, L.; Chen, H.; Wang, X.; Zhao, Y.; Yao, X.; Xiong, B.; Deng, Y.; Zhao, D. Genome-level diversification of eight ancient tea populations in the Guizhou and Yunnan regions identifies candidate genes for core agronomic traits. Hortic. Res. 2021, 8, 2559–2568. [Google Scholar] [CrossRef]
- Niu, S.; Koiwa, H.; Song, Q.; Qiao, D.; Chen, J.; Zhao, D.; Chen, Z.; Wang, Y.; Zhang, T. Development of core-collections for Guizhou tea genetic resources and GWAS of leaf size using SNP developed by genotyping-by-sequencing. PeerJ 2020, 8, e8572. [Google Scholar] [CrossRef] [PubMed]
- Kaundun, S.S.; Park, Y.G. Genetic Structure of Six Korean Tea Populations as Revealed by RAPD-PCR Markers. Crop Sci. 2002, 42, 594–601. [Google Scholar]
- Matsumoto, S.; Kiriiwa, Y.; Takeda, Y. Differentiation of Japanese green tea cultivars as revealed by RFLP analysis of phenylalanine ammonia-lyase DNA. Theor. Appl. Genet. 2002, 104, 998–1002. [Google Scholar] [CrossRef]
- Mishra, R.K.; Chaudhury, S.; Ahmad, A.; Pradhan, M.; Siddiqi, T.O. Molecular analysis of tea clones (Camellia sinensis) using AFLP markers. Int. J. Integr. Biol 2009, 5, 130–136. [Google Scholar]
- Fang, W.; Cheng, H.; Duan, Y.; Jiang, X.; Li, X. Genetic diversity and relationship of clonal tea (Camellia sinensis) cultivars in China as revealed by SSR markers. Plant Syst. Evol. 2012, 298, 469–483. [Google Scholar] [CrossRef]
- Wang, J.B. ISSR markers and their applications in plant genetics. Yi Chuan 2002, 24, 613–616. [Google Scholar]
- Zhen, L.; Xin-Chao, W.; Liping, Z. Genetic diversity and relationship analysis of tea germplasms originated from South Western China based on EST-SSR. Mol. Plant Breed. 2008, 6, 100–110. [Google Scholar]
- Sun, X.; Liu, D.; Zhang, X.; Li, W.; Liu, H.; Hong, W.; Jiang, C.; Guan, N.; Ma, C.; Zeng, H. SLAF-seq: An efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 2013, 8, e58700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L.; Xin, H.; Li, D.; Ma, C.; Ding, X.; Hong, W.; Zhang, X. Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol. 2013, 13, 1086. [Google Scholar] [CrossRef] [PubMed]
- Degen, B.; Yanbaev, Y.A.; Ianbaev, R.Y.; Bakhtina, S.Y.; Gabitova, A.A.; Tagirova, A.A. Genetic diversity and differentiation of northern populations of pedunculate oak based on analysis of new SNP markers. Russ. J. Genet. 2021, 57, 374–378. [Google Scholar] [CrossRef]
- Wang, Y.; Jiao, Z.; Zheng, J.; Zhou, J.; Wang, B.; Zhuge, Q.; He, X. Population genetic diversity and structure of an endangered salicaceae species in Northeast China: Chosenia arbutifolia (Pall.) A. Skv. Forests 2021, 12, 1282. [Google Scholar] [CrossRef]
- Jue, D.; Liu, L.; Sang, X.; Shu, B.; Wang, J.; Wang, Y.; Zhang, C.; Shi, S. SNP-based high-density genetic map construction and candidate gene identification for fruit quality traits of Dimocarpus longan Lour. Sci. Hortic. Amsterdam 2021, 284, 110086. [Google Scholar] [CrossRef]
- Lyu, Y.; Dong, X.; Huang, L.; Zheng, J.; He, X.; Sun, H.; Jiang, Z. SLAF-seq uncovers the genetic diversity and adaptation of Chinese Elm (Ulmus parvifolia) in Eastern China. Forests 2020, 11, 80. [Google Scholar] [CrossRef]
- Liu, J.; Li, X.; Liu, Y.; Xing, C.; Xie, Y.; Cai, G.; Lu, J. Evaluation of genetic diversity and development of core collections of industrial brewing yeast using ISSR markers. Arch. Microbiol. 2021, 203, 1001–1008. [Google Scholar] [CrossRef]
- Lin, Y.; Yu, W.; Zhou, L.; Fan, X.; Wang, F.; Wang, P.; Fang, W.; Cai, C.; Ye, N. Genetic diversity of oolong tea (Camellia sinensis) germplasms based on the nanofluidic array of single-nucleotide polymorphism (SNP) markers. Tree Genet. Genomes 2020, 16, 1–14. [Google Scholar] [CrossRef]
- Li, Y.; Zeng, X.; Li, G.; Zhao, Y.; Li, J.; Zhao, D. Population structure analysis and genome-wide association study of rice landraces from Qiandongnan, China, using specific-locus amplified fragment sequencing. Int. J. Agric. Biol. 2021, 25, 388–396. [Google Scholar] [CrossRef]
- Li, Y.; Zeng, X.; Zhao, Y.; Li, J.; Zhao, D. Identification of a new rice low-tiller mutant and association analyses based on the SLAF-seq method. Plant Mol. Biol. Rep. 2017, 35, 72–82. [Google Scholar] [CrossRef]
- Jia, Y.; Westcott, S.; He, T.; Mcfawn, L.A.; Angessa, T.; Hill, C.; Tan, C.; Zhang, X.; Zhou, G.; Li, C. Genome-wide association studies reveal QTL hotspots for grain brightness and black point traits in barley. Crop J. 2021, 9, 154–167. [Google Scholar] [CrossRef]
- Thabet, S.G.; Moursi, Y.S.; Sallam, A.; Karam, M.A.; Alqudah, A.M. Genetic associations uncover candidate SNP markers and genes associated with salt tolerance during seedling developmental phase in barley. Environ. Exp. Bot. 2021, 188, 104499. [Google Scholar] [CrossRef]
- Shahzad, A.; Qian, M.; Sun, B.; Mahmood, U.; Li, S.; Fan, Y.; Chang, W.; Dai, L.; Zhu, H.; Li, J. Genome-wide association study identifies novel loci and candidate genes for drought stress tolerance in rapeseed. Oil Crop Sci. 2021, 6, 12–22. [Google Scholar] [CrossRef]
- Wang, X.; Long, Y.; Wang, N.; Zou, J.; Ding, G.; Broadley, M.R.; White, P.J.; Yuan, P.; Zhang, Q.; Luo, Z. Breeding histories and selection criteria for oilseed rape in Europe and China identified by genome wide pedigree dissection. Sci. Rep. UK 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Guo, Y.; Chen, Y.; Zhang, H.; El-Kassaby, Y.A.; Li, W. Genome wide association study identifies candidate genes related to the earlywood tracheid properties in Picea crassifolia Kom. Forests 2022, 13, 332. [Google Scholar] [CrossRef]
- Yang, Q.; Jin, H.; Yu, X.; Fu, X.; Zhi, H.; Yuan, F. Rapid identification of soybean resistance genes to soybean mosaic virus by SLAF-seq bulked segregant analysis. Plant Mol. Biol. Rep. 2020, 38, 666–675. [Google Scholar] [CrossRef]
- Ren, H.; Han, J.; Wang, X.; Zhang, B.; Yu, L.; Gao, H.; Hong, H.; Sun, R.; Tian, Y.; Qi, X. QTL mapping of drought tolerance traits in soybean with SLAF sequencing. Crop J. 2020, 8, 977–989. [Google Scholar] [CrossRef]
- Chen, L.; Yang, Y.J.; Yu, F.L. Tea Germplasm Resources Description Specification and Data Standard, 1st ed.; China Agriculture Press: Beijing, China, 2005. [Google Scholar]
- Li, B.Y.; Pan, B.; Cheng, W.; Han, J.; Qi, D.; Zhu, C. Research on geomorphological regionalization of China. Acta Geogr. Sin. 2013, 68, 291–306. [Google Scholar]
- Chen, S.; Borza, T.; Byun, B.; Coffin, R.; Coffin, J.; Peters, R.; Wang-Pruski, G. DNA markers for selection of late blight resistant potato breeding lines. Am. J. Plant Sci. 2017, 8, 1197–1209. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microb. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Mckenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Lippert, C.; Listgarten, J.; Liu, Y.; Kadie, C.M.; Davidson, R.I.; Heckerman, D. FaST linear mixed models for genome-wide association studies. Nat. Methods 2011, 8, 833–835. [Google Scholar] [CrossRef]
- Sul, J.H.; Eskin, E. Mixed models can correct for population structure for genomic regions under selection. Nat. Rev. Genet. 2013, 14, 300. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.; Shen, S. Transcriptomic and metabolomic analyses reveal the altitude adaptability and evolution of different-colored flowers in alpine Rhododendron species. Tree Physiol. 2022, 42, 1100–1113. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cao, H.; Qian, W.; Yao, L.; Hao, X.; Li, N.; Yang, Y.; Wang, X. Identification of a novel bZIP transcription factor in Camellia sinensis as a negative regulator of freezing tolerance in transgenic arabidopsis. Ann. Bot. London 2017, 119, 1195–1209. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Hao, X.; Cao, H.; Ding, C.; Yang, Y.; Wang, L.; Wang, X. ABA-dependent bZIP transcription factor, CsbZIP18, from Camellia sinensis negatively regulates freezing tolerance in Arabidopsis. Plant Cell Rep. 2020, 39, 553–565. [Google Scholar] [CrossRef] [PubMed]
- Xi, X.; Wang, P.; Wang, Z.; Yu, R. The microtubule-associated protein WDL3 mediates ABA-induced stomatal closure in Arabidopsis. Chin. Sci. Bull. 2018, 64, 95–106. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Y.; Aslam, M.; Jakada, B.H.; Qin, Y.; Cai, H. The glycine-rich domain protein GRDP2 regulates ovule development via the auxin pathway in Arabidopsis. Front. Plant Sci. 2021, 12, 698487. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, P.; Wu, J.; Qiao, L.; Zhao, G.; Jia, J.; Gao, L.; Wang, J. Identification of a novel ERF gene, TaERF8, associated with plant height and yield in wheat. BMC Plant Biol. 2020, 20, 263. [Google Scholar] [CrossRef]
- Teplyakova, S.; Lebedeva, M.; Ivanova, N.; Horeva, V.; Voytsutskaya, N.; Kovaleva, O.; Potokina, E. Impact of the 7-bp deletion in HvGA20ox2 gene on agronomic important traits in barley (Hordeum vulgare L.). BMC Plant Biol. 2017, 17, 1989. [Google Scholar] [CrossRef]
- Nguyen, H.P.; Jeong, H.Y.; Jeon, S.H.; Kim, D.; Lee, C. Rice pectin methylesterase inhibitor28 (OsPMEI28) encodes a functional PMEI and its overexpression results in a dwarf phenotype through increased pectin methylesterification levels. J. Plant Physiol. 2017, 208, 17–25. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, D.; Tang, M.; Li, D.; Zhu, Y.; Zhu, L.; Chen, C. THIS1 is a putative lipase that regulates tillering, plant height, and spikelet fertility in rice. J. Exp. Bot. 2013, 64, 4389–4402. [Google Scholar] [CrossRef]
- He, Y.; Shi, Y.; Zhang, X.; Xu, X.; Wang, H.; Li, L.; Zhang, Z.; Shang, H.; Wang, Z.; Wu, J. The OsABCI7 transporter interacts with OsHCF222 to stabilize the thylakoid membrane in rice. Plant Physiol. 2020, 184, 283–299. [Google Scholar] [CrossRef]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix–loop–helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wei, J.; Ahammed, G.J.; Zhang, L.; Li, Y.; Yan, P.; Zhang, L.; Han, W. Brassinosteroids attenuate moderate high temperature-caused decline in tea quality by enhancing theanine biosynthesis in Camellia sinensis L. Front. Plant Sci. 2018, 9, 1016. [Google Scholar] [CrossRef] [PubMed]
- Xiang, P.; Wilson, I.W.; Huang, J.; Zhu, Q.; Tan, M.; Lu, J.; Liu, J.; Gao, S.; Zheng, S.; Lin, D. Co-regulation of catechins biosynthesis responses to temperature changes by shoot growth and catechin related gene expression in tea plants (Camellia sinensis L.). J. Hortic. Sci. Biotechnol. 2021, 96, 228–238. [Google Scholar] [CrossRef]
- Fu, X.; Chen, Y.; Mei, X.; Katsuno, T.; Kobayashi, E.; Dong, F.; Watanabe, N.; Yang, Z. Regulation of formation of volatile compounds of tea (Camellia sinensis) leaves by single light wavelength. Sci. Rep. UK 2015, 5, 16858. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zeng, X.; Lin, N.; Yu, S.; Fernie, A.; Zhao, J. CsbZIP1-CsMYB12 mediates the production of bitter-tasting flavonols in tea plants (Camellia sinensis) through a coordinated activator-repressor network. Hortic. Res. 2021, 8, 110. [Google Scholar] [CrossRef]
- Wu, Z.; Li, X.; Liu, Z.; Li, H.; Wang, Y.; Zhuang, J. Transcriptome-based discovery of AP2/ERF transcription factors related to temperature stress in tea plant (Camellia sinensis). Funct. Integr. Genomic. 2015, 15, 741–752. [Google Scholar] [CrossRef]
- Liu, C.; Ou, S.; Mao, B.; Tang, J.; Wang, W.; Wang, H.; Cao, S.; Schläppi, M.R.; Zhao, B.; Xiao, G. Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Ma, H.; Liu, C.; Li, Z.; Ran, Q.; Xie, G.; Wang, B.; Fang, S.; Chu, J.; Zhang, J. ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development. Plant Physiol. 2018, 178, 753–770. [Google Scholar] [CrossRef]
- Wang, W.; Xin, H.; Wang, M.; Ma, Q.; Wang, L.; Kaleri, N.A.; Wang, Y.; Li, X. Transcriptomic analysis reveals the molecular mechanisms of drought-stress-induced decreases in Camellia sinensis leaf quality. Front. Plant Sci. 2016, 7, 385. [Google Scholar] [CrossRef]
- Sun, Y.; Zhou, J.; Guo, J. Advances in the knowledge of adaptive mechanisms mediating abiotic stress responses in Camellia sinensis. Front. Biosci.-Landmrk. 2021, 26, 1714–1722. [Google Scholar] [CrossRef]
- Körner, C.; Life, A.P. Functional plant ecology of high mountain ecosystems. Alp. Plant Life 2003, 3, 202. [Google Scholar]
- Barry, R.G. Mountain Weather and Climate, 3rd ed.; Cambridge University Press: Cambridge, UK, 1992. [Google Scholar]
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Hollender, C.A.; Dardick, C. Molecular basis of angiosperm tree architecture. New Phytol. 2015, 206, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, K.; Li, Y.; Cai, W.; Zhao, Y.; Yu, B.; Zhu, Y. Genome-wide identification of the Aux/IAA family genes (MdIAA) and functional analysis of MdIAA18 for apple tree ideotype. Biochem. Genet. 2019, 57, 709–733. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Yu, H.; Xiong, X.; Yu, Y.; Yue, X.; Liu, J.; Cao, J. Genome-wide identification and characterization of pectin methylesterase inhibitor genes in Brassica oleracea. Int. J. Mol. Sci. 2018, 19, 3338. [Google Scholar] [CrossRef]
- Hongo, S.; Sato, K.; Yokoyama, R.; Nishitani, K. Demethylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the Arabidopsis stem. Plant Cell 2012, 24, 2624–2634. [Google Scholar] [CrossRef]
- Reca, I.B.; Lionetti, V.; Camardella, L.; D Avino, R.; Giardina, T.; Cervone, F.; Bellincampi, D. A functional pectin methylesterase inhibitor protein (SolyPMEI) is expressed during tomato fruit ripening and interacts with PME-1. Plant Mol. Biol. 2012, 79, 429–442. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, X.; Xu, B.; Li, Y.; Ma, Y.; Wang, G. Phenotype and transcriptome analysis reveals chloroplast development and pigment biosynthesis together influenced the leaf color formation in mutants of Anthurium andraeanum ‘Sonate’. Front. Plant Sci. 2015, 6, 139. [Google Scholar] [CrossRef]
- Shimoni Shor, E.; Hassidim, M.; Yuval Naeh, N.; Keren, N. Disruption of Nap14, a plastid-localized non-intrinsic ABC protein in Arabidopsis thaliana results in the over-accumulation of transition metals and in aberrant chloroplast structures. Plant Cell Environ. 2010, 33, 1029–1038. [Google Scholar] [CrossRef]
- Voith Von Voithenberg, L.; Park, J.; Stübe, R.; Lux, C.; Lee, Y.; Philippar, K. A novel prokaryote-type ECF/ABC transporter module in chloroplast metal homeostasis. Front. Plant Sci. 2019, 10, 1264. [Google Scholar] [CrossRef]
- Wang, L.; Pan, D.; Liang, M.; Abubakar, Y.S.; Li, J.; Lin, J.; Chen, S.; Chen, W. Regulation of anthocyanin biosynthesis in purple leaves of zijuan tea (Camellia sinensis var. kitamura). Int. J. Mol. Sci. 2017, 18, 833. [Google Scholar] [CrossRef] [PubMed]

| Sample Number | Collection Location | Altitude(m) | Tree Shape | Leaf Color |
|---|---|---|---|---|
| GQ-B-1 | Guqi | 680 | shrub | yellow-green |
| GQ-B-3 | Guqi | 680 | shrub | dark green |
| GQ-B-4 | Guqi | 680 | shrub | dark green |
| GQ-B-5 | Guqi | 690 | shrub | dark green |
| GQ-B-7 | Guqi | 680 | shrub | green |
| GQ-B-9 | Guqi | 630 | shrub | dark green |
| GQ-B-11 | Guqi | 690 | shrub | dark green |
| GQ-B-13 | Guqi | 680 | shrub | dark green |
| GQ-B-14 | Guqi | 700 | shrub | green |
| GQ-B-16 | Guqi | 690 | shrub | green |
| GQ-B-18 | Guqi | 710 | shrub | green |
| GQ-B-19 | Guqi | 710 | shrub | green |
| GQ-B-20 | Guqi | 710 | shrub | dark green |
| GQ-B-21 | Guqi | 710 | shrub | green |
| LD-B-1 | Landong | 970 | shrub | dark green |
| LD-B-2 | Landong | 990 | shrub | dark green |
| LD-B-3 | Landong | 990 | shrub | dark green |
| LD-B-4 | Landong | 990 | shrub | dark green |
| LD-B-5 | Landong | 1000 | shrub | dark green |
| LD-B-6 | Landong | 1000 | shrub | dark green |
| LD-B-7 | Landong | 1000 | shrub | dark green |
| LD-B-8 | Landong | 990 | shrub | dark green |
| LD-B-9 | Landong | 990 | shrub | dark green |
| LD-B-10 | Landong | 990 | shrub | green |
| LD-B-12 | Landong | 990 | shrub | green |
| LD-B-13 | Landong | 980 | shrub | dark green |
| LD-B-14 | Landong | 980 | shrub | dark green |
| LD-B-15 | Landong | 980 | shrub | dark green |
| YM-T-1 | Yangmeng | 890 | shrub | dark green |
| YM-T-3 | Yangmeng | 890 | shrub | green |
| YM-T-4 | Yangmeng | 880 | shrub | dark green |
| YM-T-5 | Yangmeng | 880 | shrub | dark green |
| YM-T-7 | Yangmeng | 880 | shrub | dark green |
| YM-T-9 | Yangmeng | 880 | shrub | dark green |
| YM-B-10 | Yangmeng | 880 | tree | dark green |
| LD-T-1 | Landong | 980 | small tree | yellow-green |
| LD-T-2 | Landong | 960 | small tree | yellow-green |
| LD-T-4 | Landong | 1010 | tree | green |
| LD-T-6 | Landong | 1000 | small tree | dark green |
| LD-T-7 | Landong | 980 | small tree | green |
| LD-T-9 | Landong | 980 | small tree | yellow-green |
| LD-T-10 | Landong | 980 | small tree | green |
| LD-T-11 | Landong | 1000 | tree | green |
| LD-T-12 | Landong | 990 | tree | green |
| LD-T-13 | Landong | 930 | tree | yellow-green |
| LD-T-16 | Landong | 920 | tree | green |
| LD-T-18 | Landong | 910 | tree | green |
| LD-T-19 | Landong | 910 | tree | yellow-green |
| LD-T-20 | Landong | 920 | tree | green |
| LD-T-22 | Landong | 920 | tree | green |
| LD-T-23 | Landong | 900 | tree | yellow-green |
| LD-T-25 | Landong | 920 | tree | yellow-green |
| LD-T-28 | Landong | 860 | tree | green |
| LD-T-29 | Landong | 850 | tree | green |
| LD-T-30 | Landong | 870 | tree | yellow-green |
| LD-T-32 | Landong | 850 | tree | green |
| LD-T-34 | Landong | 870 | tree | green |
| LD-T-36 | Landong | 830 | tree | green |
| LD-T-38 | Landong | 840 | small tree | green |
| LD-T-39 | Landong | 860 | small tree | yellow-green |
| LD-T-40 | Landong | 800 | tree | dark green |
| LD-T-41 | Landong | 830 | tree | yellow-green |
| LD-T-42 | Landong | 830 | tree | yellow-green |
| LD-T-44 | Landong | 810 | tree | yellow-green |
| ZY-T-A | Zenya | 1320 | tree | dark green |
| ZY-T-C | Zenya | 1310 | tree | dark green |
| ZY-T-E | Zenya | 1310 | tree | dark green |
| ZY-T-H | Zenya | 1300 | tree | green |
| ZY-T-J | Zenya | 1326 | tree | green |
| ZY-T-L | Zenya | 1340 | tree | dark green |
| ZY-T-O | Zenya | 1300 | tree | dark green |
| ZY-T-P | Zenya | 1300 | small tree | green |
| ZY-T-R | Zenya | 1290 | tree | green |
| ZY-T-T | Zenya | 1290 | tree | green |
| ZY-T-W | Zenya | 1250 | tree | yellow-green |
| ZY-T-X | Zenya | 1250 | tree | yellow-green |
| ZY-T-1 | Zenya | 1250 | tree | green |
| ZY-T-4 | Zenya | 1280 | tree | green |
| ZY-T-8 | Zenya | 1270 | tree | yellow-green |
| ZY-T-10 | Zenya | 1270 | tree | green |
| ZY-T-13 | Zenya | 1270 | tree | dark green |
| ZY-T-15 | Zenya | 1300 | tree | dark green |
| ZY-T-31 | Zenya | 1300 | tree | green |
| ZY-T-35 | Zenya | 1300 | tree | dark green |
| ZY-T-37 | Zenya | 1330 | tree | green |
| ZY-T-40 | Zenya | 1300 | small tree | dark green |
| PA-T-1-R | Pu’an | 1210 | tree | yellow-green |
| PA-T-2N-R | Pu’an | 1230 | tree | yellow-green |
| TS-B-29-R | Tuanshan | 1117 | shrub | dark green |
| TS-B-46-R | Tuanshan | 1192 | shrub | green |
| TS-B-50-R | Tuanshan | 1190 | shrub | dark green |
| SP-R | Shiping | 880 | shrub | dark green |
| DJ-R | Dejiang | 990 | shrub | yellow-green |
| GC-R | Sanqiao | 960 | shrub | green |
| 12-R | Sanqiao | 1080 | tree | green |
| LD-T-27 | Landong | 720 | tree | dark green |
| GQ-B-8 | Guqi | 680 | shrub | dark green |
| GQ-B-12 | Guqi | 680 | shrub | green |
| GQ-B-17 | Guqi | 710 | shrub | dark green |
| GQ-B-22 | Guqi | 710 | shrub | dark green |
| LD-B-11 | Landong | 990 | shrub | dark green |
| YM-T-2 | Yangmeng | 890 | tree | dark green |
| YM-T-6 | Yangmeng | 880 | tree | yellow-green |
| YM-T-8 | Yangmeng | 890 | tree | yellow-green |
| LD-T-3 | Landong | 1010 | small tree | green |
| LD-T-5 | Landong | 1010 | small tree | yellow-green |
| LD-T-8 | Landong | 980 | tree | green |
| LD-T-15 | Landong | 990 | small tree | dark green |
| LD-T-21 | Landong | 900 | tree | green |
| LD-T-24 | Landong | 940 | small tree | dark green |
| LD-T-31 | Landong | 850 | tree | dark green |
| LD-T-37 | Landong | 840 | tree | dark green |
| LD-T-43 | Landong | 820 | tree | yellow-green |
| ZY-T-G | Zenya | 1300 | tree | green |
| ZY-T-Y | Zenya | 1250 | tree | green |
| ZY-T-Z | Zenya | 1250 | tree | yellow-green |
| ZY-T-2 | Zenya | 1280 | tree | green |
| ZY-T-12 | Zenya | 1270 | tree | green |
| ZY-T-18 | Zenya | 1380 | tree | dark green |
| ZY-T-19 | Zenya | 1380 | tree | green |
| ZY-T-20 | Zenya | 1380 | tree | yellow-green |
| ZY-T-22 | Zenya | 1300 | tree | green |
| ZY-T-26 | Zenya | 1300 | tree | yellow-green |
| ZY-T-50 | Zenya | 1300 | tree | yellow-green |
| Chr | Pos | Gene ID | −log10(P) | Distance | Allele |
|---|---|---|---|---|---|
| NW_021024511.1 | 1773825 | gene-LOC114306566 | 7.89 | 3′_7454 | G/A |
| NW_021024511.1 | 1773907 | gene-LOC114306051 | 7.57 | 3′_6491 | C/T |
| NW_021024514.1 | 254831 | gene-LOC114307084 | 8.11 | 3′_42943 | C/A |
| NW_021024514.1 | 254900 | gene-LOC114307084 | 8.20 | 3′_43012 | G/A |
| NW_021024776.1 | 114996 | gene-LOC114306513 | 6.28 | 3′_96230 | A/G |
| NW_021024867.1 | 495842 | gene-LOC114309362 | 9.23 | 5′_37266 | C/T |
| NW_021024867.1 | 495876 | gene-LOC114309362 | 7.86 | 5′_37300 | C/G |
| NW_021024952.1 | 420845 | gene-LOC114311926 | 7.10 | 5′_37614 | C/T |
| NW_021025062.1 | 271971 | gene-LOC114315782 | 6.43 | 5′_28419 | G/A |
| NW_021025062.1 | 271975 | gene-LOC114315782 | 6.03 | 5′_28423 | T/C |
| NW_021025419.1 | 765188 | gene-LOC114256993 | 6.85 | 5′_39798 | T/C |
| NW_021025653.1 | 768001 | gene-LOC114262091 | 6.99 | 5′_33301 | A/G |
| NW_021025692.1 | 1703135 | gene-LOC114262751 | 6.13 | 3′_28405 | T/A |
| NW_021025778.1 | 70220 | gene-LOC114264616 | 8.28 | 5′_88872 | C/T |
| NW_021026109.1 | 1237022 | gene-LOC114271509 | 8.73 | 3′_66332 | G/A |
| NW_021028633.1 | 425237 | gene-LOC114298253 | 9.09 | 5′_58469 | G/A |
| NW_021028891.1 | 183040 | gene-LOC114299252 | 6.23 | 3′_41369 | C/T |
| Candidate Transcript | Corresponding Candidate Gene | SwissProt Annotation | Species Name |
|---|---|---|---|
| gene-LOC114307084 | ERF4 | Ethylene-responsive transcription factor 4 | Nicotiana tabacum |
| gene-LOC114306513 | WDL3 | Protein Wave-dampende 2-like 3 | Arabidopsis thaliana |
| gene-LOC114309362 | MMK2 | Mitogen-activated protein kinase homolog MMK2 | Medicago sativa |
| gene-LOC114262751 | bZIP18 | bZIP transcription factor 18 | Arabidopsis thaliana |
| gene-LOC114271509 | MYBP1 | Transcription factor MYBP1 | Oryza sativa subsp. japonica |
| gene-LOC114298253 | GTS | Glutathione S-transferase | Arabidopsis thaliana |
| gene-LOC114299252 | HSP70 | Heat shock protein 70 | Helianthus annuus |
| Chr | Pos | Gene ID | −log10(P) | Distance | Allele |
|---|---|---|---|---|---|
| NW_021024493.1 | 2051435 | gene-LOC114313670 | 7.23 | 5′_17193 | C/T |
| NW_021024659.1 | 976560 | gene-LOC114280177 | 7.55 | 3′_18973 | C/T |
| NW_021024683.1 | 2219147 | gene-LOC114287420 | 7.10 | 3′_97898 | C/T |
| NW_021024972.1 | 976930 | gene-LOC114312788 | 8.23 | 5′_29338 | C/G |
| NW_021025124.1 | 1428384 | gene-LOC114317453 | 6.01 | 3′_83111 | C/T |
| NW_021025474.1 | 716828 | gene-LOC114258021 | 7.02 | 5′_74073 | G/A |
| NW_021025626.1 | 1177505 | gene-LOC114261209 | 6.27 | 5′_94084 | C/A |
| NW_021025626.1 | 1177505 | gene-LOC114261210 | 6.27 | 5′_38692 | C/A |
| NW_021025626.1 | 1177505 | gene-LOC114261212 | 6.27 | 5′_33377 | C/A |
| NW_021025987.1 | 742920 | gene-LOC114269018 | 7.70 | 3′_3237 | G/A |
| NW_021026130.1 | 1127861 | gene-LOC114271953 | 7.21 | 5′_91916 | G/A |
| NW_021026834.1 | 1782507 | gene-LOC114283035 | 6.66 | 5′_3360 | T/C |
| NW_021028036.1 | 538795 | gene-LOC114295371 | 6.16 | 3′_58473 | C/T |
| NW_021028084.1 | 154517 | gene-LOC114295649 | 6.53 | 3′_65378 | G/A |
| NW_021026995.1 | 2509736 | gene-LOC114285221 | 6.43 | 3′_16206 | G/A |
| Candidate Transcript | Corresponding Candidate Gene | SwissProt Annotation | Species Name |
|---|---|---|---|
| gene-LOC114280177 | GLP1 | Germin-like protein1 | Oryza sativa |
| gene-LOC114261209 | THIS1 | A putative lipase | Oryza sativa |
| gene-LOC114261210 | SWC4 | SWR1-complex protein 4 | Arabidopsis thaliana |
| gene-LOC114261212 | GA20ox2 | Gibberellin 20-dioxygenase 2 | Pisum sativum |
| gene-LOC114271953 | PMEI28 | Pectinesterase inhibitor 28 | Arabidopsis thaliana |
| gene-LOC114283035 | SGL | Kinesin-like protein SGL | Gossypium hirsutum |
| gene-LOC114295649 | GRDP2 | Glycine-rich domain protein 2 | Arabidopsis thaliana |
| gene-LOC114285221 | ERF8 | Ethylene-responsive transcription factor 8 | Arabidopsis thaliana |
| Chr | Pos | Gene ID | −log10(P) | Distance | Allele |
|---|---|---|---|---|---|
| NW_021024756.1 | 56383 | gene-LOC114306063 | 6.08 | 3′_13784 | G/A |
| NW_021025169.1 | 2005114 | gene-LOC114318750 | 6.08 | 5′_88588 | C/T |
| NW_021026114.1 | 386575 | gene-LOC114271560 | 6.13 | 5′_94608 | G/T |
| NW_021026114.1 | 386575 | gene-LOC114271592 | 6.13 | 3′_5585 | G/T |
| NW_021026114.1 | 386575 | gene-LOC114271595 | 6.13 | 3′_79759 | G/T |
| Candidate Transcript | Corresponding Candidate Gene | SwissProt Annotation | Species Name |
|---|---|---|---|
| gene-LOC114271560 | ABCI7 | Protein ABC transporter I7 | Arabidopsis thaliana |
| gene-LOC114271592 | bHLH | Basic helix-loop-helix type transcription factor | Oryza sativa |
| gene-LOC114271595 | SIG2A | Nuclear-encoded sigma (σ) factor 2A | Oryza sativa |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, L.; Dong, X.; Liu, Q.; Wang, R.; Li, Y.; Huang, X.; Zhao, Y. SLAF-Seq Technology-Based Genome-Wide Association and Population Structure Analyses of Ancient Camellia sinensis (L.) Kuntze in Sandu County, China. Forests 2022, 13, 1885. https://doi.org/10.3390/f13111885
Cheng L, Dong X, Liu Q, Wang R, Li Y, Huang X, Zhao Y. SLAF-Seq Technology-Based Genome-Wide Association and Population Structure Analyses of Ancient Camellia sinensis (L.) Kuntze in Sandu County, China. Forests. 2022; 13(11):1885. https://doi.org/10.3390/f13111885
Chicago/Turabian StyleCheng, Linan, Xuan Dong, Qing Liu, Runying Wang, Yan Li, Xiaozhen Huang, and Yichen Zhao. 2022. "SLAF-Seq Technology-Based Genome-Wide Association and Population Structure Analyses of Ancient Camellia sinensis (L.) Kuntze in Sandu County, China" Forests 13, no. 11: 1885. https://doi.org/10.3390/f13111885
APA StyleCheng, L., Dong, X., Liu, Q., Wang, R., Li, Y., Huang, X., & Zhao, Y. (2022). SLAF-Seq Technology-Based Genome-Wide Association and Population Structure Analyses of Ancient Camellia sinensis (L.) Kuntze in Sandu County, China. Forests, 13(11), 1885. https://doi.org/10.3390/f13111885






