Bayesian Approach for Optimizing Forest Inventory Survey Sampling with Remote Sensing Data
Abstract
1. Introduction
2. Materials
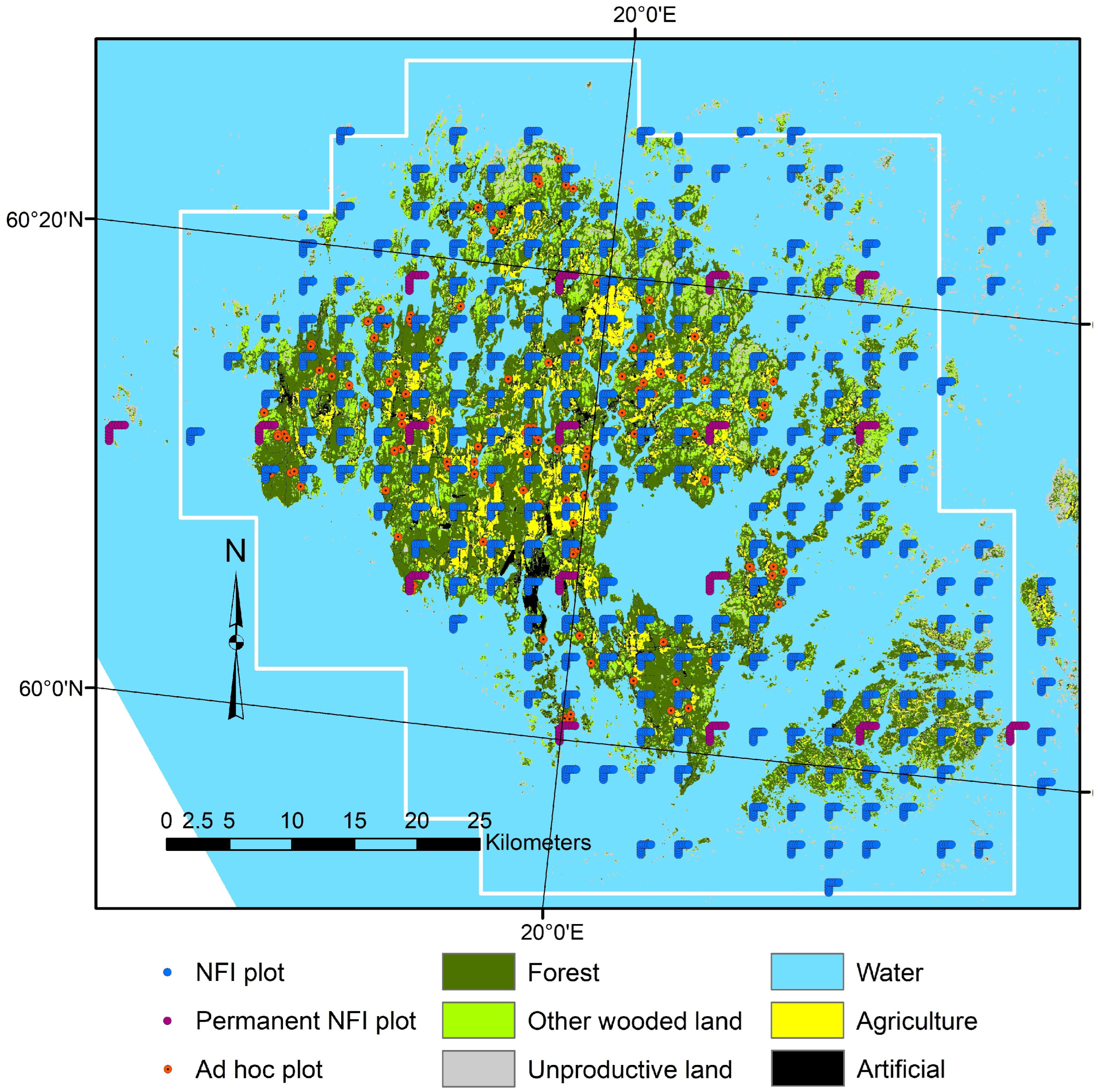
2.1. Study Area and Field Data
2.2. Remote Sensing Data
- 1
- Average, standard deviation and coefficient of variation of height above ground (H) for canopy returns, separately from first (f) and last (l) returns (havg[f/l], hstd[f/l], hcv[f/l]).
- 2
- H at which p% of cumulative sum of H of canopy returns is achieved () (hp[f/l], p is one of 0, 5, 10, 20, 30, 40, 50, 60, 70, 80, 85, 90, 95 and 100).
- 3
- Percentage of canopy returns having than corresponding (p[f/l], p is one of 20, 40, 60, 80, 95).
- 4
- Canopy densities corresponding to the proportions of points above fraction no. 0, 1, ..., 9 to a total number of points (d0, d1, ..., d9).
- 5
- (a) Ratio of first canopy returns to all first returns (vegf), and (b) Ratio of last canopy returns to all last returns (vegl).
- 6
- Ratio of intensity percentile p to the median of intensity for canopy returns (ip[f/l], p is one of 20, 40, 60 and 80).
- 1
- Average, standard deviation (std) and coefficient of variation (cv) from each of the four image bands: near-infrared (nir), red (r), green (g), blue (b).
- 2
- The following multiband transformations Normalized difference vegetation index, NDVI. See, e.g., [41]: NDVI as (nir − r)/(nir + r), modified NDVI as (nir − g)/(nir + g), nir/r, nir/g.
- 3
- Haralick textural features [42] based on co-occurrence matrices of image band values: angular second moment (ASM), contrast (Contr), correlation (Corr), variance (Var), inverse difference moment (IDM), sum average (SA), sum variance (SV), sum entropy (SE), entropy (Entr), difference variance (DV), difference entropy (DE).
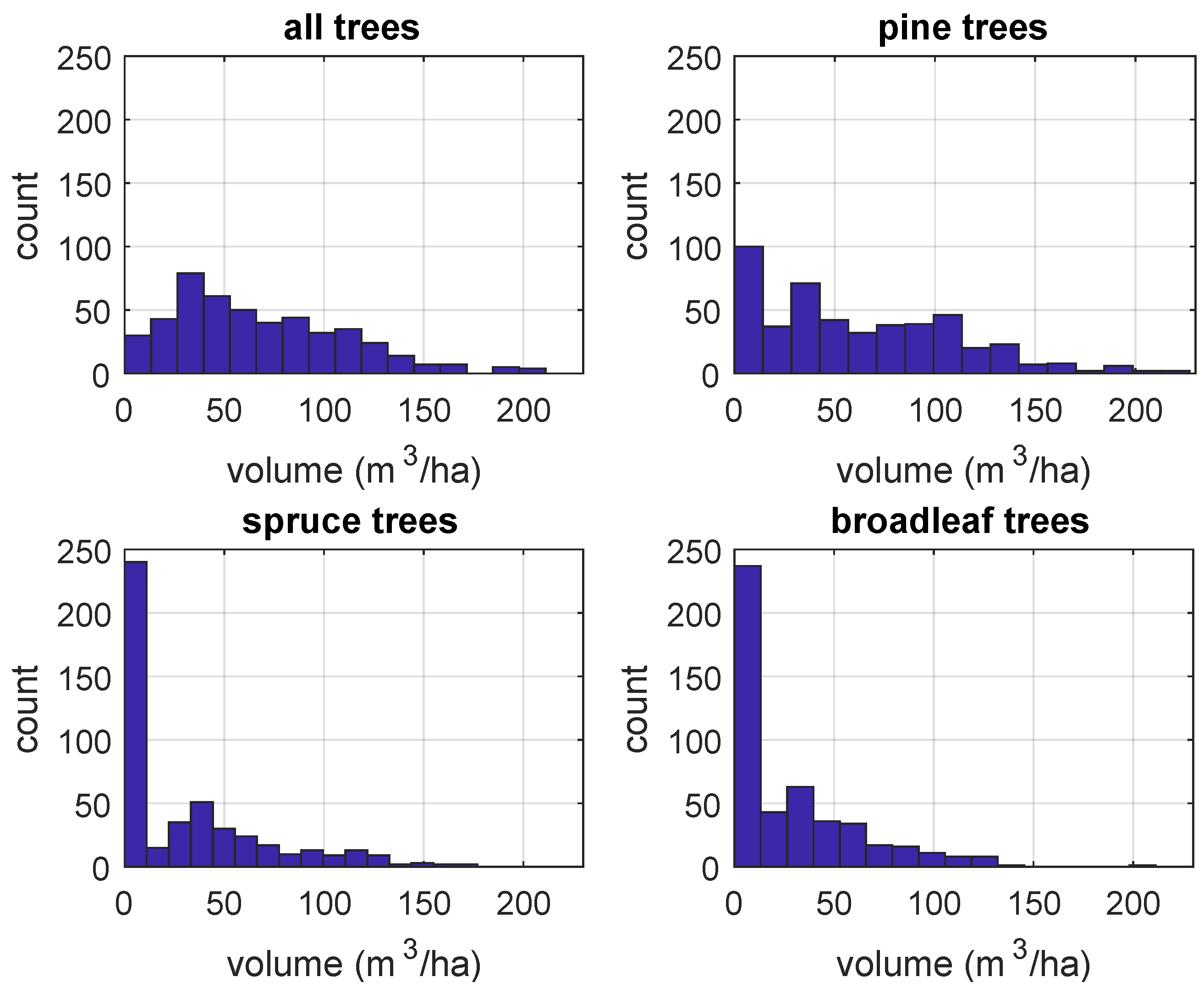
2.3. Target Population
3. Methods
3.1. Population Parameter Estimators
3.2. Simple Random and Local Pivotal Method Sampling
- 1
- Randomly choose a data point with uniform probability.
- 2
- Find the nearest neighbor (i.e., nearest in, e.g., Euclidean distance e sense) of in the feature space .
- 3
- If data point has two neighbors equally close in the feature space, then randomly with equal probability select either of the two neighbors.
- 4
- Update the inclusion probability pair using the rules found in Algorithm 1.
- 5
- Remove the data point in the pair for which the inclusion probability is either 0 or 1 from further consideration.
- 6
- If all the inclusion probabilities in set have or , then stop the algorithm and include data points with into . Otherwise, repeat from step 1.
| Algorithm 1 Pseudocode for LPM |
Require: ▹ The population data and set of initial inclusion probabilities Ensure: ▹ The returned sample data 1: set and 2: while do ▹ Repeat until sampling decision is made for all the data 3: Randomly select a data point from set with uniform probability 4: Set ▹ find the nearest neighbor 5: Update the inclusion probabilities using the rules: 6:
7: Set ▹ Remove samples with decision 8: end while 9: Set ▹ Data points with positive sampling decision 10:
return |
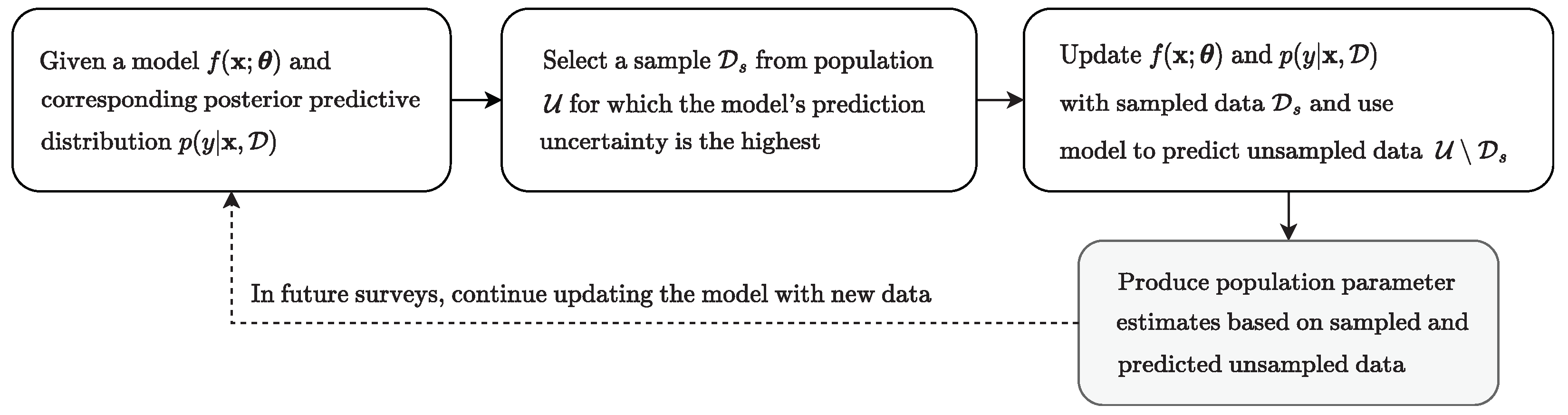
3.3. Data Sampling Via Bayesian Optimization
| Algorithm 2 Pseudocode for BMVI |
Require: ▹ Prior data set, population and sample size Ensure: ▹ Sample data set 1: Set 2: Calculate using prior data set ▹ Note 3: for to kdo ▹ Select k data points 4: Set ▹ Data point with max. uncertainty 5: Set ▹ Include data point into sample 6: Set ▹ Remove sampled point from population 7: end for 8: return ▹ Return sample of size k |
3.4. Prediction Models
3.4.1. Ridge Regression
3.4.2. Multilayer Perceptron
3.5. Implementation Details of the Empirical Analysis
| Algorithm 3 Procedure used for obtaining the empirical results |
Require: ▹ Population data, sample fraction vector and prediction model Ensure: 1: Set ▹ Sets of squared error values 2: Set 3: for to 100 do ▹ Repeat 100 times to produce averaged results 4: Select a random prior sample set from according to 5: for do ▹ Do sampling with all methods 6: Select a sample from using and 7: Set ▹ Combine prior and sampled data 8: Set ▹ Use the remaining unsampled data for testing 9: Train a prediction model f using data set 10: Set estimator 11: Set estimator 12: Set ▹ Error between estimate and true value 13: Set 14: end for 15: end for 16: for do ▹ Calculate MSEs for all methods 17: Set 18: Set 19: end for ▹ Lastly return all MSE values for all methods 20: return |
4. Results
Volume of Growing Stock Data
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A. Derivations and Proofs
References
- Särndal, C.E.; Swensson, B.; Wretman, J. Model Assisted Survey Sampling (Springer Series in Statistics); Springer: New York, NY, USA, 1992. [Google Scholar]
- Fuller, W.A. Sampling Statistics, 1st ed.; John Wiley & Sons, Inc.: Hoboken, United States, 2009; pp. 16–29. [Google Scholar] [CrossRef]
- Kangas, A.; Maltamo, M. Forest Inventory Methodology and Applications, 1st ed.; Springer: Dordrecht, The Netherlands, 2006. [Google Scholar] [CrossRef]
- Cochran, W.G. Sampling Techniques, 3rd ed.; John Wiley: Hoboken, NJ, USA, 1977. [Google Scholar]
- Loetsch, F.; Haller, K.E. Forest Inventory Vol. 1, Statistics of Forest Inventory and Information from Aerial Photographs; BLV Verlagsgesellschaft: Munich, Germany, 1964. [Google Scholar]
- Kondo, M.C.; Bream, K.D.; Barg, F.K.; Branas, C.C. A random spatial sampling method in a rural developing nation. BMC Public Health 2014, 14, 338. [Google Scholar] [CrossRef] [PubMed]
- Pennanen, O.; Mäkelä, O. Raakapuukuljetusten Kelirikkohaittojen Vähentäminen, Metsätehon Raportti; Technical Report 153; Metsäteho Ltd.: Vantaa, Finland, 2003. [Google Scholar]
- Pohjankukka, J.; Nevalainen, P.; Pahikkala, T.; Hyvönen, E.; Sutinen, R.; Hänninen, P.; Heikkonen, J. Arctic soil hydraulic conductivity and soil type recognition based on aerial gamma-ray spectroscopy and topographical data. In Proceedings of the 22nd International Conference on Pattern Recognition (ICPR 2014), Stockholm, Sweden, 24–28 August 2014; Borga, M., Heyden, A., Laurendeau, D., Felsberg, M., Boyer, K., Eds.; pp. 1822–1827. [Google Scholar] [CrossRef]
- Pohjankukka, J.; Nevalainen, P.; Pahikkala, T.; Hyvönen, E.; Middleton, M.; Hänninen, P.; Ala-Ilomäki, J.; Heikkonen, J. Predicting Water Permeability of the Soil Based on Open Data. In Proceedings of the 10th International Conference on Artificial Intelligence Applications and Innovations (AIAI 2014), Rhodes, Greece, 19–21 September 2014; Lazaros, I., Ilias, M., Harris, P., Eds.; IFIP Advances in Information and Communication Technology. Springer: Berlin/Heidelberg, Germany, 2014; Volume 436, pp. 436–446. [Google Scholar] [CrossRef]
- Pohjankukka, J.; Riihimäki, H.; Nevalainen, P.; Pahikkala, T.; Ala-Ilomäki, J.; Hyvönen, E.; Varjo, J.; Heikkonen, J. Predictability of Boreal Forest Soil Bearing Capacity by Machine Learning. J. Terramech. 2016, 68, 1–8. [Google Scholar] [CrossRef]
- Tomppo, E.; Katila, M.; Mäkisara, K.; Peräsaari, J. Multi-Source National Forest Inventory—Methods and Applications; Managing Forest Ecosystems; Springer: Berlin, Germany, 2008; Volume 18. [Google Scholar] [CrossRef]
- Wallner, A.; Elatawneh, A.; Schneider, T.; Kindu, M.; Ossig, B.; Knoke, T. Remotely sensed data controlled forest inventory concept. Eur. J. Remote. Sens. 2018, 51, 75–87. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Tomppo, E.O. Remote sensing support for national forest inventories. ForestSAT Special Issue. Remote. Sens. Environ. 2007, 110, 412–419. [Google Scholar] [CrossRef]
- Puliti, S.; Ene, L.T.; Gobakken, T.; Næsset, E. Use of partial-coverage UAV data in sampling for large scale forest inventories. Remote. Sens. Environ. 2017, 194, 115–126. [Google Scholar] [CrossRef]
- Abegg, M.; Kükenbrink, D.; Zell, J.; Schaepman, M.E.; Morsdorf, F. Terrestrial Laser Scanning for Forest Inventories—Tree Diameter Distribution and Scanner Location Impact on Occlusion. Forests 2017, 8, 184. [Google Scholar] [CrossRef]
- Kangas, A.; Astrup, R.; Breidenbach, J.; Fridman, J.; Gobakken, T.; Korhonen, K.T.; Maltamo, M.; Nilsson, M.; Nord-Larsen, T.; Næsset, E.; et al. Remote sensing and forest inventories in Nordic countries–roadmap for the future. Scand. J. For. Res. 2018, 33, 397–412. [Google Scholar] [CrossRef]
- White, J.; Coops, N.; Wulder, M.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote Sensing Technologies for Enhancing Forest Inventories: A Review. Can. J. Remote. Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef]
- Saukkola, A.; Melkas, T.; Riekki, K.; Sirparanta, S.; Peuhkurinen, J.; Holopainen, M.; Hyyppä, J.; Vastaranta, M. Predicting Forest Inventory Attributes Using Airborne Laser Scanning, Aerial Imagery, and Harvester Data. Remote Sens. 2019, 11, 797. [Google Scholar] [CrossRef]
- Saad, R.; Wallerman, J.; Holmgren, J.; Lämås, T. Local pivotal method sampling design combined with micro stands utilizing airborne laser scanning data in a long term forest management planning setting. Silva Fenn. 2016, 50, 1414. [Google Scholar] [CrossRef][Green Version]
- Grafström, A.; Lundström, N.L.; Schelin, L. Spatially Balanced Sampling through the Pivotal Method. Biometrics 2012, 68, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Grafström, A.; Ringvall, A.H. Improving forest field inventories by using remote sensing data in novel sampling designs. Can. J. For. Res. 2013, 43, 1015–1022. [Google Scholar] [CrossRef]
- Grafström, A.; Schelin, L. How to Select Representative Samples. Scand. J. Stat. 2014, 41, 277–290. [Google Scholar] [CrossRef]
- Grafström, A.; Zhao, X.; Nylander, M.; Petersson, H. A new sampling strategy for forest inventories applied to the temporary clusters of the Swedish national forest inventory. Can. J. For. Res. 2017, 47, 1161–1167. [Google Scholar] [CrossRef]
- Räty, M.; Heikkinen, J.; Kangas, A. Assessment of sampling strategies utilizing auxiliary information in large-scale forest inventory. Can. J. For. Res. 2018, 48, 749–757. [Google Scholar] [CrossRef]
- Räty, M.; Kangas, A.S. Effect of permanent plots on the relative efficiency of spatially balanced sampling in a national forest inventory. Ann. For. Sci. 2019, 76, 20. [Google Scholar] [CrossRef]
- Katila, M.; Heikkinen, J. Reducing error in small-area estimates of multi-source forest inventory by multi-temporal data fusion. For. Int. J. For. Res. 2020, 93, 471–480. [Google Scholar] [CrossRef]
- Ruotsalainen, R.; Pukkala, T.; Kangas, A.; Vauhkonen, J.; Tuominen, S.; Packalen, P. The effects of sample plot selection strategy and the number of sample plots on inoptimality losses in forest management planning based on airborne laser scanning data. Can. J. For. Res. 2019, 49, 1135–1146. [Google Scholar] [CrossRef]
- Räty, M.; Kuronen, M.; Myllymäki, M.; Kangas, A.; Mäkisara, K.; Heikkinen, J. Comparison of the local pivotal method and systematic sampling for national forest inventories. For. Ecosyst. 2020, 7, 54. [Google Scholar] [CrossRef]
- de Gruijter, J.; Brus, D.; Bierkens, M.; Knotters, M. Sampling for Natural Resource Monitoring; Springer: Berlin, Germany, 2006. [Google Scholar] [CrossRef]
- Brus, D. Spatial Sampling with R; Chapman and Hall/CRC: Boca Raton, FL, USA, 2022. [Google Scholar] [CrossRef]
- Rasmussen, C.E.; Williams, C.K.I. Gaussian Processes for Machine Learning (Adaptive Computation and Machine Learning); The MIT Press: Cambridge, MA, USA, 2005. [Google Scholar]
- Garnett, R.; Osborne, M.A.; Roberts, S.J. Bayesian Optimization for Sensor Set Selection. In Proceedings of the 9th ACM/IEEE International Conference on Information Processing in Sensor Networks, Stockholm, Sweden, 12–16 April 2010; Association for Computing Machinery: New York, NY, USA, 2010. IPSN ’10. pp. 209–219. [Google Scholar] [CrossRef]
- Flynn, E.B.; Todd, M.D. A Bayesian approach to optimal sensor placement for structural health monitoring with application to active sensing. Mech. Syst. Signal Process. 2010, 24, 891–903. [Google Scholar] [CrossRef]
- Liu, B. An Introduction to Bayesian Techniques for Sensor Networks. In Proceedings of the Wireless Algorithms, Systems, and Applications, Beijing, China, 15–17 August 2010; Pandurangan, G., Anil Kumar, V.S., Ming, G., Liu, Y., Li, Y., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 307–313. [Google Scholar]
- Tomppo, E.; Heikkinen, J.; Henttonen, H.; Ihalainen, A.; Katila, M.; Mäkelä, H.; Tuomainen, T.; Vainikainen, N. Designing and Conducting a Forest Inventory—Case: 9th National Forest Inventory of Finland, 1st ed.; Managing Forest Ecosystems 21; Springer: Dordrecht, The Netherlands, 2011. [Google Scholar] [CrossRef]
- Metsäntutkimuslaitos. Valtakunnan Metsien 11. Inventoinnin Maastotyöohje; Metla: Joensuu, Finland, 2009.
- Haara, A.; Kangas, A.; Tuominen, S. Economic losses caused by tree species proportions and site type errors in forest management planning. Silva Fenn. 2019, 53, 10089. [Google Scholar] [CrossRef]
- Næsset, E. Accuracy of forest inventory using airborne laser scanning: Evaluating the first nordic full-scale operational project. Scand. J. For. Res. 2004, 19, 554–557. [Google Scholar] [CrossRef]
- Packalén, P.; Maltamo, M. Predicting the Plot Volume by Tree Species Using Airborne Laser Scanning and Aerial Photographs. For. Sci. 2006, 52, 611–622. [Google Scholar] [CrossRef]
- Packalén, P.; Maltamo, M. Estimation of species-specific diameter distributions using airborne laser scanning and aerial photographs. Can. J. For. Res. 2008, 38, 1750–1760. [Google Scholar] [CrossRef]
- Yengoh, G.T.; Dent, D.; Olsson, L.; Tengberg, A.E.; Tucker, C.J. Use of the Normalized Difference Vegetation Index (NDVI) to Assess Land Degradation at Multiple Scales: Current Status, Future Trends, and Practical Considerations, 1st ed.; Springer Publishing Company, Incorporated: New York, NY, USA, 2015. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. Syst. Man Cybern. IEEE Trans. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Pohjankukka, J. Machine Learning Approaches for Natural Resource Data. Ph.D. Thesis, University of Turku, Turku, Finland, 2018. [Google Scholar]
- Pohjankukka, J.; Tuominen, S.; Pitkänen, J.; Pahikkala, T.; Heikkonen, J. Comparison of estimators and feature selection procedures in forest inventory based on airborne laser scanning and digital aerial imagery. Scand. J. For. Res. 2018, 33, 681–694. [Google Scholar] [CrossRef]
- Racine, E.; Coops, N.; Bégin, J. Tree species, crown cover, and age as determinants of the vertical distribution of airborne LiDAR returns. Trees 2021, 35, 1845–1861. [Google Scholar] [CrossRef]
- Kansanen, K.; Packalen, P.; Lähivaara, T.; Seppänen, A.; Vauhkonen, J.; Maltamo, M.; Mehtätalo, L. Refining and evaluating a horvitz-thompson-like stand density estimator in individual tree detection based on airborne laser scanning. Can. J. For. Res. 2022, 52, 527–538. [Google Scholar] [CrossRef]
- Beland, M.; Parker, G.; Sparrow, B. On promoting the use of lidar systems in forest ecosystem research. For. Ecol. Manag. 2019, 450, 117484. [Google Scholar] [CrossRef]
- Tuominen, S.; Balazs, A.; Kangas, A. Comparison of photogrammetric canopy models from archived and made-to-order aerial imagery in forest inventory. Silva Fenn. 2020, 54, 10291. [Google Scholar] [CrossRef]
- Pahikkala, T.; Airola, A.; Salakoski, T. Speeding Up Greedy Forward Selection for Regularized Least-Squares. In Proceedings of the 2010 Ninth International Conference on Machine Learning and Applications, Washington, DC, USA, 12–14 December 2010; pp. 325–330. [Google Scholar] [CrossRef][Green Version]
- Deville, J.C.; Tillé, Y. Unequal probability sampling without replacement through a splitting method. Biometrika 1998, 85, 89–101. [Google Scholar] [CrossRef]
- Bishop, C.M. Neural Networks for Pattern Recognition; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- MacKay, D.J.C. Information-Based Objective Functions for Active Data Selection. Neural Comput. 1992, 4, 590–604. [Google Scholar] [CrossRef]
- MacKay, D.J.C. The Evidence Framework Applied to Classification Networks. Neural Comput. 1992, 4, 720–736. [Google Scholar] [CrossRef]
- MacKay, D.J.C. Bayesian Interpolation. Neural Comput. 1992, 4, 415–447. [Google Scholar] [CrossRef]
- Neal, R.M. Bayesian Learning for Neural Networks; Springer: Berlin/Heidelberg, Germany, 1996. [Google Scholar] [CrossRef]
- Xia, G.; Miranda, M.L.; Gelfand, A.E. Approximately optimal spatial design approaches for environmental health data. Environmetrics 2006, 17, 363–385. [Google Scholar] [CrossRef]
- Chipeta, M.; Terlouw, D.; Phiri, K.; Diggle, P. Inhibitory geostatistical designs for spatial prediction taking account of uncertain covariance structure. Environmetrics 2017, 28, e2425. [Google Scholar] [CrossRef]
- Müller, W.G. Collecting Spatial Data: Optimum Design of Experiments for Random Fields, 3rd ed.; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar] [CrossRef]
- Zhu, Z.; Stein, M.L. Spatial sampling design for prediction with estimated parameters. J. Agric. Biol. Environ. Stat. 2006, 11, 24–44. [Google Scholar] [CrossRef]
- Diggle, P.; Lophaven, S. Bayesian Geostatistical Design. Scand. J. Stat. 2006, 33, 53–64. [Google Scholar] [CrossRef]
- Snoek, J.; Larochelle, H.; Adams, R.P. Practical Bayesian Optimization of Machine Learning Algorithms. In Proceedings of the 25th International Conference on Neural Information Processing Systems—Volume 2, Lake Tahoe, NV, USA, 3–6 December 2012; Curran Associates Inc.: Red Hook, NY, USA, 2012. NIPS’12. pp. 2951–2959. [Google Scholar]
- Osborne, M.A. Bayesian Gaussian Processes for Sequential Prediction, Optimisation and Quadrature. Ph.D. Thesis, Oxford University, Oxford, UK, 2010. [Google Scholar]
- Werner, J.; Müller, G. Spatio-Temporal Design; John Wiley & Sons, Ltd.: Hoboken, United States, 2012. [Google Scholar]
- BMVI. Bayesian Maximum Variance Inclusion—Python Implementation. 2019. Available online: https://github.com/jjepsuomi/Bayesian-maximum-variance-inclusion (accessed on 23 September 2019).
- Bishop, C.M. Pattern Recognition and Machine Learning (Information Science and Statistics); Springer: Secaucus, NJ, USA, 2006. [Google Scholar]
- Vapnik, V.N. Statistical Learning Theory; Wiley-Interscience: Hoboken, NJ, USA, 1998; Volume 1. [Google Scholar]
- Murphy, K.P. Machine Learning: A Probabilistic Perspective; The MIT Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Gelman, A.; Carlin, J.B.; Stern, H.S.; Dunson, D.B.; Vehtari, A.; Rubin, D.B. Bayesian Data Analysis, 3rd ed.; Chapman & Hall/CRC Texts in Statistical Science, Taylor & Francis: Boca Raton, FL, USA, 2013. [Google Scholar]
- Nabney, I.T. NETLAB: Algorithms for Pattern Recognition; Springer: London, UK, 2004. [Google Scholar]
- Bazaraa, M.S. Nonlinear Programming: Theory and Algorithms, 3rd ed.; Wiley Publishing: Hoboken, NJ, USA, 2013. [Google Scholar] [CrossRef]
| Volume All Trees |
|---|
| texture feature, sum average, ALS based canopy height |
| percentage of last canopy returns above 20% height limit |
| percentage of first canopy returns above 90% height limit |
| texture feature, entropy, ALS based canopy height |
| texture feature, angular second moment, ALS based canopy height |
| texture feature, inverse difference moment, ALS based canopy height |
| H at which 100% of cumulative sum of last canopy returns is achieved () |
| gndvi, transformation from band averages within the pixel windows: nir − g/nir + g |
| percentage of last canopy returns having than corresponding |
| coefficient of determination of first returned canopy returns |
| Volume Pine Trees |
| percentage of last canopy returns above 70% height limit |
| texture feature, angular second moment, ALS based intensity |
| texture feature, contrast, near-infrared band of CIR imagery |
| transformation from band averages within the pixel windows: nir/r |
| texture feature, sum average, ALS based canopy height |
| texture feature, sum average, ALS based intensity |
| texture feature, difference variance, blue band of RGB imagery |
| ratio of last canopy returns to all last returns |
| ratio of first canopy returns to all first returns |
| Volume Spruce Trees |
| ratio of intensity percentile 20 to the median of intensity for last canopy returns |
| percentage of last canopy returns above 30% height limit |
| ratio of intensity percentile 60 to the median of intensity for last canopy returns |
| ratio of intensity percentile 80 to the median of intensity for first canopy returns |
| texture feature, difference variance, blue band of RGB imagery |
| texture feature, coefficient of determination, near-infrared band of CIR imagery |
| percentage of first canopy returns above 30% height limit |
| coefficient of determination of last returned canopy returns |
| texture feature, coefficient of determination, red band of CIR imagery |
| texture feature, contrast, blue band of RGB imagery |
| Volume Broadleaf Trees |
| texture feature, sum average, ALS based intensity |
| ratio of intensity percentile 40 to the median of intensity for first canopy returns |
| ratio of intensity percentile 20 to the median of intensity for first canopy returns |
| ratio of intensity percentile 40 to the median of intensity for last canopy returns |
| texture feature, entropy, ALS based intensity |
| texture feature, variance, ALS based intensity |
| texture feature, inverse difference moment, ALS based intensity |
| percentage of first canopy returns having than corresponding |
| percentage of first canopy returns above 20% height limit |
| H at which 5% of cumulative sum of last canopy returns is achieved () |
| Regularized Least Squares | Multilayer Perceptron | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SRS | 1.510 | 2.829 | 1.794 | 1.399 | 1.778 | 3.176 | 2.732 | 1.911 | |
| 0.1/0.6/0.3 | LPM | 1.198 | 2.595 | 1.382 | 1.639 | 1.973 | 2.361 | 2.735 | 3.757 |
| BMVI | 0.832 | 2.036 | 20.065 | 6.502 | 11.086 | 16.368 | 6.977 | 2.036 | |
| SRS | 1.353 | 3.072 | 1.642 | 1.583 | 1.913 | 2.349 | 2.180 | 2.333 | |
| 0.2/0.5/0.3 | LPM | 1.229 | 2.044 | 1.307 | 1.384 | 1.585 | 2.181 | 2.276 | 2.202 |
| BMVI | 0.796 | 1.430 | 19.881 | 6.796 | 7.030 | 16.576 | 2.054 | 1.561 | |
| SRS | 1.536 | 2.502 | 2.111 | 1.731 | 2.323 | 2.416 | 2.047 | 2.226 | |
| 0.3/0.4/0.3 | LPM | 1.733 | 2.589 | 1.851 | 1.747 | 2.610 | 2.439 | 2.303 | 2.774 |
| BMVI | 1.046 | 2.158 | 17.148 | 5.848 | 11.222 | 9.252 | 1.800 | 1.659 | |
| SRS | 1.381 | 2.083 | 2.085 | 1.579 | 1.935 | 2.077 | 2.612 | 2.378 | |
| 0.4/0.3/0.3 | LPM | 1.241 | 2.416 | 1.648 | 1.414 | 2.006 | 1.852 | 2.472 | 2.248 |
| BMVI | 0.945 | 2.037 | 12.111 | 6.148 | 3.024 | 5.594 | 2.332 | 1.853 | |
| SRS | 1.588 | 2.434 | 1.812 | 1.623 | 1.851 | 2.638 | 2.176 | 2.804 | |
| 0.5/0.2/0.3 | LPM | 1.184 | 2.195 | 1.569 | 1.853 | 1.472 | 1.955 | 2.638 | 2.104 |
| BMVI | 0.871 | 2.321 | 6.851 | 3.495 | 2.621 | 3.756 | 1.443 | 1.015 | |
| SRS | 1.211 | 3.615 | 1.613 | 1.861 | 1.824 | 2.811 | 1.795 | 2.491 | |
| 0.6/0.1/0.3 | LPM | 1.184 | 2.996 | 1.830 | 1.353 | 2.494 | 2.341 | 2.186 | 2.059 |
| BMVI | 1.094 | 1.641 | 2.761 | 1.855 | 2.782 | 4.221 | 0.844 | 0.861 | |
| Regularized Least Squares | Multilayer Perceptron | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SRS | 6.255 | 8.666 | 8.204 | 6.632 | 7.555 | 4.972 | 3.799 | 3.726 | |
| 0.1/0.6/0.3 | LPM | 7.829 | 9.072 | 8.395 | 5.202 | 9.882 | 6.821 | 3.323 | 4.655 |
| BMVI | 0.243 | 5.499 | 1.244 | 0.699 | 12.202 | 9.341 | 2.962 | 1.954 | |
| SRS | 7.293 | 8.661 | 9.115 | 6.638 | 9.562 | 5.906 | 3.606 | 4.271 | |
| 0.2/0.5/0.3 | LPM | 6.981 | 9.381 | 8.197 | 5.818 | 9.950 | 5.438 | 3.460 | 3.082 |
| BMVI | 0.184 | 5.184 | 0.892 | 0.710 | 6.899 | 7.962 | 0.379 | 0.997 | |
| SRS | 5.697 | 8.377 | 9.399 | 5.952 | 8.268 | 5.231 | 3.753 | 3.758 | |
| 0.3/0.4/0.3 | LPM | 8.497 | 8.346 | 9.627 | 5.682 | 9.737 | 5.789 | 4.097 | 4.362 |
| BMVI | 0.258 | 5.513 | 0.935 | 0.704 | 4.242 | 3.499 | 0.180 | 0.168 | |
| SRS | 4.365 | 9.241 | 9.490 | 5.282 | 7.489 | 5.902 | 4.165 | 2.660 | |
| 0.4/0.3/0.3 | LPM | 5.726 | 7.925 | 8.725 | 5.088 | 6.747 | 5.516 | 3.147 | 2.730 |
| BMVI | 0.436 | 5.255 | 2.078 | 0.683 | 3.936 | 3.598 | 0.388 | 0.247 | |
| SRS | 5.527 | 9.382 | 7.713 | 5.088 | 7.093 | 5.786 | 2.846 | 3.394 | |
| 0.5/0.2/0.3 | LPM | 5.282 | 8.416 | 8.212 | 6.579 | 8.525 | 5.609 | 3.486 | 3.807 |
| BMVI | 0.336 | 5.418 | 4.095 | 1.288 | 2.364 | 2.495 | 0.246 | 0.163 | |
| SRS | 5.263 | 8.452 | 7.706 | 5.826 | 10.022 | 5.101 | 3.254 | 3.384 | |
| 0.6/0.1/0.3 | LPM | 4.405 | 9.165 | 8.143 | 5.556 | 11.979 | 6.345 | 3.713 | 2.682 |
| BMVI | 0.469 | 6.016 | 5.155 | 1.743 | 1.698 | 2.948 | 0.223 | 0.553 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pohjankukka, J.; Tuominen, S.; Heikkonen, J. Bayesian Approach for Optimizing Forest Inventory Survey Sampling with Remote Sensing Data. Forests 2022, 13, 1692. https://doi.org/10.3390/f13101692
Pohjankukka J, Tuominen S, Heikkonen J. Bayesian Approach for Optimizing Forest Inventory Survey Sampling with Remote Sensing Data. Forests. 2022; 13(10):1692. https://doi.org/10.3390/f13101692
Chicago/Turabian StylePohjankukka, Jonne, Sakari Tuominen, and Jukka Heikkonen. 2022. "Bayesian Approach for Optimizing Forest Inventory Survey Sampling with Remote Sensing Data" Forests 13, no. 10: 1692. https://doi.org/10.3390/f13101692
APA StylePohjankukka, J., Tuominen, S., & Heikkonen, J. (2022). Bayesian Approach for Optimizing Forest Inventory Survey Sampling with Remote Sensing Data. Forests, 13(10), 1692. https://doi.org/10.3390/f13101692






