Genetic Evaluation and Combined Selection for the Simultaneous Improvement of Growth and Wood Properties in Catalpa bungei Clones
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Experimental Site
2.2. Growth Traits
2.3. Wood Sampling and Wood Property Measurement
2.3.1. Wood Hardness
2.3.2. Wood Density
2.3.3. Compression Strength Parallel to the Grain of the Wood
2.4. Statistical Analysis
2.4.1. Mixed Linear Models and Genetic Parameters
2.4.2. The Selection of Optimal Clones under Different Selection Scenarios
3. Results
3.1. Variation in Growth Traits and Wood Properties in Catalpa bungei Clones
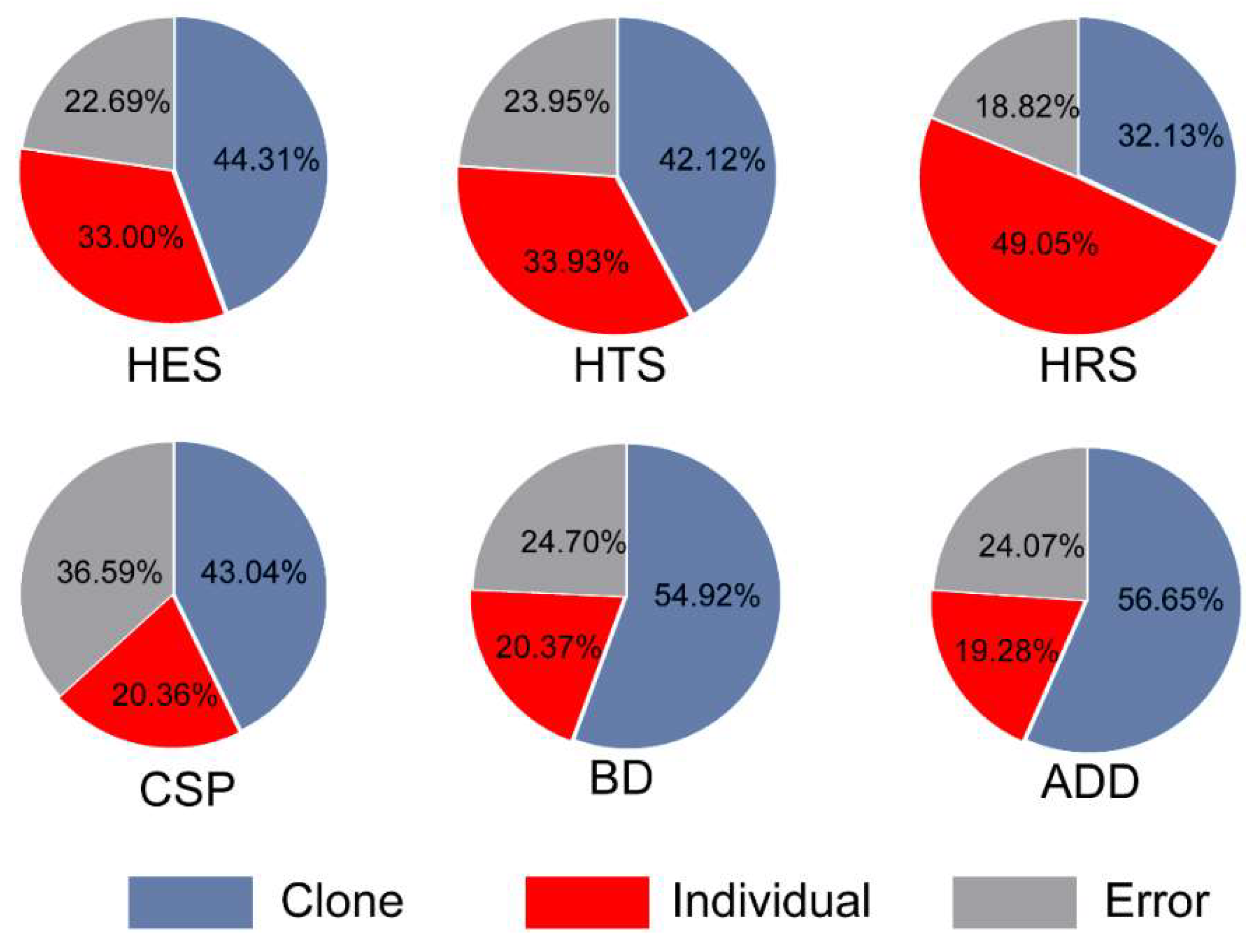
3.2. Variance Component Proportion for Wood Properties
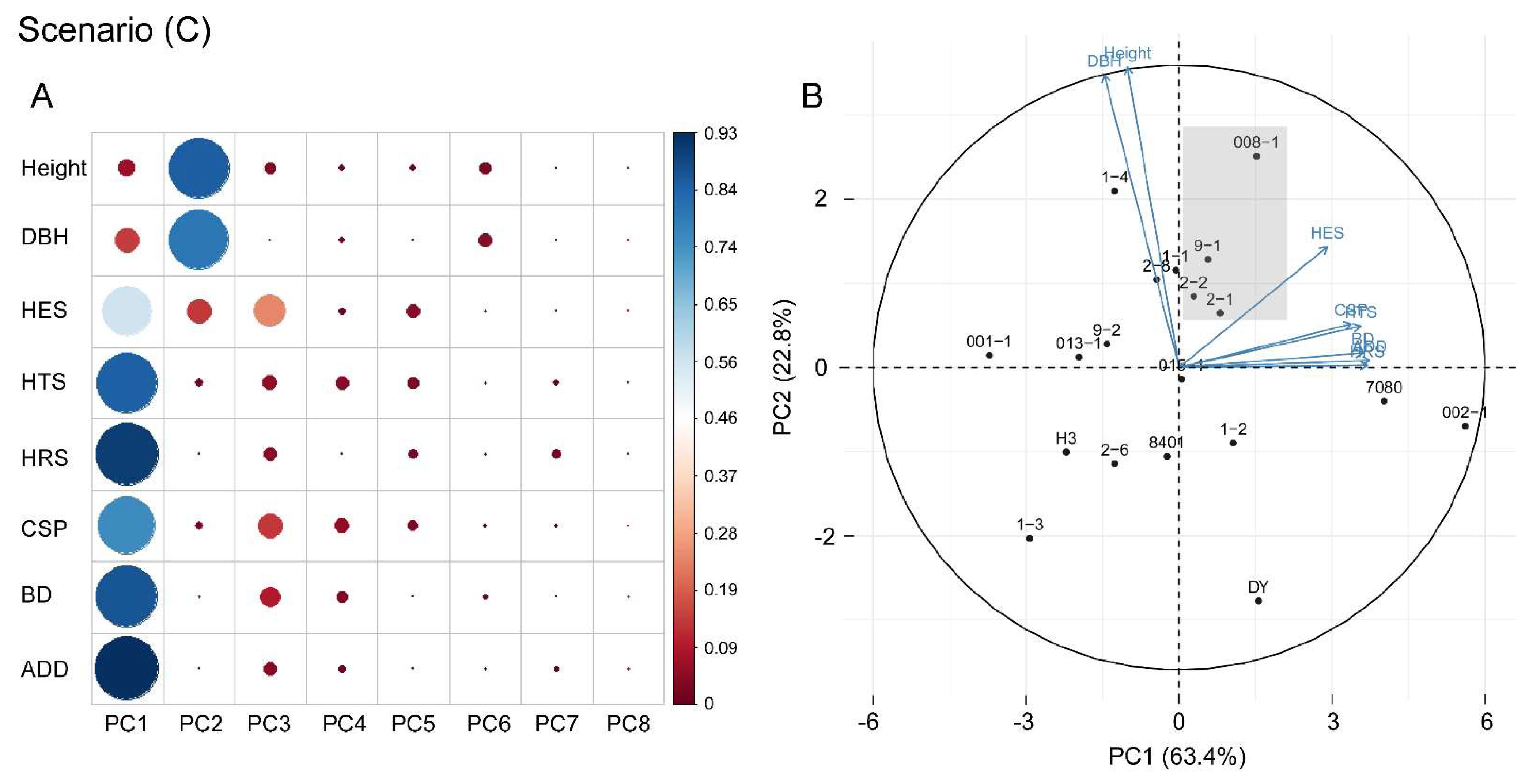
3.3. Genetic and Phenotypic Correlation Analysis
3.4. Selection of Optimal Clones
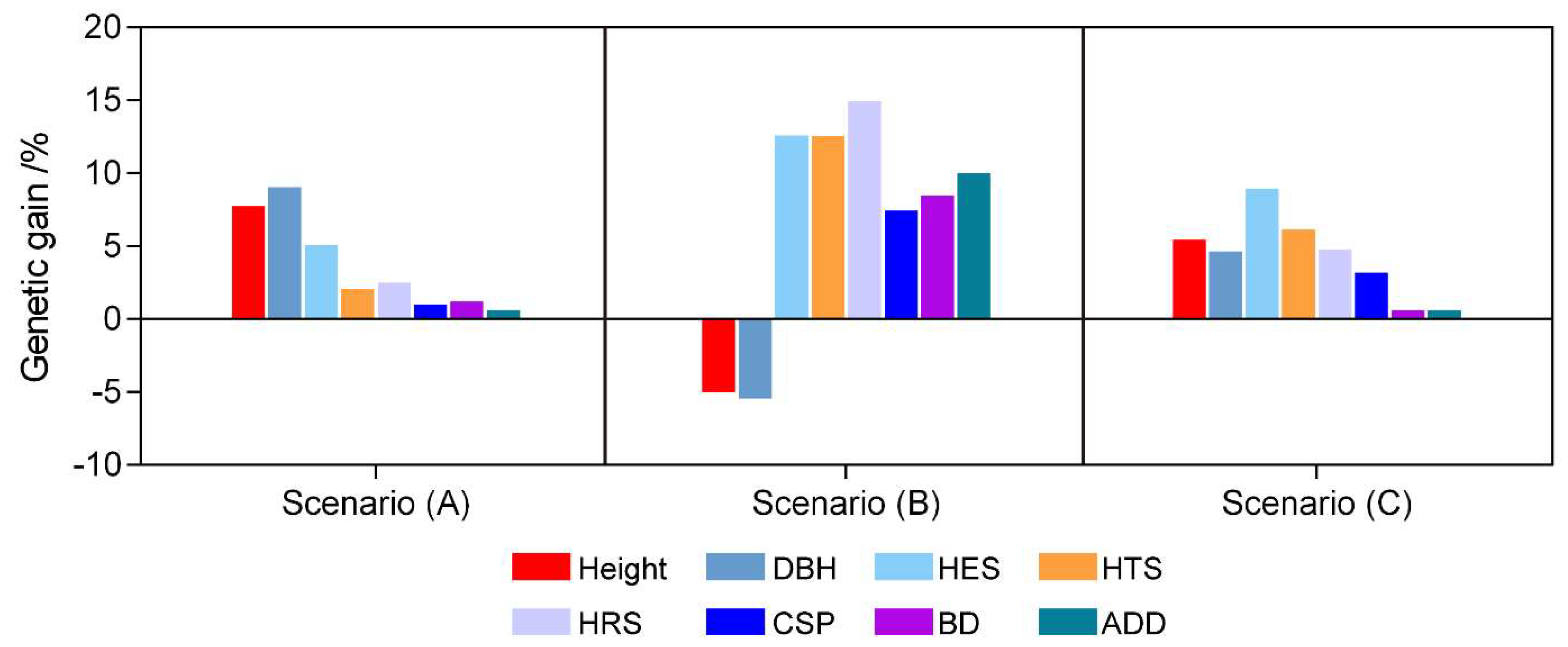
3.5. Genetic Gains under Different Selection Scenarios
4. Discussion
4.1. Genetic Variation and Genetic Parameters
4.2. Genetic Correlation between Growth Traits and Wood Properties
4.3. Different Selection Scenario for C. bungei Clones
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADD | Air-dry density |
| AWD | Area-weighted average wood density of the entire disk at breast height |
| BD | Basic density |
| CSP | Compressive strength parallel to the wood grain |
| DBH | Diameter at breast height |
| HES | Hardness of transverse section |
| HRS | Hardness of radial section |
| HTS | Hardness of tangential section |
| MFV | Membership function value method |
| MOEtof | Modulus of elasticity using time-off-light |
| PCA | Principal component analysis |
| PVC | Phenotypic variation coefficient |
| RD5 | Average wood density of outer most five rings at breast height |
References
- Ma, W.J.; Zhang, S.G.; Wang, J.H.; Zhai, W.J.; Cui, Y.Z.; Wang, Q.X. Timber Physical and Mechanical Properties of New Catalpa bungei Clones. Sci. Silvae Sin. 2013, 49, 126–134. [Google Scholar]
- Wei, X.M.; Borralho, N.M.G. Objectives and Selection Criteria for Pulp Production of Eucalyptus urophylla Plantations in South East China. For. Genet. 1999, 6, 181–190. [Google Scholar]
- Antony, F.; Schimleck, L.R.; Jordan, L.; Hornsby, B.; Dahlen, J.; Daniels, R.F.; Clark, A.; Apiolaza, L.A.; Huber, D. Growth and Wood Properties of Genetically Improved Loblolly Pine: Propagation Type Comparison and Genetic Parameters. Can. J. For. Res. 2014, 44, 263–272. [Google Scholar] [CrossRef]
- Hannrup, B.; Cahalan, C.; Chantre, G.; Grabner, M.; Karlsson, B.; Bayon, I.L.; Jones, G.L.; Müller, U.; Pereira, H.; Rodrigues, J.C.; et al. Genetic Parameters of Growth and Wood Quality Traits in Picea abies. Scand. J. For. Res. 2004, 19, 14–29. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, Q.H.; Tian, Y.G.; Yang, S.C.; Wang, H.W.; Wang, L.K.; Li, Y.L.; Zhang, P.; Zhao, X.Y. Comprehensive Assessment of Growth Traits and Wood Properties in half-sib Pinus koraiensis families. Euphytica 2018, 214, 202. [Google Scholar] [CrossRef]
- Ma, W.; Zhang, S.; Wang, J.; Zhang, J.; Zhao, K. Growth Traits of One-Year-Old Catalpa bungei Clones in Seedling Stage. For. Res. 2012, 25, 657–663. [Google Scholar]
- Xiao, Y.; Ma, W.J.; Lu, N.; Wang, Z.; Wang, N.; Zhai, W.J.; Kong, L.S.; Qu, G.Z.; Wang, Q.X.; Wang, J.H. Genetic Variation of Growth Traits and Genotype-by-Environment Interactions in Clones of Catalpa bungei and Catalpa fargesii f. duclouxii. Forests 2019, 10, 57. [Google Scholar] [CrossRef] [Green Version]
- Zhai, W.J.; Ma, W.J.; Wang, Q.X.; Wang, P.; Wang, J.H. Combined Selection of Excellent Families and Plus Trees of Catalpa bungei at Nursery Stage. J. Northwest For. Univ. 2012, 27, 68–71. [Google Scholar]
- Fukatsu, E.; Hiraoka, Y.; Matsunaga, K.; Tsubomura, M.; Nakada, R. Genetic Relationship Between Wood Properties and Growth Traits in Larix kaempferi Obtained from a Diallel Mating Test. J. Wood Sci. 2015, 61, 10–18. [Google Scholar] [CrossRef]
- Rozenberg, P.; Cahalan, C. Spruce and Wood Quality: Genetic Aspects (A review). Silvae Genet. 1997, 46, 72–82. [Google Scholar]
- Zhang, S.Y.; Yu, Q.B.; Chauret, G.; Koubaa, A. Selection for Both Growth and Wood Properties in Hybrid Poplar Clones. For. Sci. 2003, 49, 901–908. [Google Scholar]
- Burdon, R.D.; Paul, K.R.; Walker, J.C.F.; Megraw, R.A.; Robert, E.; Cown, D.J. Juvenile Versus Mature Wood: A New Concept, Orthogonal to Corewood Versus Outerwood, with Special Reference to Pinus radiata and P. taeda. For. Sci. 2004, 50, 399–415. [Google Scholar]
- Zhou, H.; Anders, F.; Wu, H.X. High Negative Genetic Correlations Between Growth Traits and Wood Properties Suggest Incorporating Multiple Traits Selection Including Economic Weights for The Future Scots pine Breeding Programs. Ann. For. Sci. 2014, 71, 463–472. [Google Scholar]
- Li, C.R.; Weng, Q.J.; Chen, J.B.; Li, M.; Zhou, C.P.; Chen, S.K.; Zhou, W.; Guo, D.Q.; Lu, C.X.; Chen, J.C.; et al. Genetic Parameters for Growth and Wood Mechanical Properties in Eucalyptus cloeziana F. Muell. New For. 2016, 48, 33–49. [Google Scholar] [CrossRef]
- Apiolaza, L.A.; Raymond, C.A.; Yeo, B.J. Genetic Variation of Physical and Chemical Wood Properties. Silvae Genet. 2005, 54, 4–5. [Google Scholar]
- Cotterill, P.P.; Dean, C.A. Successful Tree Breeding with Index Selection; CSIRO Publications: Clayton, Australia, 1990. [Google Scholar]
- Hayatgheibi, H.; Fries, A.; Kroon, J.; Wu, H.X. Estimation of Genetic Parameters, Provenance Performances, and Genotype by Environment Interactions for Growth and Stiffness in Lodgepole Pine (Pinus contorta). Scand. J. For. Res. 2018, 34, 1–11. [Google Scholar] [CrossRef]
- Li, L.; Wu, H.X. Efficiency of Early Selection for Rotation-aged Growth and Wood Density Traits in Pinus radiata. Can. J. For. Res. 2005, 35, 2019–2029. [Google Scholar] [CrossRef]
- Hayatgheibi, H.; Fries, A.; Kroon, J.; Wu, H.X. Genetic Analysis of Fiber-dimension Traits and Combined Selection for Simultaneous Improvement of Growth and Stiffness in Lodgepole Pine (Pinus contorta). Can. J. For. Res. 2019, 49, 500–509. [Google Scholar] [CrossRef]
- GB-T 1941–1991, Method of Testing in Hardness of Wood. Beijing, China. 1992. Available online: http://openstd.samr.gov.cn/bzgk/gb/newGbInfo?hcno=AABA7A552DBD6D91817E79D3F4CEE3C2 (accessed on 8 May 2021).
- Cheng, J.Q. Science of Wood, 2nd ed.; China Forestry Publishing House: Beijing, China, 1985. [Google Scholar]
- GB-T 1935–1991, Method of Testing in Compressive Strength Parallel to Grain of Wood. Beijing, China. 1992. Available online: https://www.codeofchina.com/standard/GBT1935-1991.html (accessed on 8 May 2021).
- Gilmour, A.R.; Gogel, B.; Cullis, B.; Thompson, R.; Butler, D. ASReml User Guide Release 3.0; VSN International Ltd.: Hemel Hempstead, UK, 2009. [Google Scholar]
- Chen, X.; Min, D.; Yasir, T.A.; Hu, Y.-G. Evaluation of 14 Morphological, Yield-related and Physiological Traits as Indicators of Drought Tolerance in Chinese Winter Bread Wheat Revealed by Analysis of the Membership Function Value of Drought Tolerance (MFVD). Field Crops Res. 2012, 137, 195–201. [Google Scholar] [CrossRef]
- Agrawal, A.; Chiu, A.M.; Le, M.; Halperin, E.; Sankararaman, S. Scalable Probabilistic PCA for Large-scale Genetic Variation Data. PLoS Genet. 2020, 16, e1008773. [Google Scholar] [CrossRef]
- Isik, F.; Li, B.; Frampton, J.; Goldfarb, B. Efficiency of Seedlings and Rooted Cuttings for Testing and Selection in Pinus taeda. For. Sci. 2004, 50, 44–53. [Google Scholar]
- Osorio, L.F.; White, T.L.; Huber, D.A. Age–age and Trait–trait Correlations for Eucalyptus grandis Hill ex Maiden and Their Implications for Optimal Selection Age and Design of Clonal Ttrials. Theor. Appl. Genet. 2003, 106, 735–743. [Google Scholar] [CrossRef]
- Liu, Y.M.; Zhou, L.; Zhu, Y.Q.; Liu, S.Q. Anatomical Features and Its Radial Variations among Different Catalpa bungei Clones. Forests 2020, 11, 824. [Google Scholar] [CrossRef]
- Wu, S.J.; Xu, J.M.; Li, G.Y.; Risto, V.; Du, Z.H.; Lu, Z.H.; Li, B.Q.; Wang, W. Genotypic Variation in Wood Properties and Growth Traits of Eucalyptus Hybrid Clones in Southern China. New For. 2011, 42, 35–50. [Google Scholar] [CrossRef]
- Missanjo, E.; Matsumura, J. Genetic Improvement of Wood Properties in Pinus kesiya Royle ex Gordon for Sawn Timber Production in Malawi. Forests 2016, 7, 253. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.L.; Shi, Y.; Fang, L.Q.; Liu, X.E.; Ji, C.J. Patterns and Determinants of Wood Physical and Mechanical Properties across Major Tree Species in China. Sci. China Life Sci. 2015, 58, 602–612. [Google Scholar] [CrossRef]
- Fujimoto, T.; Akutsu, H.; Nei, M.; Kita, K.; Kuromaru, M.; Oda, K. Genetic Variation in Wood Stiffness and Strength Properties of Hybrid Larch (Larix gmelinii var. japonica × L. kaempferi). J. For. Res. 2017, 11, 343–349. [Google Scholar] [CrossRef]
- Steffenrem, A.; Kvaalen, H.; H⊘ib⊘, O.A.; Edvardsen, Ø.M.; Skr⊘ppa, T. Genetic Variation of Wood Quality Traits and Relationships with Growth in Picea abies. Scand. J. For. Res. 2009, 24, 15–27. [Google Scholar] [CrossRef]
- Gapare, W.J.; Baltunis, B.S.; Ivković, M.; Wu, H.X. Genetic Correlations among Juvenile Wood Quality and Growth Traits and Implications for Selection Strategy in Pinus radiata D. Don. Ann. For. Sci. 2009, 66, 606. [Google Scholar] [CrossRef] [Green Version]
- Hayatgheibi, H.; Anders, F.; Kroon, J.; Wu, H.X. Genetic Analysis of Lodgepole Pine (Pinus contorta) Solid-wood Quality Traits. Can. J. For. Res. 2017, 47, 1303–1313. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.L.; Aitken, S.N.; Philippe, R.; Carlson, M.R. Selection for Height Growth and Pilodyn Pin Penetration in Lodgepole Pine: Effects on Growth Traits, Wood Properties, and Their Relationships. Can. J. For. Res. 1999, 29, 434–445. [Google Scholar] [CrossRef]
- Yang, H.Y.; Weng, Q.J.; Li, F.; Zhou, C.P.; Li, M.; Chen, S.K.; Ji, H.X.; Gan, S.M. Genotypic Variation and Genotype-by-Environment Interactions in Growth and Wood Properties in a Cloned Eucalyptus urophylla × E. tereticornis Family in Southern China. For. Sci. 2018, 64, 225–232. [Google Scholar] [CrossRef]
- Bradbury, G.J.; Potts, B.M.; Beadle, C.L. Genetic and Environmental Variation in Wood Properties of Acacia melanoxylon. Ann. For. Sci. 2011, 68, 1363–1373. [Google Scholar] [CrossRef] [Green Version]
- Baltunis, B.S.; Gapare, W.J.; Wu, H.X. Genetic Parameters and Genotype by Environment Interaction in Radiata Pine for Growth and Wood Quality Traits in Australia. Silvae Genet. 2010, 59, 2–3. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.X.; Ying, C.C. Genetic Parameters and Selection Efficiencies in Resistance to Western Gall Rust, Stalactiform Blister Rust, Needle Cast, and Sequoia Pitch Moth in Lodgepole Pine. For. Sci. 1997, 43, 571–581. [Google Scholar]

| No. | Clone | Parental Combination | Origin of Clone | Style of Propagation |
|---|---|---|---|---|
| 1 | 1-1 | 6-8 × CGQ | Hybrid | Cutting |
| 2 | 1-2 | 6-8 × CGQ | Hybrid | Cutting |
| 3 | 1-3 | 6-8 × CGQ | Hybrid | Cutting |
| 4 | 1-4 | 6-8 × CGQ | Hybrid | Cutting |
| 5 | 2-1 | 6-7 × CGQ | Hybrid | Cutting |
| 6 | 2-2 | 6-7 × CGQ | Hybrid | Cutting |
| 7 | 2-6 | 6-7 × CGQ | Hybrid | Cutting |
| 8 | 2-8 | 6-7 × CGQ | Hybrid | Cutting |
| 9 | 001-1 | 8-5 × CGQ | Hybrid | Cutting |
| 10 | 002-1 | 6-8 × CGQ | Hybrid | Cutting |
| 11 | 008-1 | 7-8 × 10-1 | Hybrid | Cutting |
| 12 | 013-1 | \ | Hybrid | Cutting |
| 13 | 015-1 | 7-8 × 6-7 | Hybrid | Cutting |
| 14 | 9-1 | 8-5 × 6-7 | Hybrid | Cutting |
| 15 | 9-2 | 8-5 × 6-7 | Hybrid | Cutting |
| 16 | 7080 | \ | Excellent natural individual | Cutting |
| 17 | 8401 | \ | Excellent natural individual | Cutting |
| 18 | H3 | \ | Excellent natural individual | Cutting |
| 19 | DY | \ | Excellent natural individual | Cutting |
| Traits | Block | Clone | Clone × Block | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Df | MS | F-Value | p-Value | Df | MS | F-Value | p-Value | Df | MS | F-Value | p-Value | |
| Height | 3 | 4.011 | 18.948 | <0.0001 | 18 | 3.951 | 18.664 | <0.0001 | 53 | 0.943 | 4.455 | <0.0001 |
| DBH | 3 | 26.107 | 32.520 | <0.0001 | 18 | 8.544 | 10.643 | <0.0001 | 53 | 2.158 | 2.689 | <0.0001 |
| Traits | Clone | Individual | ||||||
|---|---|---|---|---|---|---|---|---|
| Df | MS | F-Value | p-Value | Df | MS | F-Value | p-Value | |
| HES | 18 | 7,532,606.00 | 4.73 | <0.0001 | 37 | 1,594,249.00 | 23.66 | <0.0001 |
| HTS | 18 | 7,286,269.00 | 4.36 | <0.0001 | 37 | 1,672,308.00 | 23.57 | <0.0001 |
| HRS | 18 | 7,067,767.00 | 2.76 | 0.0044 | 37 | 2,561,230.00 | 41.87 | <0.0001 |
| CSP | 18 | 977.30 | 6.69 | <0.0001 | 37 | 146.10 | 19.15 | <0.0001 |
| BD | 18 | 0.09 | 9.23 | <0.0001 | 38 | 0.01 | 24.47 | <0.0001 |
| ADD | 18 | 0.15 | 9.88 | <0.0001 | 38 | 0.02 | 23.82 | <0.0001 |
| No. | Clone | Height/m | DBH/cm | HES/N | HTS/N | HRS/N | CSP/Mpa | BD/g·cm3 | ADD/g·cm3 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1-1 | 8.24 (0.70) | 11.33 (1.30) | 3407.13 (243.39) | 2991.43 (301.07) | 2331.35 (304.70) | 40.31 (2.81) | 0.40 (0.02) | 0.48 (0.02) |
| 2 | 1-2 | 7.81 (1.11) | 8.99 (0.94) | 3051.34 (285.24) | 3069.31 (415.87) | 2452.78 (309.65) | 42.77 (3.41) | 0.42 (0.03) | 0.50 (0.03) |
| 3 | 1-3 | 7.23 (0.84) | 9.17 (1.22) | 2754.22 (455.92) | 2316.38 (273.34) | 1933.20 (153.20) | 36.29 (2.42) | 0.35 (0.02) | 0.42 (0.03) |
| 4 | 1-4 | 9.46 (0.12) | 11.47 (2.52) | 3070.44 (225.58) | 2622.69 (272.88) | 2214.44 (273.42) | 39.54 (2.57) | 0.40 (0.03) | 0.47 (0.03) |
| 5 | 2-1 | 8.33 (0.37) | 10.07 (1.31) | 3681.39 (251.33) | 3102.43 (495.89) | 2641.26 (521.04) | 40.41 (2.34) | 0.40 (0.01) | 0.48 (0.02) |
| 6 | 2-2 | 8.39 (0.94) | 10.40 (0.97) | 3453.06 (295.46) | 3113.52 (319.32) | 2243.71 (459.89) | 43.57 (4.16) | 0.39 (0.01) | 0.47 (0.02) |
| 7 | 2-6 | 7.38 (1.42) | 9.49 (2.40) | 3371.86 (308.93) | 2671.43 (298.35) | 2318.53 (304.20) | 36.32 (2.48) | 0.37 (0.03) | 0.44 (0.03) |
| 8 | 2-8 | 8.58 (0.59) | 10.66 (0.65) | 3480.83 (448.58) | 2905.65 (227.99) | 2266.66 (528.99) | 39.46 (2.27) | 0.39 (0.03) | 0.47 (0.03) |
| 9 | 001-1 | 8.18 (0.60) | 10.89 (0.95) | 3110.21 (520.27) | 2195.26 (275.43) | 1815.62 (487.58) | 33.84 (4.48) | 0.35 (0.02) | 0.41 (0.03) |
| 10 | 002-1 | 7.34 (1.11) | 8.34 (0.95) | 4158.87 (207.47) | 3625.41 (549.93) | 3214.43 (539.30) | 46.00 (3.62) | 0.48 (0.02) | 0.59 (0.02) |
| 11 | 008-1 | 8.90 (0.19) | 11.39 (1.47) | 4200.94 (288.84) | 2989.05 (274.36) | 2587.08 (260.32) | 44.11 (3.54) | 0.41 (0.02) | 0.50 (0.03) |
| 12 | 013-1 | 8.26 (0.42) | 10.48 (1.03) | 2929.75 (519.98) | 2401.46 (610.65) | 1908.87 (547.79) | 40.14 (2.64) | 0.38 (0.03) | 0.45 (0.04) |
| 13 | 015-1 | 8.05 (0.72) | 10.05 (0.94) | 2934.16 (352.56) | 2694.13 (328.22) | 2300.77 (343.43) | 43.41 (3.50) | 0.41 (0.03) | 0.49 (0.04) |
| 14 | 9-1 | 8.68 (0.43) | 10.62 (0.83) | 3584.20 (261.50) | 3068.26 (260.01) | 2699.06 (331.34) | 40.93 (2.39) | 0.40 (0.02) | 0.47 (0.02) |
| 15 | 9-2 | 7.95 (0.68) | 10.70 (1.19) | 3315.36 (393.89) | 2835.67 (147.73) | 2115.97 (353.41) | 39.36 (4.09) | 0.36 (0.04) | 0.44 (0.05) |
| 16 | 7080 | 7.20 (0.77) | 9.54 (1.38) | 3770.25 (431.34) | 3530.58 (442.09) | 3150.36 (475.47) | 45.69 (4.48) | 0.45 (0.04) | 0.54 (0.05) |
| 17 | 01 | 7.69 (0.50) | 9.31 (0.94) | 2933.40 (253.03) | 2380.76 (345.48) | 2116.11 (372.63) | 42.78 (2.76) | 0.42 (0.02) | 0.49 (0.03) |
| 18 | H3 | 7.93 (0.60) | 9.31 (1.59) | 2999.67 (366.58) | 2374.22 (328.37) | 1900.33 (343.01) | 37.71 (3.15) | 0.37 (0.02) | 0.44 (0.02) |
| 19 | DY | 6.45 (1.13) | 7.86 (1.82) | 3413.84 (536.56) | 3084.87 (539.89) | 2733.25 (584.45) | 41.36 (2.97) | 0.40 (0.03) | 0.49 (0.03) |
| Mean | 8.00 | 10.01 | 3348.47 | 2840.66 | 2365.46 | 40.74 | 0.40 | 0.48 | |
| Minimum | 6.45 | 7.86 | 2754.22 | 2195.26 | 1815.62 | 33.84 | 0.35 | 0.41 | |
| Maximum | 9.46 | 11.47 | 4200.94 | 3625.41 | 3214.43 | 46.00 | 0.48 | 0.59 | |
| Phenotypic variation coefficient | 8.73% | 10.23% | 12.16% | 14.03% | 16.70% | 7.95% | 8.21% | 8.63% | |
| Repeatability | 0.76 (0.09) | 0.76 (0.09) | 0.79 (22.36) | 0.78 (25.27) | 0.66 (15.91) | 0.84 (0.06) | 0.89 (0.05) | 0.89 (0.04) | |
| Scenario (A) | Scenario (B) | ||||
|---|---|---|---|---|---|
| Rank | Clone | Average Membership Function Value | Rank | Clone | Average Membership Function Value |
| 1 | 1-4 | 1.000 | 1 | 002-1 | 0.995 |
| 2 | 008-1 | 0.896 | 2 | 7080 | 0.841 |
| 3 | 1-1 | 0.778 | 3 | 008-1 | 0.646 |
| 4 | 9-1 | 0.753 | 4 | DY | 0.533 |
| 5 | 2-8 | 0.742 | 5 | 2-1 | 0.531 |
| 6 | 001-1 | 0.707 | 6 | 9-1 | 0.520 |
| 7 | 2-2 | 0.674 | 7 | 1-2 | 0.511 |
| 8 | 013-1 | 0.664 | 8 | 2-2 | 0.487 |
| 9 | 9-2 | 0.643 | 9 | 1-1 | 0.439 |
| 10 | 2-1 | 0.618 | 10 | 015-1 | 0.415 |
| 11 | 015-1 | 0.569 | 11 | 2-8 | 0.408 |
| 12 | H3 | 0.447 | 12 | 8401 | 0.359 |
| 13 | 8401 | 0.407 | 13 | 1-4 | 0.326 |
| 14 | 1-2 | 0.382 | 14 | 9-2 | 0.296 |
| 15 | 2-6 | 0.380 | 15 | 2-6 | 0.273 |
| 16 | 7080 | 0.357 | 16 | 013-1 | 0.213 |
| 17 | 1-3 | 0.311 | 17 | H3 | 0.159 |
| 18 | 002-1 | 0.214 | 18 | 1-3 | 0.078 |
| 19 | DY | 0.000 | 19 | 001-1 | 0.041 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, Y.; Wang, J.; Yun, H.; Yang, G.; Ma, J.; Ma, W.; Qu, G. Genetic Evaluation and Combined Selection for the Simultaneous Improvement of Growth and Wood Properties in Catalpa bungei Clones. Forests 2021, 12, 868. https://doi.org/10.3390/f12070868
Xiao Y, Wang J, Yun H, Yang G, Ma J, Ma W, Qu G. Genetic Evaluation and Combined Selection for the Simultaneous Improvement of Growth and Wood Properties in Catalpa bungei Clones. Forests. 2021; 12(7):868. https://doi.org/10.3390/f12070868
Chicago/Turabian StyleXiao, Yao, Junhui Wang, Huiling Yun, Guijuan Yang, Jianwei Ma, Wenjun Ma, and Guanzheng Qu. 2021. "Genetic Evaluation and Combined Selection for the Simultaneous Improvement of Growth and Wood Properties in Catalpa bungei Clones" Forests 12, no. 7: 868. https://doi.org/10.3390/f12070868
APA StyleXiao, Y., Wang, J., Yun, H., Yang, G., Ma, J., Ma, W., & Qu, G. (2021). Genetic Evaluation and Combined Selection for the Simultaneous Improvement of Growth and Wood Properties in Catalpa bungei Clones. Forests, 12(7), 868. https://doi.org/10.3390/f12070868






