Rapid Detection of Pine Pathogens Lecanosticta acicola, Dothistroma pini and D. septosporum on Needles by Probe-Based LAMP Assays
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. DNA Extraction
2.3. LAMP Primers and Probes Design
2.4. LAMP Reactions
2.5. Detection on Naturally Infected Pine Needle Samples
3. Results
3.1. LAMP Specificity and Sensitivity
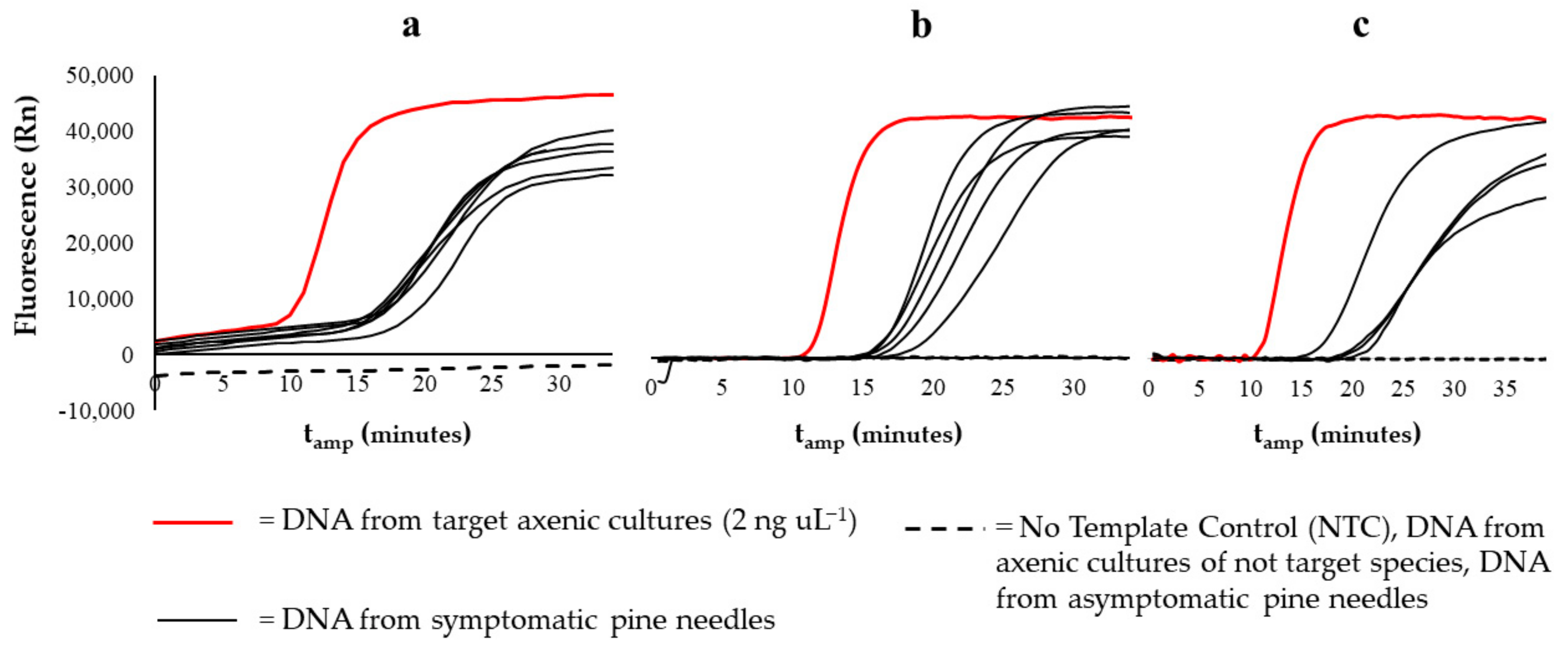
3.2. Detection on Naturally Infected Pine Needles Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pehl, L.; Cech, T.L.; Ioos, R. Lecanosticta acicola (formerly Mycosphaerella dearnessii), Dothistroma septosporum (formerly Mycosphaerella pini) and Dothistroma pini. EPPO Bull. 2015, 45, 163–182. [Google Scholar]
- Gibson, I.A.S. Dothistroma Blight of Pinus radiata. Annu. Rev. Phytopathol. 1972, 10, 51–72. [Google Scholar] [CrossRef]
- De Urbina, E.O.; Mesanza, N.; Aragonés, A.; Raposo, R.; Elvira-Recuenco, M.; Boqué, R.; Patten, C.; Aitken, J.; Iturritxa, E. Emerging Needle Blight diseases in Atlantic Pinus ecosystems of Spain. Forests 2016, 8, 18. [Google Scholar] [CrossRef]
- Ivory, M. Resistance to Dothistroma needle blight induced in Pinus radiata by maturity and shade. Trans. Br. Mycol. Soc. 1972, 59, 205–212. [Google Scholar] [CrossRef]
- Rodas, C.A.; Wingfield, M.J.; Granados, G.M.; Barnes, I. Dothistroma needle blight: An emerging epidemic caused by Dothistroma septosporum in Colombia. Plant Pathol. 2016, 65, 53–63. [Google Scholar] [CrossRef]
- EPPO Lecanosticta acicola (SCIRAC), Categorization. EPPO Global Database. 2021. Available online: https://gd.eppo.int/taxon/SCIRAC/categorization (accessed on 18 February 2021).
- EPPO Dothistroma septosporum (SCIRPI), Categorizasion. EPPO Global Database. 2021. Available online: https://gd.eppo.int/taxon/SCIRPI/categorization (accessed on 18 February 2021).
- EPPO Dothistroma pini (DOTSPI), Categorization. EPPO Global Database. 2021. Available online: https://gd.eppo.int/taxon/DOTSPI/categorization (accessed on 18 February 2021).
- European Parliament; Council of the European Union. EU Regulation 2016/2031 of the European Parliament and the Council on protective measures against pests of plants. Off. J. EU 2016, 317, 4–104. Available online: http://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:32016R2031 (accessed on 8 March 2021).
- European Commission, Directorate-General for Health and Food Safety. Commission Implementing Regulation (EU) 2019/2072 of 28 November 2019 establishing uniform conditions for the implementation of Regulation (EU) 2016/2031 of the European Parliament and the Council, as regards protective measures against pests of plants, and repealing Commission Regulation (EC) No 690/2008 and amending Commission Implementing Regulation (EU) 2018/2019. Off. J. EU 2019, 319, 1. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A32019R2072 (accessed on 8 March 2021).
- Drenkhan, R.; Tomešová-Haataja, V.; Fraser, S.; Bradshaw, R.E.; Vahalík, P.; Mullett, M.S.; Martín-García, J.; Bulman, L.S.; Wingfield, M.J.; Kirisits, T.; et al. Global geographic distribution and host range of Dothistroma species: A comprehensive review. For. Pathol. 2016, 46, 408–442. [Google Scholar] [CrossRef]
- Siggers, P.V. The brown-spot needle blight of longleaf pine seedlings. J. Forest. 1932, 30, 579–593. [Google Scholar] [CrossRef]
- Martinez, J.B. The mycoses of Pinus insignis in Guipúzcoa. Publ. Inst. For. Investig. Exp. 1942, 13, 1–72. [Google Scholar]
- Van Der Nest, A.; Wingfield, M.J.; Janoušek, J.; Barnes, I. Lecanosticta acicola: A growing threat to expanding global pine forests and plantations. Mol. Plant. Pathol. 2019, 20, 1327–1364. [Google Scholar] [CrossRef]
- Markovskaja, S.; Raitelaitytė, K.; Kačergius, A.; Kolmakov, P.; Vasilevich, V. Occurrence of Dothistroma needle blight in Lithuania and Belarus: The risk posed to native Scots Pine forests. For. Pathol. 2020, 50, e12626. [Google Scholar] [CrossRef]
- Mullett, M.S.; Adamson, K.; Bragança, H.; Bulgakov, T.S.; Georgieva, M.; Henriques, J.; Jürisoo, L.; Laas, M.; Drenkhan, R. New country and regional records of the pine needle blight pathogens Lecanosticta acicola, Dothistroma septosporum and Dothistroma pini. For. Pathol. 2018, 48, e12440. [Google Scholar] [CrossRef]
- Piškur, B.; Hauptman, T.; Jurc, D. Dothistroma Needle Blight in Slovenia is caused by two cryptic species: Dothistroma pini and Dothistroma septosporum. For. Pathol. 2013, 43, 518–521. [Google Scholar] [CrossRef]
- Piou, D.; Ioos, R. First report of Dothistroma pini, a recent agent of the Dothistroma Needle Blight, on Pinus radiata in France. Plant. Dis. 2014, 98, 841. [Google Scholar] [CrossRef]
- Ghelardini, L.; Aglietti, C.; Loria, F.; Cerboneschi, M.; Gionni, A.; Maresi, G.; Moricca, S.; Marchi, G. Dothistroma needle blight in protected pine forests in Italy. Manag. Biol. Invasions 2020, 11, 689–702. [Google Scholar] [CrossRef]
- Oskay, F.; Laas, M.; Mullett, M.; Lehtijärvi, A.; Doğmuş-Lehtijärvi, H.T.; Woodward, S.; Drenkhan, R. First report of Lecanosticta acicola on pine and non-pine hosts in Turkey. For. Pathol. 2020, 50, 12654. [Google Scholar] [CrossRef]
- Mesanza, N.; Raposo, R.; Elvira-Recuenco, M.; Barnes, I.; van der Nest, A.; Hernández, M.; Pascual, M.T.; Barrena, I.; Martín, U.S.; Cantero, A.; et al. New hosts for Lecanosticta acicola and Dothistroma septosporum in newly established arboreta in Spain. For. Pathol. 2021, 51. [Google Scholar] [CrossRef]
- Drenkhan, R.; Adamson, K.; Jürimaa, K.; Hanso, M. Dothistroma septosporum on firs (Abies spp.) in the northern Baltics. For. Pathol. 2014, 44, 250–254. [Google Scholar] [CrossRef]
- Lang, V.K.J. Dothistroma pini an jungen Fichten (Picea abies). For. Pathol. 1987, 17, 316–317. [Google Scholar] [CrossRef]
- Mullett, M.; Fraser, S. Infection of Cedrus species by Dothistroma septosporum. For. Pathol. 2015, 46, 551–554. [Google Scholar] [CrossRef]
- Woods, A.; Coates, K.D.; Hamann, A. Is an unprecedented Dothistroma needle blight epidemic related to climate change? Bioscience 2005, 55, 761–769. [Google Scholar] [CrossRef]
- Watt, M.S.; Kriticos, D.J.; Alcaraz, S.; Brown, A.V.; Leriche, A. The hosts and potential geographic range of Dothistroma needle blight. For. Ecol. Manag. 2009, 257, 1505–1519. [Google Scholar] [CrossRef]
- Woods, A.J.; Martín-García, J.; Bulman, L.; Vasconcelos, M.W.; Boberg, J.; La Porta, N.; Peredo, H.; Vergara, G.; Ahumada, R.; Brown, A.; et al. Dothistroma needle blight, weather and possible climatic triggers for the disease’s recent emergence. For. Pathol. 2016, 46, 443–452. [Google Scholar] [CrossRef]
- Barnes, I.; Wingfield, M.J.; Carbone, I.; Kirisits, T.; Wingfield, B.D. Population structure and diversity of an invasive pine needle pathogen reflects anthropogenic activity. Ecol. Evol. 2014, 4, 3642–3661. [Google Scholar] [CrossRef]
- Bulman, L.S.; Bradshaw, R.E.; Fraser, S.D.; Martín-García, J.; Barnes, I.; Musolin, D.L.; La Porta, N.; Woods, A.J.; Diez-Casero, J.; Koltay, A.; et al. A worldwide perspective on the management and control of Dothistroma needle blight. For. Pathol. 2016, 46, 472–488. [Google Scholar] [CrossRef]
- Capron, A.; Feau, N.; Heinzelmann, R.; Barnes, M.I.; Benowicz, A.; Bradshaw, R.E.; Dale, A.L.; Lewis, K.J.; Owen, T.J.; Reich, R.; et al. Signatures of post-glacial genetic isolation and human-driven migration in the Dothistroma needle Blight pathogen in Western Canada. Phytopathology 2021, 111, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Mullett, M.; Drenkhan, R.; Adamson, K.; Boroń, P.; Lenart-Boroń, A.; Barnes, I.; Tomšovský, M.; Jánošíková, Z.; Adamčíková, K.; Ondrušková, E.; et al. Worldwide genetic structure elucidates the Eurasian origin and invasion pathways of Dothistroma septosporum, causal agent of Dothistroma Needle Blight. J. Fungi 2021, 7, 111. [Google Scholar] [CrossRef]
- Dale, A.L.; Lewis, K.J.; Murray, B.W. Sexual reproduction and gene flow in the pine pathogen Dothistroma septosporum in British Columbia. Phytopathology 2011, 101, 68–76. [Google Scholar] [CrossRef]
- Sadiković, D.; Piškur, B.; Barnes, I.; Hauptman, T.; Diminić, D.; Wingfield, M.J.; Jurc, D. Genetic diversity of the pine pathogen Lecanosticta acicola in Slovenia and Croatia. Plant. Pathol. 2019, 68, 1120–1131. [Google Scholar] [CrossRef]
- Drenkhan, R.; Hantula, J.; Vuorinen, M.; Jankovský, L.; Müller, M.M. Genetic diversity of Dothistroma septosporum in Estonia, Finland and Czech Republic. Eur. J. Plant. Pathol. 2012, 136, 71–85. [Google Scholar] [CrossRef]
- Feau, N.; Ramsfield, T.D.; Myrholm, C.L.; Tomm, B.; Cerezke, H.F.; Benowicz, A.; Samis, E.; Romano, A.; Dale, A.L.; Capron, A.; et al. DNA-barcoding identification of Dothistroma septosporum on Pinus contorta var. latifolia, P. banksiana and their hybrid in northern Alberta, Canada. Can. J. Plant. Pathol. 2020, 1–8. [Google Scholar] [CrossRef]
- Bradshaw, R.E.; Sim, A.D.; Chettri, P.; Dupont, P.; Guo, Y.; Hunziker, L.; McDougal, R.L.; Van Der Nest, A.; Fourie, A.; Wheeler, D.; et al. Global population genomics of the forest pathogen Dothistroma septosporum reveal chromosome duplications in high dothistromin-producing strains. Mol. Plant. Pathol. 2019, 20, 784–799. [Google Scholar] [CrossRef]
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Lau, H.Y.; Botella, J.R. Advanced DNA-based point-of-care diagnostic methods for plant diseases detection. Front. Plant. Sci. 2017, 8, 2016. [Google Scholar] [CrossRef] [PubMed]
- Barnes, I.; Van Der Nest, A.; Mullett, M.S.; Crous, P.W.; Drenkhan, R.; Musolin, D.L.; Wingfield, M.J. Neotypification of Dothistroma septosporum and epitypification of D. pini, causal agents of Dothistroma needle blight of pine. For. Pathol. 2016, 46, 388–407. [Google Scholar] [CrossRef]
- Millberg, H.; Hopkins, A.J.M.; Boberg, J.; Davydenko, K.; Stenlid, J. Disease development of Dothistroma needle blight in seedlings of Pinus sylvestris and Pinus contorta under Nordic conditions. For. Pathol. 2015, 46, 515–521. [Google Scholar] [CrossRef]
- Moraal, L.G. Bionomics of Haematoloma dorsatum (Hom., Cercopidae) in relation to needle damage in pine forests. Anzeiger für Schädlingskunde Pflanzen und Umweltschutz 1996, 69, 114–118. [Google Scholar] [CrossRef]
- Covassi, M.; Roversi, P.F.; Toccafondi, P. Danni da Haematoloma dorsatum (Ahrens) su conifere (Homoptera, Cercopidae). I. Alterazioni macroscopiche degli apparati fogliari. [Damages caused by Haematoloma dorsatum (Ahrens) on conifers. I. Macro-scopical alterations of leaves]. J. Zool. 1989, 72, 259–275. [Google Scholar]
- Castro-Valderrama, U.; Romero-Nápoles, J.; Peck, D.C.; Valdez-Carrasco, J.M.; Llanderal-Cázares, C.; Bravo-Mojica, H.; Hernández-Rosas, F.; Cibrián-Llanderal, V.D. First report of spittlebug species (Hemiptera: Cercopidae) associated with Pinus species (Pinaceae) in Mexico. Fla. Èntomol. 2017, 100, 206–208. [Google Scholar] [CrossRef]
- Matsiakh, I.; Avtzis, D.N.; Adamson, K.; Augustin, S.; Beram, R.C.; Cech, T.; Drenkhan, R.; Kirichenko, N.; Maresi, G.; Morales-Rodríguez, C.; et al. Damage to foliage of coniferous woody plants. In Field Guide for the Identification of Damage on Woody Sentinel Plants; Roques, A., Cleary, M., Matsiakh, I., Eschen, R., Eds.; CABI Publishing: Wallingford, UK, 2017; pp. 167–188. [Google Scholar] [CrossRef]
- Groenewald, M.; Barnes, I.; Bradshaw, R.E.; Brown, A.V.; Dale, A.; Groenewald, J.Z.; Lewis, K.J.; Wingfield, B.D.; Wingfield, M.J.; Crous, P.W. Characterization and distribution of mating type genes in the Dothistroma needle blight pathogens. Phytopathology 2007, 97, 825–834. [Google Scholar] [CrossRef]
- Ioos, R.; Fabre, B.; Saurat, C.; Fourrier, C.; Frey, P.; Marçais, B. Development, comparison, and validation of real-time and conventional PCR tools for the detection of the fungal pathogens causing Brown Spot and Red Band Needle Blights of Pine. Phytopathology 2010, 100, 105–114. [Google Scholar] [CrossRef]
- Langrell, S.R.H. Nested polymerase chain reaction–based detection of Dothistroma septosporum, red band needle blight of pine, a tool in support of phytosanitary regimes. Mol. Ecol. Resour. 2011, 11, 749–752. [Google Scholar] [CrossRef] [PubMed]
- Janousek, J.; Krumböck, S.; Kirisits, T.; Bradshaw, R.E.; Barnes, I.; Jankovský, L.; Stauffer, C. Development of microsatellite and mating type markers for the pine needle pathogen Lecanosticta acicola. Australas. Plant. Pathol. 2014, 43, 161–165. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchai, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, 63. [Google Scholar] [CrossRef] [PubMed]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, H.; Kawana, T.; Fukushima, E.; Suzutani, T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 2007, 70, 499–501. [Google Scholar] [CrossRef] [PubMed]
- Mikita, K.; Maeda, T.; Yoshikawa, S.; Ono, T.; Miyahira, Y.; Kawana, A. The Direct Boil-LAMP method: A simple and rapid diagnostic method for cutaneous leishmaniasis. Parasitol. Int. 2014, 63, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, J.A.; Ostoja-Starzewska, S.; Webb, K.; Cole, J.A.; Barnes, A.V.; Dickinson, M.; Boonham, N. A loop-mediated isothermal amplification-based method for confirmation of Guignardia citricarpa in citrus black spot lesions. Eur. J. Plant. Pathol. 2013, 136, 217–224. [Google Scholar] [CrossRef]
- Hamilton, J.L.; Workman, J.N.; Nairn, C.J.; Fraedrich, S.W.; Villari, C. Rapid detection of Raffaelea lauricola directly from host plant and beetle vector tissues using loop-mediated isothermal amplification. Plant. Dis. 2020, 104, 3151–3158. [Google Scholar] [CrossRef]
- Njiru, Z.K. Loop-mediated isothermal amplification technology: Towards point of care diagnostics. PLoS Negl. Trop. Dis. 2012, 6, e1572. [Google Scholar] [CrossRef] [PubMed]
- Aglietti, C.; Luchi, N.; Pepori, A.L.; Bartolini, P.; Pecori, F.; Raio, A.; Capretti, P.; Santini, A. Real-time loop-mediated isothermal amplification: An early-warning tool for quarantine plant pathogen detection. AMB Express 2019, 9, 50. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Ye, J.; Zhang, L. The application prospect of loop-mediated isothermal amplification method in forest disease detection. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2012, 36, 135–139. [Google Scholar]
- Sillo, F.; Giordano, L.; Gonthier, P. Fast and specific detection of the invasive forest pathogen Heterobasidion irregulare through a Loop-mediated isothermal AMPlification (LAMP) assay. For. Pathol. 2017, 48, e12396. [Google Scholar] [CrossRef]
- Stehlíková, D.; Luchi, N.; Aglietti, C.; Pepori, A.L.; Diez, J.J.; Santini, A. Real-time loop-mediated isothermal amplification assay for rapid detection of Fusarium circinatum. Biotechniques 2020, 69, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Myrholm, C.; Tomm, B.; Heinzelmann, R.; Feau, N.; Hamelin, R.; McDougal, R.; Winkworth, R.; Ramsfield, T. Development of a rapid loop-mediated isothermal amplification assay for the detection of Dothistroma septosporum. Forests 2021, 12, 362. [Google Scholar] [CrossRef]
- Mullett, M.S.; Barnes, I. Dothistroma isolation and molecular identification methods. In COST ACTION FP1102 Determining Invasiveness and Risk of Dothistroma; Training School Detection and Diagnostics of Dothistroma: Brno, Czech Republic, 2012. Available online: https://www.forestresearch.gov.uk/documents/305/DIAROD_052012_Isolation_and_indentification_97fNCCI.pdf (accessed on 9 March 2021).
- Quaedvlieg, W.; Groenewald, J.; Yáñez-Morales, M.D.J.; Crous, P. DNA barcoding of Mycosphaerella species of quarantine importance to Europe. Pers. Mol. Phylogeny Evol. Fungi 2012, 29, 101–115. [Google Scholar] [CrossRef]
- Van der Nest, A.; Wingfield, M.J.; Ortiz, P.C.; Barnes, I. Biodiversity of Lecanosticta pine-needle blight pathogens suggests a Mesoamerican centre of origin. IMA Fungus 2019, 10, 1–28. [Google Scholar] [CrossRef]
- Kwok, S.; Kellogg, D.E.; McKinney, N.; Spasic, D.; Goda, L.; Levenson, C.; Sninsky, J.J. Effects of primer-template mismatches on the polymerase chain reaction: Human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 1990, 18, 999–1005. [Google Scholar] [CrossRef]
- Kubota, R.; Alvarez, A.M.; Su, W.W.; Jenkins, D.M. FRET-based assimilating probe for sequence-specific real-time monitoring of loop-mediated isothermal alification (LAMP). Biol. Eng. Trans. 2011, 4, 81–100. [Google Scholar] [CrossRef]
- Laas, M.; Adamson, K.; Drenkhan, R. A look into the genetic diversity of Lecanosticta acicola in northern Europe. Fungal Biol. 2019, 123, 773–782. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Plant Health (PLH). Scientific Opinion on the risk to plant health posed by Dothistroma septosporum (Dorog.) M. Morelet (Mycosphaerella pini E. Rostrup, syn. Scirrhia pini) and Dothistroma pini Hulbary to the EU territory with the identification and evaluation of risk reduct. EFSA J. 2013, 11, 3026. [Google Scholar] [CrossRef]
- Thiessen, L.D.; Keune, J.A.; Neill, T.M.; Turechek, W.W.; Grove, G.G.; Mahaffee, W.F. Development of a grower-conducted inoculum detection assay for management of grape powdery mildew. Plant. Pathol. 2016, 65, 238–249. [Google Scholar] [CrossRef]
| Fungal Species | Isolate ID (Mating Type) | Host | Locality | Collector/Collection | LAMP Detection Results | ||
|---|---|---|---|---|---|---|---|
| L. acicola Assay | D. pini Assay | D. septosporum Assay | |||||
| D. septosporum1 | DS 3212 (MAT2) | P. sylvestris | Võru County, Estonia | R. Drenkhan | - | - | + |
| D. septosporum1 | Ds 57 | P. contorta | Pärnu County, Estonia | R. Drenkhan | - | - | + |
| D. septosporum2 | DSEP_KC_19_Ne1_TAIGA_504 (MAT2) | P. contorta var. latifolia | British Columbia, Canada | R. Hamelin | - | - | + |
| D. septosporum2 | DSEP_CLG_22_TAIGA_601 (MAT1) | P. contorta var. latifolia | British Columbia, Canada | R. Hamelin | - | - | + |
| D. septosporum2 | DSEP_PGTIS_P3_P16_Ne2_TAIGA_460 (MAT1) | P. contorta var. latifolia | British Columbia, Canada | R. Hamelin | - | - | + |
| D. septosporum2 | DSEP_WC_27_Ne1_TAIGA_626 (MAT2) | P. contorta var. latifolia | British Columbia, Canada | R. Hamelin | - | - | + |
| D. septosporum2 | DSEP_FLNRO2_19M_Ne1_TAIGA_486 (MAT1) | P. contorta var. latifolia | British Columbia, Canada | R. Hamelin | - | - | + |
| D. septosporum2 | DSEP_SM_1_4_Ne1_TAIGA_484 (MAT2) | P. contorta var. latifolia | British Columbia, Canada | R. Hamelin | - | - | + |
| D. pini2 | CMW 10951 CBS 116487 | P. radiata | Michigan, USA | G. Adams | - | + | - |
| D. pini2 | CMW 37634 | P. cembra | North Dakota, USA | J. Walla | - | + | - |
| D. pini2 | CMW 37786 | P. nigra | Indiana, USA | J. Walla | - | + | - |
| D. pini2 | CMW 38037 | P. ponderosa | South Dakota, USA | J. Walla | - | + | - |
| D. pini2 | CMW 42947 | P. nigra subsp. pallasiana | Kherson, Ukraine | K. Davydenko | - | + | - |
| D. pini2 | CMW 43903 | P. nigra subsp. laricio | La Ferte Imbault, France | I. Barnes | - | + | - |
| D. pini2 | CMW 29366 | P. pallasiana | Tarasovsky, Russia | S.B. Timur | - | + | - |
| D. pini2 | CMW 37633 | P. ponderosa | North Dakota, USA | J. Walla | - | + | - |
| D. pini2 | CMW 41496 | P. nigra | France | I. Barnes | - | + | - |
| D. pini2 | CMW 50237 | Pinus sp. | Arkansas, USA | M.S. Mullett | - | + | - |
| D. pini2 | A10 | P. nigra | Ontario, Canada | S. McGowan | - | + | - |
| D. pini2 | A11 | P. nigra | Ontario, Canada | S. McGowan | - | + | - |
| D. pini2 | A12 | P. nigra | Ontario, Canada | S. McGowan | - | + | - |
| D. pini2 | A13 | P. nigra | Ontario, Canada | S. McGowan | - | + | - |
| D. pini2 | A14 | P. nigra | Ontario, Canada | S. McGowan | - | + | - |
| D. pini2 | A20 | P. nigra | Ontario, Canada | S. McGowan | - | + | - |
| D. pini1 | E18/63-6 | Pinus sp. | Slovenia | B. Piškur | - | + | - |
| L. acicola2 | CV2019013 | P. palustris | Georgia, USA | C. Villari | + | - | - |
| L. acicola1 | 8496 (MAT1) | P. sylvestris | Tartu County, Estonia | R. Drenkhan | + | - | - |
| L. acicola1 | B1599 (MAT1) | P. radiata | France | R. Ioos | + | - | - |
| L. acicola1 | B1569 (MAT11) | P. radiata | France | R. Ioos | + | - | - |
| L. acicola3 | CMW 45427 CBS 133791 | P. strobus | New Hampshire, USA | B. Ostrofsky | + | - | - |
| L. acicola3 | CMW 45428 CBS 322.33 | P. palustris | USA | P.V. Siggers | + | - | - |
| L. acicola3 | MX7 | P. halepensis | Nuevo León, Mexico | J.G. Marmolejo | + | - | - |
| L. brevispora3 | CMW 45424 CBS 133601 | Pinus sp. | Mexico | M. de Jesús Yáñez-Morales | - | - | - |
| L. brevispora3 | CMW 46502 | P. pseudostrobus | Chimaltenango, Guatemala | I. Barnes | - | - | - |
| L. gloeospora3 | CMW 42645 IMI 283812 | P. pseudostrobus | Nuevo León, Mexico | H.C. Evans | - | - | - |
| L. guatemalensis3 | CMW 42206 IMI 281598 | P. oocarpa | Guatemala | H.C. Evans | - | - | - |
| L. guatemalensis3 | CMW 43892 | P. oocarpa | Chiquimula, Guatemala | I. Barnes | - | - | - |
| L. jani3 | CMW 38958 CBS 144456 | P. oocarpa | Jalapa, Guatemala | I. Barnes | - | - | - |
| L. jani3 | CMW 48831 CBS 144447 | P. oocarpa | Alta Verapaz, Guatemala | I. Barnes | - | - | - |
| L. longispora3 | CMW 45429 CBS 133602 | Pinus sp. | Mexico | M. de Jesús Yáñez-Morales | - | - | - |
| L. longispora3 | CMW 45430 | Pinus sp. | Mexico | M. de Jesús Yáñez-Morales | - | - | - |
| L. pharomachri3 | CMW 37134 | P. tecunumanii | Baja Verapaz, Guatemala | I. Barnes | - | - | - |
| L. pharomachri3 | CMW 37136 CBS 144448 | P. tecunumanii | Baja Verapaz, Guatemala | I. Barnes | - | - | - |
| L. tecunumanii3 | CMW 46805 CBS 144450 | P. tecunumanii | Baja Verapaz, Guatemala | I. Barnes | - | - | - |
| L. tecunumanii3 | CMW 49403 CBS 144451 | P. tecunumanii | Baja Verapaz, Guatemala | I. Barnes | - | - | - |
| L. variabilis3 | CMW 42205 CBS144453 | P. caribaea | Santa Barbara, Honduras | H.C. Evans | - | - | - |
| L. variabilis3 | MX1 | P. arizonica var. stormiae | Nuevo León, Mexico | J.G. Marmolejo | - | - | - |
| Leptographium profanum2 | CV20170072 | P. taeda | Georgia, USA | C. Villari | - | - | - |
| Leptographium procerum2 | CV2017311 | P. taeda | Georgia, USA | C. Villari | - | - | - |
| Leptographium sp. 2 | CV20170049 | P. taeda | Georgia, USA | C. Villari | - | - | - |
| Rhizosphaera sp. 2 | CV2018024 | P. taeda | Georgia, USA | C. Villari | - | - | - |
| Cladosporium sp. 2 | CV2018023 | P. taeda | Georgia, USA | C. Villari | - | - | - |
| Alternaria tenuissima2 | CV2018022 | P. taeda | Georgia, USA | C. Villari | - | - | - |
| Dothideomycetes sp. 2 | CV2018020 | P. taeda | Georgia, USA | C. Villari | - | - | - |
| Leotiomycetes sp. 2 | CV2018019 | P. taeda | Georgia, USA | C. Villari | - | - | - |
| Nigrospora oryzae2 | CV2018018 | P. taeda | Georgia, USA | C. Villari | - | - | - |
| Lophodermium conigeum2 | CV2018002 | P. taeda | Georgia, USA | C. Villari | - | - | - |
| Lophodermium australe2 | CV2018001 | P. taeda | Georgia, USA | C. Villari | - | - | - |
| Plant Species | Locality | Symptoms on Needles | LAMP Results (min) [Total Reaction Time 35 min] | qPCR Results (min/Ct) [Total Reaction Time 1 h 30 min] | ||||
|---|---|---|---|---|---|---|---|---|
| L. acicola | D. pini | D. septosporum | L. acicola | D. pini | D. septosporum | |||
| Pinus cembra | Val Sarentino, Bolzano, Italy | + | - | - | 15 | - | - | 64/26.95 |
| P. cembra | Val Sarentino, Bolzano, Italy | + | - | - | 20 | - | - | 67/28.53 |
| P. cembra | Val Sarentino, Bolzano, Italy | + | - | - | - | - | - | - |
| P. mugo | Val Sarentino, Bolzano, Italy | + | - | - | - | - | - | - |
| P. mugo | Auronzo di Cadore, Belluno, Italy | ++ | - | - | 20 | - | - | 75/32.73 |
| P. mugo | Paluzza, Udine, Italy | ++ | 20 | - | - | 71/30.58 | - | - |
| P. mugo | Gardone, Brescia, Italy | + | - | - | - | - | - | - |
| P. nigra var. laricio | La Sila, Cosenza, Italy | + | - | - | 15 | - | - | 73/31.66 |
| P. nigra var. laricio | La Sila, Cosenza, Italy | ++ | - | - | 14 | - | - | 63/26.74 |
| P. palustris | Newton, Georgia, USA | + | 20 | - | - | 69/29.61 | - | - |
| P. palustris | Newton, Georgia, USA | ++ | 20 | - | - | 72/30.88 | - | - |
| P. palustris | Newton, Georgia, USA | + | 20 | - | - | 75/32.76 | - | - |
| P. radiata | La Sila, Cosenza, Italy | + | - | - | 12 | - | - | 67/28.71 |
| P. taeda | Athens, Georgia, USA | N | - | - | - | - | - | - |
| Pinus sp. | Slovenia | + | - | 20 | - | - | 71/30.45 | - |
| Pinus sp. | Slovenia | + | - | 20 | - | - | 72/31.33 | - |
| Pinus sp. | Slovenia | ++ | - | 15 | - | - | 62/25.90 | - |
| Pinus sp. | Slovenia | + | - | 20 | - | - | 70/29.90 | - |
| Pinus sp. | Slovenia | ++ | - | 15 | - | - | 65/27.54 | - |
| Primers | Sequence 5′->3′ |
|---|---|
| LAMP primers—Lecanosticta acicola | |
| La_F3 | GTACGCATGGGTCCTCGA |
| La_B3 | GAAATCACGGTGACCAGGAG |
| LA_FIP | CGTACAGTTACGTAATATGAGCGTGAGCGTGGTATC |
| LA_BIP | GGACTCTTCGCTGCCGCCCGATGACCTTTCACGGGTTA |
| LA_LoopB | TCGCTGTCGCAACACCC |
| LAMP primers—Dothistroma pini | |
| Dp_F3 | GTTGGGATGTATGTGGTGTTA |
| Dp_B3 | CTCCATCGACATCTCCAAGA |
| Dp_FIP | GAAGTAAACATTCAACCGCTCGCACTCGTGAAGAAAGCTTGTG |
| Dp_BIP | CGAGGTACGGACTTCACTTCACAGTAAAGTGATGCTGTGCTG |
| Dp_LoopF | CCTCGTATCTGCGAGTCTTC |
| LAMP primers—Dothistroma septosporum | |
| Ds_F3 | TTTCTGGCAGACCATTTCTG |
| Ds_B3 | ACGGCTCTTTCAAATGACTT |
| Ds_FIP | GTGCCTTCGTATCTGCATTTCATCCAGGACAGTATGTGGAATCC |
| Ds_BIP | CGAGAGCGACTGAGTGTCTATTTCGCATAGTGTTGAAGCACTGG |
| Ds_LoopB | GATGAGGTAGGTGCTCCTCT |
| Assimilating sequence-specific probes | |
| LA_LFPr 1—Lecanosticta acicola | FAM 2-ACGCTGAGGACCCGGATGCGAATGCGGATGCGGATGCCGAGGCGTTTCAAACTTCCACAGAG |
| DP_LBPr 1—Dothistroma pini | FAM 2-ACGCTGAGGACCCGGATGCGAATGCGGATGCGGATGCCGATTCCAGTGTGCTATGGCAAT |
| DS_LFPr 1—Dothistroma septosporum | FAM 2-ACGCTGAGGACCCGGATGCGAATGCGGATGCGGATGCCGAAGTACGAATCTGCATGACGC |
| Quencher strand 3 | TCGGCATCCGCATCCGCATTCGCATCCGGGTCCTCAGCGT-BHQ 4 |
| Target DNA Concentration (pg µL−1) | LAMP Results (min) [Total Reaction Time 35 min] | qPCR Results (min/Ct) [Total Reaction Time 1 h 30 min] | ||||
|---|---|---|---|---|---|---|
| L. acicola | D. pini | D. septosporum | L. acicola | D. pini | D. septosporum | |
| 10,000 | 10 | 10 | 10 | 40/15.10 | 37/13.51 | 42/16.23 |
| 2000 | 11 | 12 | 11 | 45/17.48 | 40/15.15 | 47/18.54 |
| 400 | 12 | 14 | 13 | 54/22.23 | 47/18.50 | 53/21.36 |
| 80 | 13 | 16 | 14 | 59/24.30 | 51/20.55 | 56/23.22 |
| 16 | 15 | 18 | 18 | 62/25.94 | 56/23.20 | 61/25.61 |
| 3.2 | 16 | 20 | 21 | 68/28.97 | 61/25.37 | 71/30.47 |
| 0.64 | 18 | 22 | - | 72/31.21 | 68/29.22 | 74/32.15 |
| 0.128 | 22 | - | - | 77/33.78 | 74/32.16 | 80/35.00 |
| 0.02 | - | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aglietti, C.; Meinecke, C.D.; Ghelardini, L.; Barnes, I.; van der Nest, A.; Villari, C. Rapid Detection of Pine Pathogens Lecanosticta acicola, Dothistroma pini and D. septosporum on Needles by Probe-Based LAMP Assays. Forests 2021, 12, 479. https://doi.org/10.3390/f12040479
Aglietti C, Meinecke CD, Ghelardini L, Barnes I, van der Nest A, Villari C. Rapid Detection of Pine Pathogens Lecanosticta acicola, Dothistroma pini and D. septosporum on Needles by Probe-Based LAMP Assays. Forests. 2021; 12(4):479. https://doi.org/10.3390/f12040479
Chicago/Turabian StyleAglietti, Chiara, Colton D. Meinecke, Luisa Ghelardini, Irene Barnes, Ariska van der Nest, and Caterina Villari. 2021. "Rapid Detection of Pine Pathogens Lecanosticta acicola, Dothistroma pini and D. septosporum on Needles by Probe-Based LAMP Assays" Forests 12, no. 4: 479. https://doi.org/10.3390/f12040479
APA StyleAglietti, C., Meinecke, C. D., Ghelardini, L., Barnes, I., van der Nest, A., & Villari, C. (2021). Rapid Detection of Pine Pathogens Lecanosticta acicola, Dothistroma pini and D. septosporum on Needles by Probe-Based LAMP Assays. Forests, 12(4), 479. https://doi.org/10.3390/f12040479







