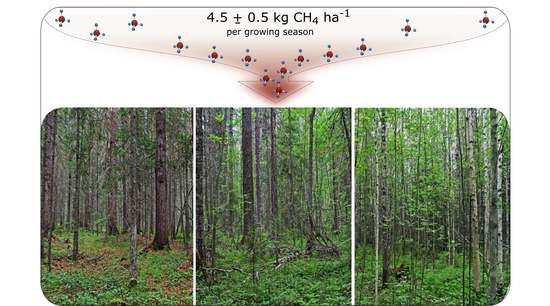
Atmospheric Methane Consumption and Methanotroph Communities in West Siberian Boreal Upland Forest Ecosystems
Abstract
:1. Introduction
2. Materials and Methods
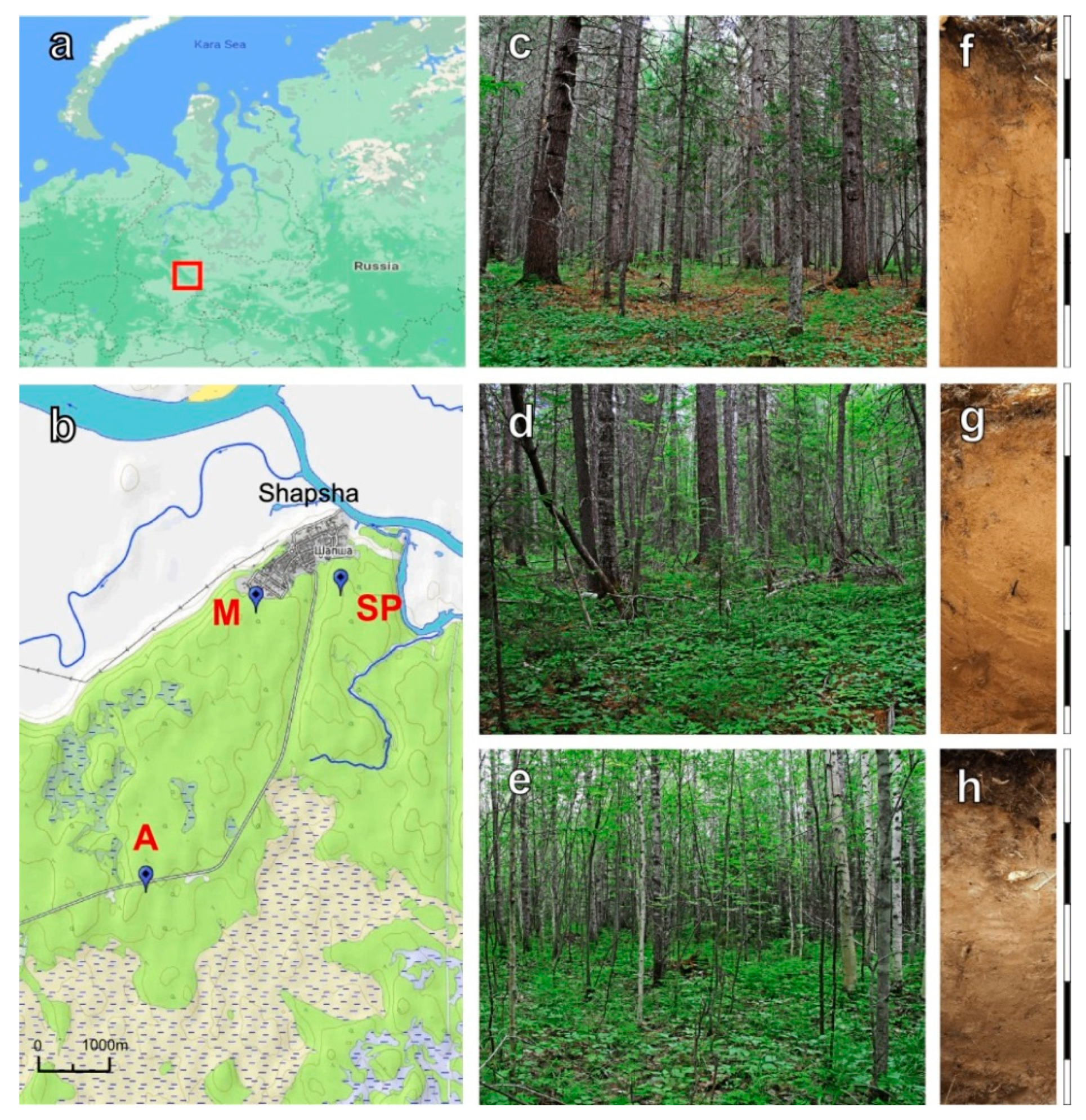
2.1. Site Description
2.2. CH4 Flux Measurements
2.3. Determination of the Soil Methane Oxidation Rate
2.4. Methane Concentration
2.5. Soil Chemical and Physical Properties
2.6. Statistical Analysis
2.7. Soil DNA Extraction
2.8. Illumina Sequencing and Analysis of 16S rRNA Gene Fragments
2.9. Illumina Sequencing and Analysis of pmoA Gene Fragments
3. Results and Discussion
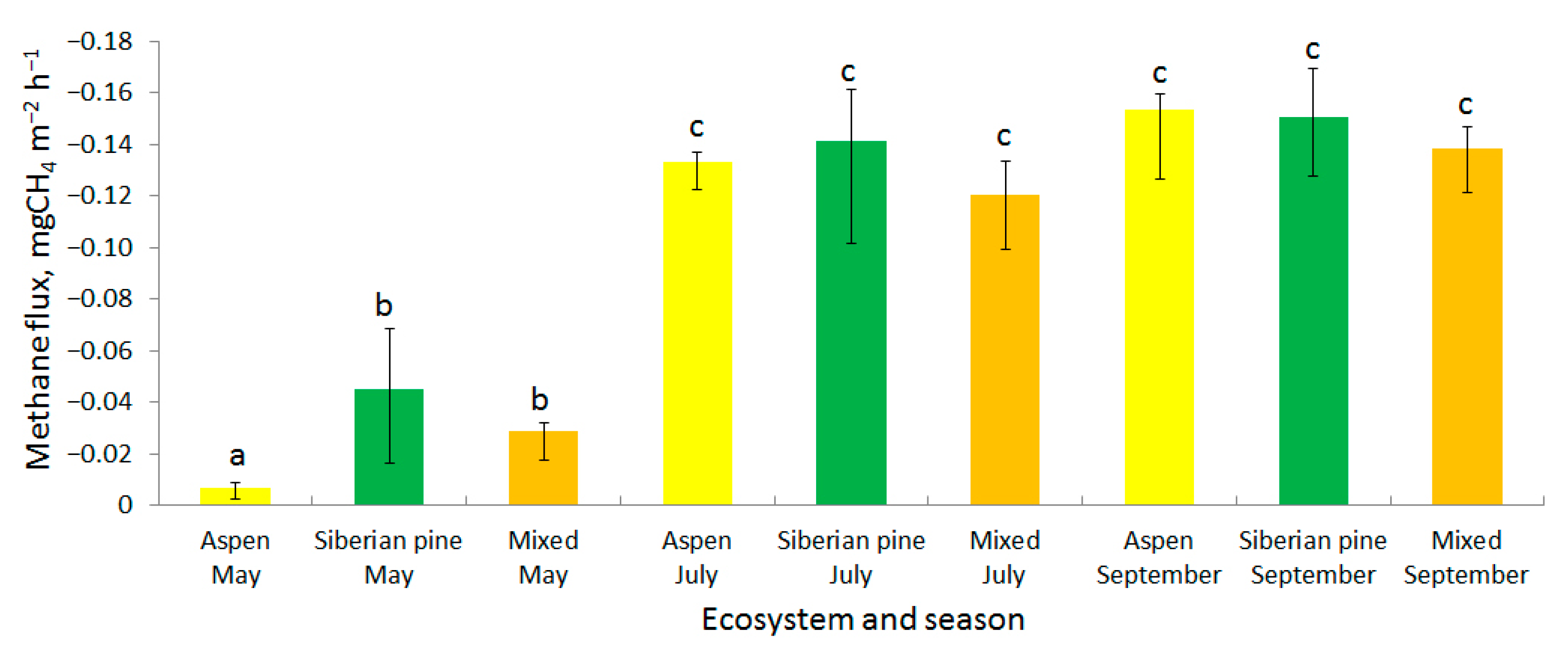
3.1. Methane Fluxes
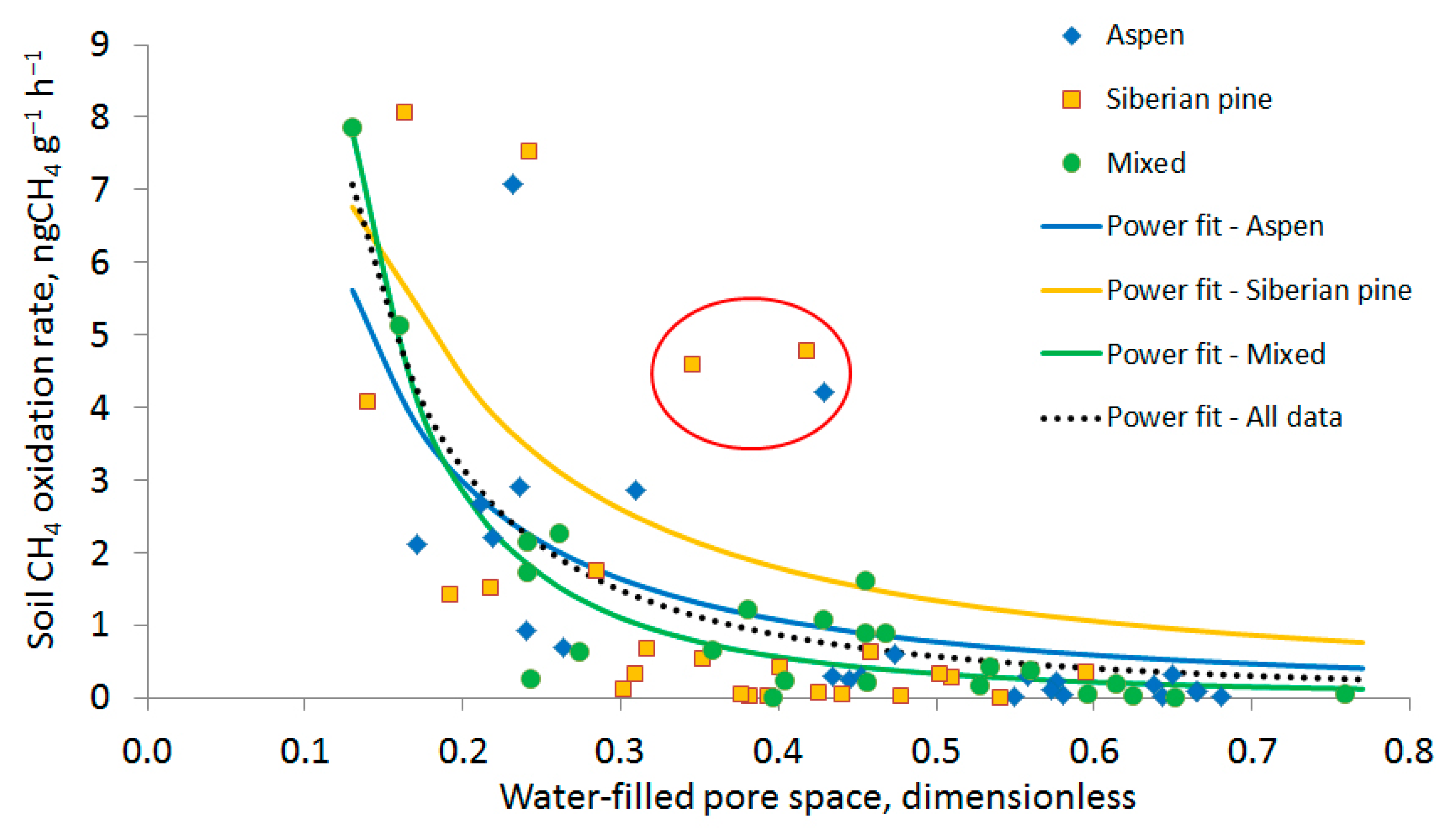
3.2. Soil Methane Oxidation Rates
3.3. Prokaryote Diversity Patterns in Forest Soils
3.4. Identification of Methanotrophs Based on 16S rRNA Gene Analysis
3.5. PmoA-Based Identification of Methanotrophic Bacteria
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fletcher, S.E.M.; Schaefer, H. Rising methane: A new climate challenge. Science 2019, 364, 932–933. [Google Scholar] [CrossRef] [PubMed]
- Saunois, M.; Stavert, A.R.; Poulter, B.; Bousquet, P.; Canadell, J.G.; Jackson, R.B.; Raymond, P.A.; Dlugokencky, E.J.; Houweling, S.; Patra, P.K.; et al. The global methane budget 2000–2017. Earth Syst. Sci. Data 2020, 12, 1561–1623. [Google Scholar] [CrossRef]
- Rosentreter, J.A.; Borges, A.V.; Deemer, B.R.; Holgerson, M.A.; Liu, S.; Song, C.; Melack, J.; Raymond, P.A.; Duarte, C.M.; Allen, G.H.; et al. Half of global methane emissions come from highly variable aquatic ecosystem sources. Nat. Geosci. 2021, 14, 225–230. [Google Scholar] [CrossRef]
- Lan, X.; Basu, S.; Schwietzke, S.; Bruhwiler, L.M.P.; Dlugokencky, E.J.; Michel, S.E.; Sherwood, O.A.; Tans, P.P.; Thoning, K.; Etiope, G.; et al. Improved constraints on global methane emissions and sinks using δ 13C-CH4. Glob. Biogeochem. Cycles 2021, 35, e2021GB007000. [Google Scholar] [CrossRef] [PubMed]
- Dutaur, L.; Verchot, L.V. A global inventory of the soil CH4 sink. Glob. Biogeochem. Cycles 2007, 21, 4013. [Google Scholar] [CrossRef]
- Degelmann, D.M.; Borken, W.; Kolb, S. Methane oxidation kinetics differ in European beech and Norway spruce soils. Eur. J. Soil Sci. 2009, 60, 499–506. [Google Scholar] [CrossRef]
- Yu, L.; Huang, Y.; Zhang, W.; Li, T.; Sun, W. Methane uptake in global forest and grassland soils from 1981 to 2010. Sci. Total Environ. 2017, 607, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Global Forest Resources Assessment 2020; Food and Agriculture Organization of the United Nations: Italy, Rome, 2020.
- Nakano, T.; Inoue, G.; Fukuda, M. Methane consumption and soil respiration by a birch forest soil in West Siberia. Tellus B Chem. Phys. Meteorol. 2004, 56, 223–229. [Google Scholar] [CrossRef]
- Kizilova, A.; Yurkov, A.; Kravchenko, I. Aerobic methanotrophs in natural and agricultural soils of European Russia. Diversity 2013, 5, 541–556. [Google Scholar] [CrossRef]
- Semenov, V.M.; Kravchenko, I.K.; Kuznetsova, T.V.; Semenova, N.A.; Bykova, S.A.; Dulov, L.E.; Gal’chenko, V.F.; Pardini, G.; Gispert, M.; Boeckx, P.; et al. Seasonal dynamics of atmospheric methane oxidation in gray forest soils. Microbiology 2004, 73, 356–362. [Google Scholar] [CrossRef]
- Kravchenko, I.; Sukhacheva, M. Methane oxidation and diversity of aerobic methanotrophs in forest and agricultural soddy–podzolic soils. Appl. Soil Ecol. 2017, 119, 267–274. [Google Scholar] [CrossRef]
- Menyailo, O.V.; Hungate, B.A.; Abraham, W.R.; Conrad, R. Changing land use reduces soil CH4 uptake by altering biomass and activity but not composition of high-affinity methanotrophs. Glob. Chang. Biol. 2008, 14, 2405–2419. [Google Scholar] [CrossRef]
- Belova, S.E.; Danilova, O.V.; Ivanova, A.A.; Merkel, A.Y.; Dedysh, S.N. Methane-oxidizing communities in lichen-dominated forested tundra are composed exclusively of high-affinity USCα methanotrophs. Microorganisms 2020, 8, 2047. [Google Scholar] [CrossRef]
- Takakai, F.; Desyatkin, A.R.; Lopez, C.M.L.; Fedorov, A.N.; Desyatkin, R.V.; Hatano, R. CH4 and N2O emissions from a forest-alas ecosystem in the permafrost taiga forest region, eastern Siberia, Russia. J. Geophys. Res. Biogeosci. 2008, 113, 2002. [Google Scholar] [CrossRef]
- Sabrekov, A.F.; Glagolev, M.V.; Fastovets, I.A.; Smolentsev, B.A.; Il’yasov, D.V.; Maksyutov, S.S. Relationship of methane consumption with the respiration of soil and grass-moss layers in forest ecosystems of the southern taiga in Western Siberia. Eurasian Soil Sci. 2015, 48, 841–851. [Google Scholar] [CrossRef]
- Sabrekov, A.F.; Glagolev, M.V.; Alekseychik, P.K.; Smolentsev, B.A.; Terentieva, I.E.; Krivenok, L.A.; Maksyutov, S.S. A process-based model of methane consumption by upland soils. Environ. Res. Lett. 2016, 11, 075001. [Google Scholar] [CrossRef]
- Yu Mochenov, S.; Churkina, A.I.; Sabrekov, S.F.; Glagolev, M.V.; Il’Yasov, D.V.; Terentieva, I.E.; Maksyutov, S.S. Soils in seasonally flooded forests as methane sources: A case study of West Siberian South taiga. IOP Conf. Ser. Earth Environ. Sci. 2018, 138, 012012. [Google Scholar] [CrossRef]
- Schneider, J.; Ťupek, B.; Lukasheva, M.; Gudyrev, V.; Miglovets, M.; Jungkunst, H.F. Methane emissions from paludified boreal soils in European Russia as measured and modelled. Ecosystems 2017, 21, 827–838. [Google Scholar] [CrossRef]
- Masyagina, O.V.; Menyailo, O.V. The impact of permafrost on carbon dioxide and methane fluxes in Siberia: A meta-analysis. Environ. Res. 2020, 182, 109096. [Google Scholar] [CrossRef]
- Feng, H.; Guo, J.; Han, M.; Wang, W.; Peng, C.; Jin, J.; Song, X.; Yu, S. A review of the mechanisms and controlling factors of methane dynamics in forest ecosystems. For. Ecol. Manag. 2020, 455, 117702. [Google Scholar] [CrossRef]
- Gatica, G.; Fernández, M.E.; Juliarena, M.P.; Gyenge, J. Environmental and anthropogenic drivers of soil methane fluxes in forests: Global patterns and among-biomes differences. Glob. Chang. Biol. 2020, 26, 6604–6615. [Google Scholar] [CrossRef]
- Kirpotin, S.N.; Callaghan, T.V.; Peregon, A.M.; Babenko, A.S.; Berman, D.I.; Bulakhova, N.A.; Byzaakay, A.A.; Chernykh, T.M.; Chursin, V.; Interesova, E.A.; et al. Impacts of environmental change on biodiversity and vegetation dynamics in Siberia. Ambio 2021, 50, 1926–1952. [Google Scholar] [CrossRef] [PubMed]
- Kharuk, V.I.; Ponomarev, E.I.; Ivanova, G.A.; Dvinskaya, M.L.; Coogan, S.C.P.; Flannigan, M.D. Wildfires in the Siberian taiga. Ambio 2021, 50, 1953–1974. [Google Scholar] [CrossRef] [PubMed]
- Filippova, N.V.; Bulyonkova, T.M. The diversity of larger fungi in the vicinities of Khanty-Mansiysk (middle taiga of West Siberia). Environ. Dyn. Glob. Clim. Chang. 2017, 8, P13–P24. [Google Scholar] [CrossRef]
- Dabros, A.; Pyper, M.; Castilla, G. Seismic lines in the boreal and arctic ecosystems of North America: Environmental impacts, challenges, and opportunities. Environ. Rev. 2018, 26, 214–229. [Google Scholar] [CrossRef] [Green Version]
- Fang, H.J.; Yu, G.R.; Cheng, S.L.; Zhu, T.H.; Wang, Y.S.; Yan, J.H.; Wang, M.; Cao, M.; Zhou, M. Effects of multiple environmental factors on CO2 emission and CH4 uptake from old-growth forest soils. Biogeosciences 2010, 7, 395–407. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Estiarte, M.; Peñuelas, J. Soil moisture as the key factor of atmospheric CH4 uptake in forest soils under environmental change. Geoderma 2019, 355, 113920. [Google Scholar] [CrossRef]
- Meier, I.C.; Leuschner, C.; Marini, E.; Fender, A.C. Species-specific effects of temperate trees on greenhouse gas exchange of forest soil are diminished by drought. Soil Biol. Biochem. 2016, 95, 122–134. [Google Scholar] [CrossRef]
- Walkiewicz, A.; Rafalska, A.; Bulak, P.; Bieganowski, A.; Osborne, B. How can litter modify the fluxes of CO2 and CH4 from forest soils? A mini-review. Forests 2021, 12, 1276. [Google Scholar] [CrossRef]
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef]
- Trotsenko, Y.A.; Murrell, J.C. Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 2008, 63, 183–229. [Google Scholar] [PubMed]
- Chistoserdova, L.; Lidstrom, M.E. Aerobic Methylotrophic Prokaryotes. In The Prokaryotes: Prokaryotic Physiology and Biochemistry; Springer: Berlin/Heidelberg, Germany, 2013; pp. 267–285. ISBN 9783642301414. [Google Scholar]
- Khmelenina, V.N.; Colin Murrell, J.; Smith, T.J.; Trotsenko, Y.A. Physiology and Biochemistry of the Aerobic Methanotrophs. In Aerobic Utilization of Hydrocarbons, Oils, and Lipids; Springer: Cham, Switzerland, 2019; pp. 1–25. [Google Scholar]
- Dedysh, S.N.; Knief, C. Diversity and Phylogeny of Described Aerobic Methanotrophs. In Methane Biocatalysis: Paving the Way to Sustainability; Springer: Cham, Switzerland, 2018; pp. 17–42. [Google Scholar]
- Knief, C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 2015, 6, 1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunfield, P.F. The Soil Methane Sink. In Greenhouse Gas Sinks; eBook: Athenaeum Press Ltd.: Gateshead, UK, 2007; pp. 152–170. [Google Scholar]
- Kolb, S. The quest for atmospheric methane oxidizers in forest soils. Environ. Microbiol. Rep. 2009, 1, 336–346. [Google Scholar] [CrossRef]
- Holmes, A.J.; Roslev, P.; McDonald, I.R.; Iversen, N.; Henriksen, K.; Murrell, J.C. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl. Environ. Microbiol. 1999, 65, 3312–3318. [Google Scholar] [CrossRef] [Green Version]
- Knief, C.; Lipski, A.; Dunfield, P.F. Diversity and activity of methanotrophic bacteria in different upland soils. Appl. Environ. Microbiol. 2003, 69, 6703–6714. [Google Scholar] [CrossRef] [Green Version]
- Henckel, T.; Jäckel, U.; Schnell, S.; Conrad, R. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl. Environ. Microbiol. 2000, 66, 1801–1808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, S.; Holmes, A.J.; Olsen, R.A.; Murrell, J.C. Detection of methane oxidizing bacteria in forest soil by monooxygenase PCR amplification. Microb. Ecol. 2000, 39, 282–289. [Google Scholar]
- Kolb, S.; Knief, C.; Dunfield, P.F.; Conrad, R. Abundance and activity of uncultured methanotrophic bacteria involved in the consumption of atmospheric methane in two forest soils. Environ. Microbiol. 2005, 7, 1150–1161. [Google Scholar] [CrossRef] [PubMed]
- Degelmann, D.M.; Borken, W.; Drake, H.L.; Kolb, S. Different atmospheric methane-oxidizing communities in european beech and norway spruce soils. Appl. Environ. Microbiol. 2010, 10, 3228–3235. [Google Scholar] [CrossRef] [Green Version]
- Dörr, N.; Glaser, B.; Kolb, S. Methanotrophic communities in brazilian ferralsols from naturally forested, afforested, and agricultural Sites. Appl. Environ. Microbiol. 2010, 76, 1307–1310. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Zhou, X.; Shi, L.; Jia, Z. Atmospheric methane oxidizers are dominated by Upland Soil Cluster Alpha in 20 forest soils of China. Microb. Ecol. 2020, 80, 859–871. [Google Scholar] [CrossRef] [PubMed]
- Pratscher, J.; Vollmers, J.; Wiegand, S.; Dumont, M.G.; Kaster, A.K. Unravelling the identity, metabolic potential and global biogeography of the atmospheric methane-oxidizing upland soil cluster α. Environ. Microbiol. 2018, 20, 1016–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tveit, A.T.; Hestnes, A.G.; Robinson, S.L.; Schintlmeister, A.; Dedysh, S.N.; Jehmlich, N.; von Bergen, M.; Herbold, C.; Wagner, M.; Richter, A.; et al. Widespread soil bacterium that oxidizes atmospheric methane. Proc. Natl. Acad. Sci. USA 2019, 10, 589. [Google Scholar] [CrossRef] [Green Version]
- Hutchinson, G.L.; Mosier, A.R. Improved soil cover method for field measurement of nitrous oxide fluxes. Soil Sci. Soc. Am. J. 1981, 45, 311–316. [Google Scholar] [CrossRef]
- Milliken, G.; Johnson, D. Analysis of Messy Data-Volume 1: Designed Experiments; CRC Press: Boca Raton, FL, USA, 1992. [Google Scholar]
- Gohl, D.; Gohl, D.M.; MacLean, A.; Hauge, A.; Becker, A.; Walek, D.; Beckman, K.B. An optimized protocol for high-throughput amplicon-based microbiome profiling. Protoc. Exch. 2016. [Google Scholar] [CrossRef]
- Caporaso, J.; Kuczynski, J.; Stombaugh, J. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Holmes, A.J.; Costello, A.; Lidstrom, M.E.; Murrell, J.C. Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 1995, 132, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Bourne, D.G.; McDonald, I.R.; Murrell, J.C. Comparison of pmoA PCR primer sets as tools for investigating methanotroph diversity in three Danish soils. Appl. Environ. Microbiol. 2001, 67, 3802–3809. [Google Scholar] [CrossRef] [Green Version]
- Dumont, M.G.; Lüke, C.; Deng, Y.; Frenzel, P. Classification of pmoA amplicon pyrosequences using BLAST and the lowest common ancestor method in MEGAN. Front. Microbiol. 2014, 5, 34. [Google Scholar] [CrossRef]
- Wen, X.; Yang, S.; Liebner, S. Evaluation and update of cutoff values for methanotrophic pmoA gene sequences. Arch. Microbiol. 2016, 198, 629–636. [Google Scholar] [CrossRef] [Green Version]
- Sabrekov, A.F.; Semenov, M.V.; Terent’eva, I.E.; Litti, Y.V.; Il’yasov, D.V.; Glagolev, M.V. The link between soil methane oxidation rate and abundance of methanotrophs estimated by quantitative PCR. Microbiology 2020, 89, 182–191. [Google Scholar] [CrossRef]
- Crill, P. Seasonal patterns of methane uptake and carbon dioxide release by a temperate woodland soil. Glob. Biogeochem. Cycles 1991, 5, 319–334. [Google Scholar] [CrossRef]
- Ullah, S.; Moore, T.R. Biogeochemical controls on methane, nitrous oxide, and carbon dioxide fluxes from deciduous forest soils in eastern Canada. J. Geophys. Res. Biogeosci. 2011, 116, 3010. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.P.; Zhang, W.J.; Hu, C.S.; Tang, X.G. Soil greenhouse gas fluxes from different tree species on Taihang Mountain, North China. Biogeosciences 2014, 11, 1649–1666. [Google Scholar] [CrossRef] [Green Version]
- Borken, W.; Beese, F. Methane and nitrous oxide fluxes of soils in pure and mixed stands of European beech and Norway spruce. Eur. J. Soil Sci. 2006, 57, 617–625. [Google Scholar] [CrossRef]
- Christiansen, J.R.; Gundersen, P. Stand age and tree species affect N2O and CH4 exchange from afforested soils. Biogeosciences 2011, 8, 2535–2546. [Google Scholar] [CrossRef] [Green Version]
- Reay, D.S.; Radajewski, S.; Murrell, J.C.; McNamara, N.; Nedwell, D.B. Effects of land-use on the activity and diversity of methane oxidizing bacteria in forest soils. Soil Biol. Biochem. 2001, 33, 1613–1623. [Google Scholar] [CrossRef]
- Christiansen, J.R.; Gundersen, P.; Frederiksen, P.; Vesterdal, L. Influence of hydromorphic soil conditions on greenhouse gas emissions and soil carbon stocks in a Danish temperate forest. For. Ecol. Manag. 2012, 284, 185–195. [Google Scholar] [CrossRef]
- Täumer, J.; Kolb, S.; Boeddinghaus, R.S.; Wang, H.; Schöning, I.; Schrumpf, M.; Urich, T.; Marhan, S. Divergent drivers of the microbial methane sink in temperate forest and grassland soils. Glob. Chang. Biol. 2021, 27, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Whalen, S.C.; Reeburgh, W.S.; Barber, V.A. Oxidation of methane in boreal forest soils: A comparison of seven measures. Biogeochemistry 1992, 16, 181–211. [Google Scholar] [CrossRef] [Green Version]
- Kawamoto, K.; Moldrup, P.; Schjønning, P.; Iversen, B.V.; Komatsu, T.; Rolston, D.E. Gas transport parameters in the vadose zone: Development and tests of power-law models for air permeability. Vadose Zone J. 2006, 5, 1205–1215. [Google Scholar] [CrossRef]
- Moldrup, P.; Deepagoda, T.K.K.C.; Hamamoto, S.; Komatsu, T.; Kawamoto, K.; Rolston, D.E.; Jonge, L.W. de Structure-dependent water-induced linear reduction model for predicting gas diffusivity and tortuosity in repacked and intact soil. Vadose Zone J. 2013, 12, vzj2013-01. [Google Scholar]
- Lind, S.E.; Virkajärvi, P.; Hyvönen, N.P.; Maljanen, M.; Kivimäenpää, M.; Jokinen, S.; Antikainen, S.; Latva, M.; Räty, M.; Martikainen, P.J.; et al. Carbon dioxide and methane exchange of a perennial grassland on a boreal mineral soil. Boreal Environ. Res. 2020, 25, 1–17. [Google Scholar]
- Bender, M.; Conrad, R. Kinetics of CH4 oxidation in oxic soils exposed to ambient air or high CH4 mixing ratios. FEMS Microbiol. Ecol. 1992, 10, 261–269. [Google Scholar] [CrossRef]
- Pratscher, J.; Dumont, M.G.; Conrad, R. Assimilation of acetate by the putative atmospheric methane oxidizers belonging to the USCα clade. Environ. Microbiol. 2011, 13, 2692–2701. [Google Scholar] [CrossRef]
- Göttlein, A.; Matzner, E. Microscale heterogeneity of acidity related stress-parameters in the soil solution of a forested cambic podzol. Plant Soil 1997, 192, 95–105. [Google Scholar] [CrossRef]
- Lange, B.; Lüescher, P.; Germann, P.F. Significance of tree roots for preferential infiltration in stagnic soils. Hydrol. Earth Syst. Sci. 2009, 13, 1809–1821. [Google Scholar] [CrossRef] [Green Version]
- Bundt, M.; Widmer, F.; Pesaro, M.; Zeyer, J.; Blaser, P. Preferential flow paths: Biological “hot spots” in soils. Soil Biol. Biochem. 2001, 33, 729–738. [Google Scholar] [CrossRef]
- Llado, S.; Lopez-Mondejar, R.; Baldrian, P. Forest soil bacteria: Diversity, involvement in ecosystem processes, and response to global change. Microbiol. Mol. Biol. Rev. 2017, 81, e00063-16. [Google Scholar] [CrossRef] [Green Version]
- Brofft, J.E.; McArthur, J.V.; Shimkets, L.J. Recovery of novel bacterial diversity from a forested wetland impacted by reject coal. Environ. Microbiol. 2002, 4, 764–769. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, A.A.; Zhelezova, A.D.; Chernov, T.I.; Dedysh, S.N. Linking ecology and systematics of acidobacteria: Distinct habitat preferences of the Acidobacteriia and Blastocatellia in tundra soils. PLoS ONE 2020, 15, e0230157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nogales, B.; Moore, E.R.; Llobet-Brossa, E.; Rossello-Mora, R.; Amann, R.; Timmis, K.N. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 2001, 67, 1874–1884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengtson, P.; Basiliko, N.; Dumont, M.G.; Hills, M.; Murrell, J.C.; Roy, R.; Grayston, S.J. Links between methanotroph community composition and CH4 oxidation in a pine forest soil. FEMS Microbiol. Ecol. 2009, 70, 356–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]

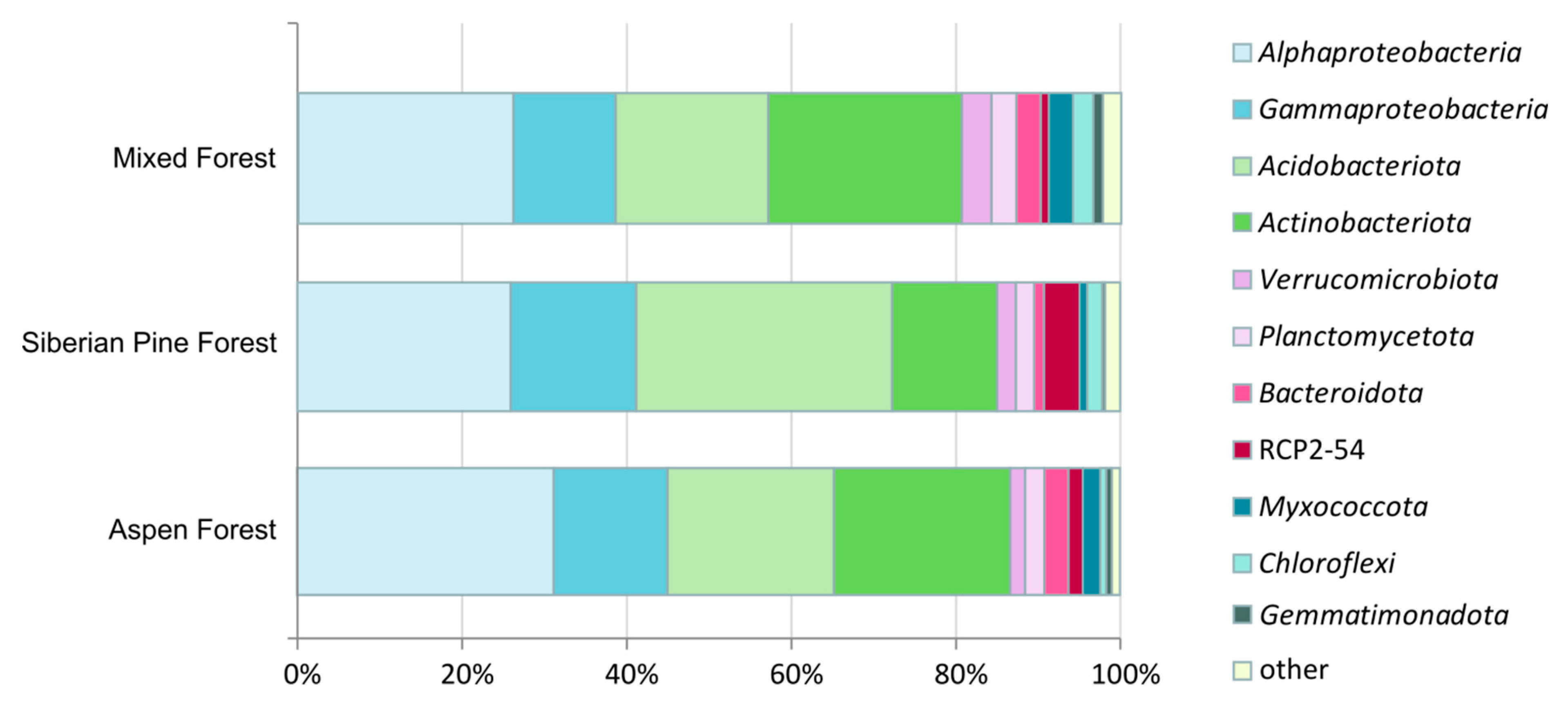
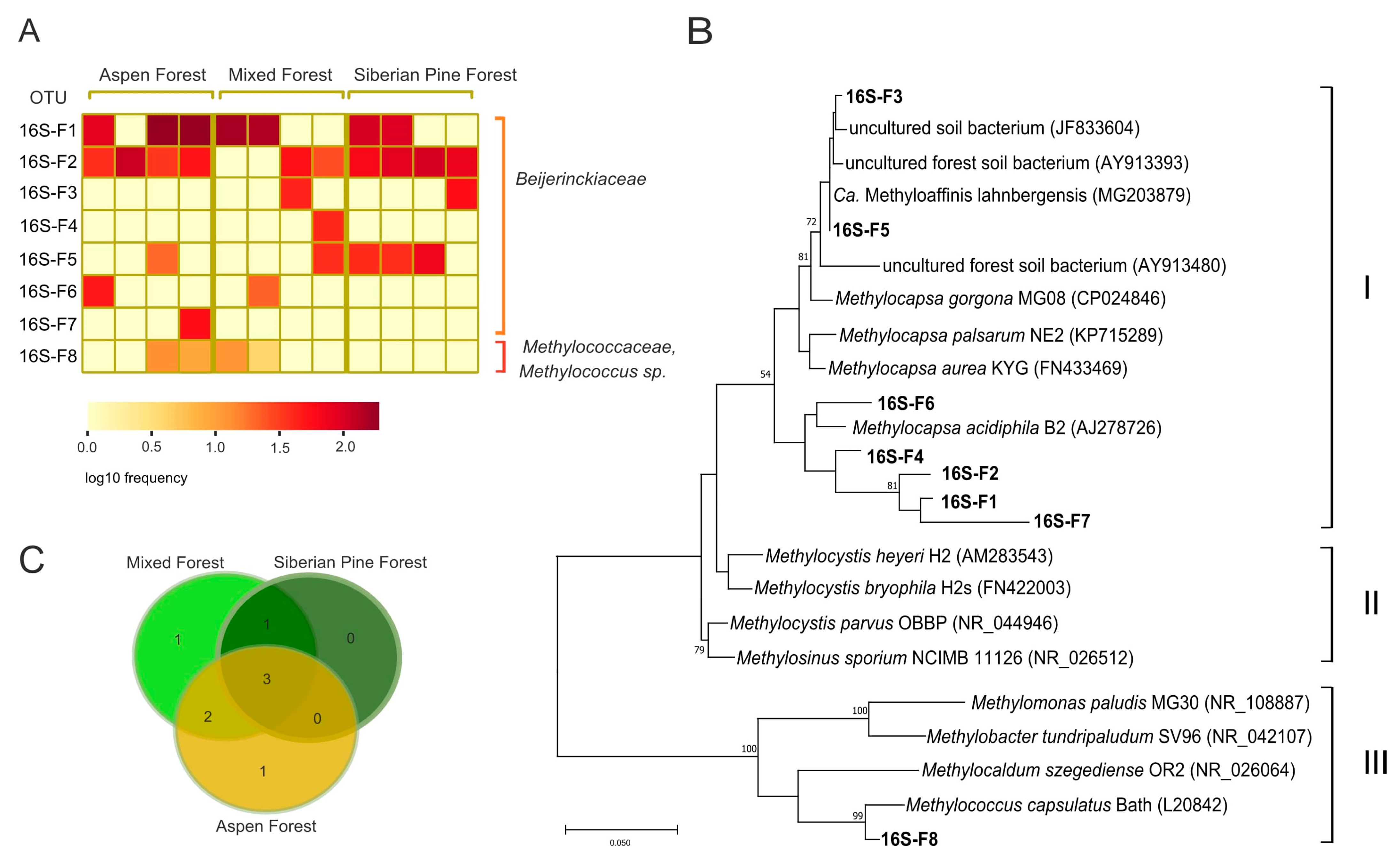
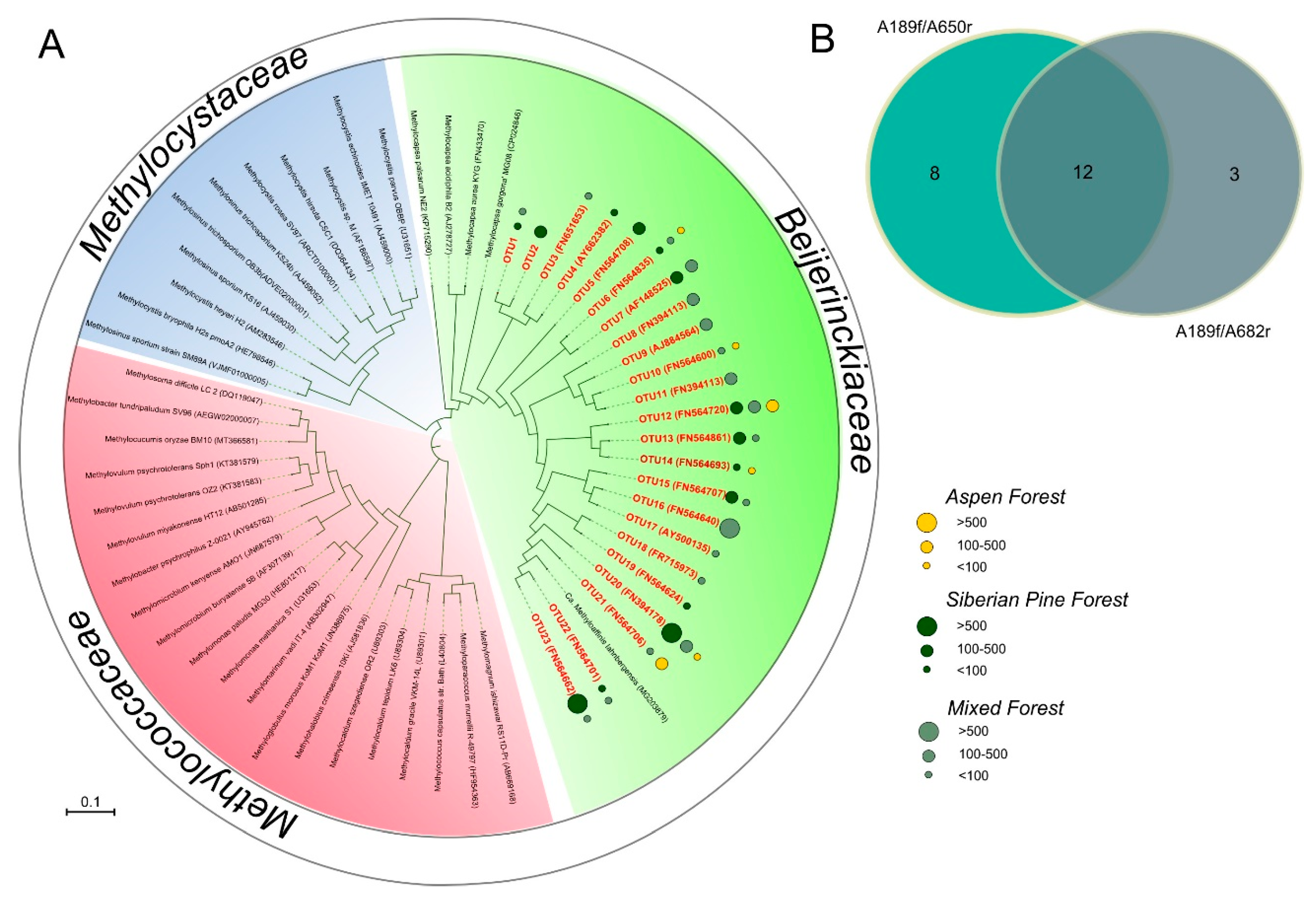
| Forest Ecosystem | Coordinates | Bulk Density, g cm−3 | Litter Thickness, cm | pH | Corg, % | NH4+, µgN gDW−1 | NO3−, µgN gDW−1 |
|---|---|---|---|---|---|---|---|
| Aspen | 61.05623° N 69.42942° E | 0.84 ± 0.08 | 2 ± 1 | 5.8 ± 0.2 | 2.5 ± 0.4 | 7.1 ± 1.4 | 121.0 ± 9.8 |
| Siberian pine | 61.08571° N 69.46918° E | 0.99 ± 0.05 | 1 ± 1 | 5.1 ± 0.2 | 1.4 ± 0.4 | 12.9 ± 6.1 | 93.2 ± 23.6 |
| Mixed | 61.08301° N 69.45383° E | 1.05 ± 0.06 | 1 ± 1 | 5.4 ± 0.2 | 1.8 ± 0.3 | 9.8 ± 3.9 | 151.4 ± 68.9 |
| Parameter | Season | Ecosystem | Depth |
|---|---|---|---|
| Flux | 0.72 *** | 0.03 ** | NM |
| CH4 oxidation rate | 0.29 *** | NS | 0.48 *** |
| Soil moisture (by volume) | 0.46 *** | 0.05 *** | 0.08 *** |
| Water-filled pore space | 0.27 *** | 0.03 ** | 0.39 *** |
| CH4 concentration | 0.25 *** | 0.02 ** | 0.45 *** |
| Soil Temperature | 0.90 *** | 0.01 *** | 0.05 *** |
| pH | NS | 0.17 ** | NS |
| NH4+ | 0.24 ** | NS | 0.16 ** |
| NO3− | 0.40 *** | 0.12 ** | NS |
| Methanotrophs abundance a | 0.04 * | NS | 0.53 *** |
| OTU ID | Relative Abundance (%) | Taxonomy | Closest Silva Match | Habitat | |
|---|---|---|---|---|---|
| Siberian Pine Forest | SF-16S-1 | 5.71 | Bradyrhizobium sp. 175LB2PYPT | HQ698291 | Root nodules of Acacia pycnantha |
| SF-16S-2 | 3.92 | Uncultured WD260 group of Gammaproteobacteria | EU150272 | Boreal pine forest soil | |
| SF-16S-3 | 3.02 | Uncultured RCP2-54 bacterium | DQ451494 | Forest soil | |
| SF-16S-4 | 2.75 | Uncultured WD260 group of Gammaproteobacteria | EU150268 | Soil from Niwot Ridge LTER | |
| SF-16S-5 | 2.73 | Mycobacterium celatum MB72 | AJ416914 | AIDS patients | |
| SF-16S-6 | 2.58 | Acidobacteriales (Sd 1) uncultured | HQ598256 | Woodland soil | |
| SF-16S-7 | 2.43 | Acidobacteriales (Sd 1) uncultured | FJ624925 | Boreal pine forest soil | |
| SF-16S-8 | 2.29 | Acidobacteriales (Sd 1) uncultured | AY963436 | Soil of evergreen broad-leaved forest | |
| SF-16S-9 | 2.13 | Bradyrhizobium sp. | JX644393 | Earthworm nephridia | |
| SF-16S-10 | 2.13 | Acidobacteriae (Sd 2) uncultured | FJ466148 | Acidic fen soil | |
| Mixed Forest | MF-16S-1 | 6.22 | Bradyrhizobium sp. 175LB2PYPT | HQ698291 | Root nodules of Acacia pycnantha |
| MF-16S-2 | 5.43 | Solirubrobacteraceae uncultured | HM270154 | Tobacco rhizosphere | |
| MF-16S-3 | 2.54 | Uncultured WD260 group of Gammaproteobacteria | EU150268 | Soil from Niwot Ridge LTER | |
| MF-16S-4 | 2.02 | Mycobacterium sp. MPLK-65 | KX689762 | Subterranean mine | |
| MF-16S-5 | 1.80 | Uncultured WD260 group of Gammaproteobacteria | AB991083 | Temperate highland grassland | |
| MF-16S-6 | 1.79 | Mycobacterium celatum MB72 | AJ416914 | AIDS patients | |
| MF-16S-7 | 1.27 | Conexibacter sp. uncultured | HM263196 | Grassland soil | |
| MF-16S-8 | 1.09 | Xanthobacteraceae uncultured | AY963361 | Poplar tree rhizosphere | |
| MF-16S-9 | 1.01 | Roseiarcus sp. | AY425766 | Tobacco rhizosphere | |
| MF-16S-10 | 0.99 | Uncultured WD260 group of Gammaproteobacteria | EU150272 | Boreal pine forest soil | |
| Aspen Forest | AF-16S-1 | 9.10 | Bradyrhizobium sp. 175LB2PYPT | HQ698291 | Root nodules of Acacia pycnantha |
| AF-16S-2 | 3.86 | Mycobacterium sp. MPLK-65 | KX689762 | Subterranean mine | |
| AF-16S-3 | 3.15 | Uncultured WD260 group of Gammaproteobacteria | AB991083 | Temperate highland grassland | |
| AF-16S-4 | 2.91 | Mycobacterium celatum MB72 | AJ416914 | AIDS patients | |
| AF-16S-5 | 2.32 | Uncultured WD260 group of Gammaproteobacteria | EU150268 | Soil from Niwot Ridge LTER | |
| AF-16S-6 | 1.67 | Acidobacteriae (Sd 2) uncultured | AY963303 | Soil of evergreen broad-leaved forest | |
| AF-16S-7 | 1.58 | Roseiarcus sp. | AY425766 | Tobacco rhizosphere | |
| AF-16S-8 | 1.39 | Mycobacterium conspicuum JCM14738 | X88922 | Patients with disseminated infections | |
| AF-16S-9 | 1.35 | Granulicella sp. | JN023575 | Temperate highland grassland | |
| AF-16S-10 | 1.32 | Solirubrobacteraceae uncultured | KM200386 | Tobacco rhizosphere |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabrekov, A.F.; Danilova, O.V.; Terentieva, I.E.; Ivanova, A.A.; Belova, S.E.; Litti, Y.V.; Glagolev, M.V.; Dedysh, S.N. Atmospheric Methane Consumption and Methanotroph Communities in West Siberian Boreal Upland Forest Ecosystems. Forests 2021, 12, 1738. https://doi.org/10.3390/f12121738
Sabrekov AF, Danilova OV, Terentieva IE, Ivanova AA, Belova SE, Litti YV, Glagolev MV, Dedysh SN. Atmospheric Methane Consumption and Methanotroph Communities in West Siberian Boreal Upland Forest Ecosystems. Forests. 2021; 12(12):1738. https://doi.org/10.3390/f12121738
Chicago/Turabian StyleSabrekov, Aleksandr F., Olga V. Danilova, Irina E. Terentieva, Anastasia A. Ivanova, Svetlana E. Belova, Yuri V. Litti, Mikhail V. Glagolev, and Svetlana N. Dedysh. 2021. "Atmospheric Methane Consumption and Methanotroph Communities in West Siberian Boreal Upland Forest Ecosystems" Forests 12, no. 12: 1738. https://doi.org/10.3390/f12121738
APA StyleSabrekov, A. F., Danilova, O. V., Terentieva, I. E., Ivanova, A. A., Belova, S. E., Litti, Y. V., Glagolev, M. V., & Dedysh, S. N. (2021). Atmospheric Methane Consumption and Methanotroph Communities in West Siberian Boreal Upland Forest Ecosystems. Forests, 12(12), 1738. https://doi.org/10.3390/f12121738