Endophytic Community Composition and Genetic-Enzymatic Features of Cultivable Bacteria in Vaccinium myrtillus L. in Forests of the Baltic-Nordic Region
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Plant Material
2.2. Isolation of Endophytic Bacteria
2.3. Classification of Isolated Endophytic Bacteria
2.4. Determination of the Enzymatic Activity of the Bacterial Endophytes
2.5. Identification of Genes Involved in Plant Growth Promotion
3. Results
3.1. Biodiversity of Endophytic Bacteria in Bilberry Leaves
3.2. Enzymatic-Genetic Features of Endophytic Bacteria in Bilberry Leaves
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Santoyo, G.; Moreno-Hagelsieb, G.; del Carmen Orozco-Mosqueda, M.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xing, K.; Jiang, J.H.; Xu, L.H.; Li, W.J. Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Adv. Appl. Microbiol. 2011, 89, 457–473. [Google Scholar] [CrossRef]
- Afzal, I.; Shinwaria, Z.K.; Sikandarb, S.; Shahzadc, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Coombs, J.T.; Franco, C.M.M. Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl. Environ. Microbiol. 2003, 69, 5603–5608. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef]
- Mora, Y.; Díaz, R.; Vargas-Lagunas, C.; Peratta, H.; Guerrero, G.; Aguilar, A.; Encarnación, S.; Girard, L.; Mora, L. Nitrogen-fixing rhizobial strains isolated from common bean seeds: Phylogeny, physiology, and genome analysis. Appl. Environ. Microbiol. 2014, 80, 5644–5654. [Google Scholar] [CrossRef] [PubMed]
- Gouda, S.; Das, G.; Sen, S.K.; Shin, H.S.; Patra, J.K. Endophytes: A Treasure House of Bioactive Compounds of Medicinal Importance. Front. Microbiol. 2016, 7, 1538. [Google Scholar] [CrossRef] [PubMed]
- Rosenblueth, M.; Martínez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 2006, 19, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Miliute, I.; Buzaite, O.; Baniulis, D.; Stanys, V. Bacterial endophytes in agricultural crops and their role in stress tolerance: A review. Zemdirbyste 2015, 102, 465–478. Available online: https://hdl.handle.net/20.500.12259/52375 (accessed on 26 November 2021). [CrossRef]
- Anyasi, R.O.; Atagana, H.I. Endophyte: Understanding the microbes and its applications. Pak. J. Biol. Sci. 2019, 22, 154–167. Available online: https://scialert.net/abstract/?doi=pjbs.2019.154.167 (accessed on 26 November 2021). [CrossRef] [PubMed]
- Xu, W.; Wang, F.; Zhang, M.; Ou, T.; Wang, R.; Strobel, G.; Xiang, Z.; Zhou, Z.; Xie, J. Diversity of cultivable endophytic bacteria in mulberry and their potential for antimicrobial and plant growth-promoting activities. Microbiol. Res. 2019, 229, 126328. [Google Scholar] [CrossRef]
- Senthilkumar, M.; Anandham, R.; Madhaiyan, M.; Venkateswaran, V.; Sa, T. Endophytic 895 bacteria: Perspectives and applications in agricultural crop production. In Bacteria in Agrobiology: Crop 896 Ecosystems; Springer: Berlin/Heidelberg, Germany, 2011; pp. 61–96. [Google Scholar]
- Dastogeer, K.M.G.; Tumpa, F.H.; Sultana, A.; Akter, M.A.; Chakraborty, A. Plant microbiome–an account of the factors that shape community composition and diversity. Curr. Plant Biol. 2020, 23, 100161. [Google Scholar] [CrossRef]
- Bujor, O.; Le, B.C.; Volf, I.; Popa, V.I.; Dufour, C. Seasonal variations of the phenolic constituents in bilberry (Vaccinium myrtillus L.) leaves, stems and fruits, and their antioxidant activity. Food Chem. 2016, 213, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Ziemlewska, A.; Zagórska-Dziok, M.; Nizioł-Łukaszewska, Z. Assessment of cytotoxicity and antioxidant properties of berry leaves as by-products with potential application in cosmetic and pharmaceutical products. Sci. Rep. 2021, 11, 3240. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; van Overbeek, L.S.; Elsas, J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008, 16, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Company, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Kumar, A.; Singh, R.; Pandey, K.D. Endophytic bacteria: A new source of bioactive compounds. 3 Biotech. 2017, 7, 315. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Y.; Shao, Z.; Li, J.; Zan, S.; Zhou, S.; Yang, R. Two selenium tolerant Lysinibacillus sp. strains are capable of reducing selenite to elemental Se efficiently under aerobic conditions. J. Environ. Sci. 2019, 77, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef]
- Fouda, A.; Eid, A.M.; Elsaied, A.; El-Belely, E.F.; Barghoth, M.G.; Azab, E.; Gobouri, A.A.; Hassan, S.E. Plant Growth-Promoting Endophytic Bacterial Community Inhabiting the Leaves of Pulicaria incisa (Lam.) DC Inherent to Arid Regions. Plants 2021, 10, 76. [Google Scholar] [CrossRef]
- Lyngwi, N.A.; Nongkhlaw, M.; Kalita, D.; Joshi, S.R. Bioprospecting of Plant Growth Promoting Bacilli and Related Genera Prevalent in Soils of Pristine Sacred Groves: Biochemical and Molecular Approach. PLoS ONE 2016, 11, e0152951. [Google Scholar] [CrossRef] [PubMed]
- Myers, M.D.; Leake, J.R. Phosphodiesters as mycorrhizal P sources. II. Ericoid mycorrhiza and the utilization of nuclei as a phosphorus and nitrogen source by Vaccinium macrocarpon. New Phytologist. 1996, 132, 445–451. [Google Scholar] [CrossRef]
- Papik, J.; Folkmanova, M.; Polivkova-Majorova, M.; Suman, J.; Uhlik, O. The invisible life inside plants: Deciphering the riddles of endophytic bacterial diversity. Biotechnol. Adv. 2020, 44, 107614. [Google Scholar] [CrossRef]
- Radha, S.; Ashok, D.K. Diversity and Applications of Endophytic Actinobacteria of Plants in Special and Other Ecological Niches. Front. Microbiol. 2018, 9, 1767. [Google Scholar] [CrossRef]
- Ek-Ramos, M.J.; Gomez-Flores, R.; Orozco-Flores, A.A.; Rodríguez-Padilla, C.; González-Ochoa, G.; Tamez-Guerra, P. Bioactive Products From Plant-Endophytic Gram-Positive Bacteria. Front. Microbiol. 2019, 10, 463. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-Y.; Liu, X.-Y.; Liu, S.-J. Agrococcus terreus sp. nov. and Micrococcus terreus sp. nov., isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2010, 60, 1897–1903. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Janarthine, S.R.S.; Eganathan, P. Plant Growth Promoting of Endophytic Sporosarcina aquimarina SjAM16103 Isolated from the Pneumatophores of Avicennia marina L. Int. J. Microbiol. 2012, 2012, 532060. [Google Scholar] [CrossRef] [PubMed]
- Kukkurainen, S.; Leino, A.; Vähämiko, S.; Kärkkäinen, H.R.; Ahanen, K.; Sorvari, S.; Rugienius, R.; Toldi, O. Occurrence and Location of Endophytic Bacteria in Garden and Wild Strawberry. HortScience 2005, 40, 348–352. [Google Scholar] [CrossRef]
- Thomas, P.; Soly, T.A. Endophytic Bacteria Associated with Growing Shoot Tips of Banana (Musa sp.) cv. Grand Naine and the Affinity of Endophytes to the Host. Microb. Ecol. 2009, 58, 952–964. Available online: http://www.jstor.org/stable/27770582 (accessed on 26 November 2021). [CrossRef] [PubMed]
- Xia, Y.; Greissworth, E.; Mucci, C.; Williams, M.A.; Debolt, S. Characterization of culturable bacterial endophytes of switchgrass (Panicum virgatum L.) and their capacity to influence plant growth. Glob. Change Biol. Bioenergy 2013, 5, 674–682. [Google Scholar] [CrossRef]
- Pacifico, D.; Squartini, A.; Crucitti, D.; Barizza, E.; Lo Schiavo, F.; Muresu, R.; Carimi, F.; Zottini, M. The Role of the Endophytic Microbiome in the Grapevine Response to Environmental Triggers. Front. Plant Sci. 2019, 10, 1256. [Google Scholar] [CrossRef] [PubMed]
- Wani, Z.A.; Ashraf, N.; Mohiuddin, T.; Riyaz-Ul-Hassan, S. Plant-endophyte symbiosis, an eco-logical perspective. Appl. Microbiol. Biotechnol. 2015, 99, 2955–2965. [Google Scholar] [CrossRef]
- Miliute, I.; Buzaite, O.; Gelvonauskiene, D.; Sasnauskas, A.; Stanys, V.; Baniulis, D. Plant growth promoting and antagonistic properties of endophytic bacteria isolated from domestic apple. Zemdirbyste 2016, 103, 77–82. [Google Scholar] [CrossRef]
- Ponnuraj, K.; Nessi, C.; Setlow, P.; Jedrzejas, M.J. Structural studies of a novel germination protease from spores of Bacillus megaterium. J. Struct. Biol. 1999, 125, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Kamasaka, H.; Sagimoto, K.; Takata, H.; Nishimura, T.; Kuirk, T. Bacillus stearothermophilus Neopullulase selective hydrolysis of amulose to maltose in the presence of amylopectin. Appl. Environ. Microbiol. 2002, 6, 1658–1664. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Traving, S.J.; Thygesen, U.H.; Riemann, L.; Stedmon, C.A. A model of extracellular enzymes in free-living microbes: Which strategy pays off? Appl. Environ. Microbiol. 2015, 81, 7385–7393. [Google Scholar] [CrossRef]
- Sturz, A.V.; Christie, B.R.; Nowak, J. Bacterial endophytes: Potential role in developing sustain-able systems of crop production. CRC Crit Rev Plant Sci 2000, 19, 1–30. [Google Scholar] [CrossRef]
- Vijayalakshmi, R.; Kairunnisa1, K.; Sivvaswamy, N.S.; Dharan, S.S.; Natarajan, S. Enzyme production and antimicrobial activity of endophytic bacteria isolated from medicinal plants. Indian J. Technol. 2016, 9, 1–8. [Google Scholar] [CrossRef]
- Sudewi, S.; Ala, A.; Baharuddin, F.M. The isolation, characterization endophytic bacteria from roots of local rice plant Kamba in, Central Sulawesi, Indonesia. Biodiversitas 2020, 21, 1614–1624. [Google Scholar] [CrossRef]
- Babiker, B.M.; Ahmed, M.A.E.; Ibrahim, H.M. Isolation & identification of catalase producing Bacillus spp.: A comparative study. Int. J. Adv. Res. 2016, 4, 1206–1211. Available online: https://www.journalijar.com/uploads/493_IJAR-8701.pdf (accessed on 26 November 2021).
- Bouffaud, M.L.; Renoud, S.; Dubost, A.; Moënne-Loccoz, Y.; Muller, D. Aminocyclopropane-1-carboxylate deaminase producers associated to maize and other Poaceae species. Microbiome 2018, 6, 114. [Google Scholar] [CrossRef] [PubMed]
- Zahir, Z.A.; Ghani, U.; Naveed, M.; Nadeem, S.M.; Asghar, H.N. Comparative effectiveness of Pseudomonas and Serratia sp. containing ACC-deaminase for improving growth and yield of wheat (Triticum aestivum L.) under salt-stressed conditions. Arch. Microbiol. 2009, 191, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.G.; Kim, W.T.; Yun, H.S.; Chang, S.C. Use of plant growth-promoting rhizobacteria to control stress responses of plant roots. Plant Biotechnol. Rep. 2010, 4, 179–183. [Google Scholar] [CrossRef]
- Neal, A.L.; Blackwell, M.; Akkari, E.; Guyomar, S.; Clark, I.; Hirsch, P.R. Phylogenetic distribution, biogeography and the effects of land management upon bacterial non-specific Acid phosphatase Gene diversity and abundance. Plant Soil 2018, 427, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
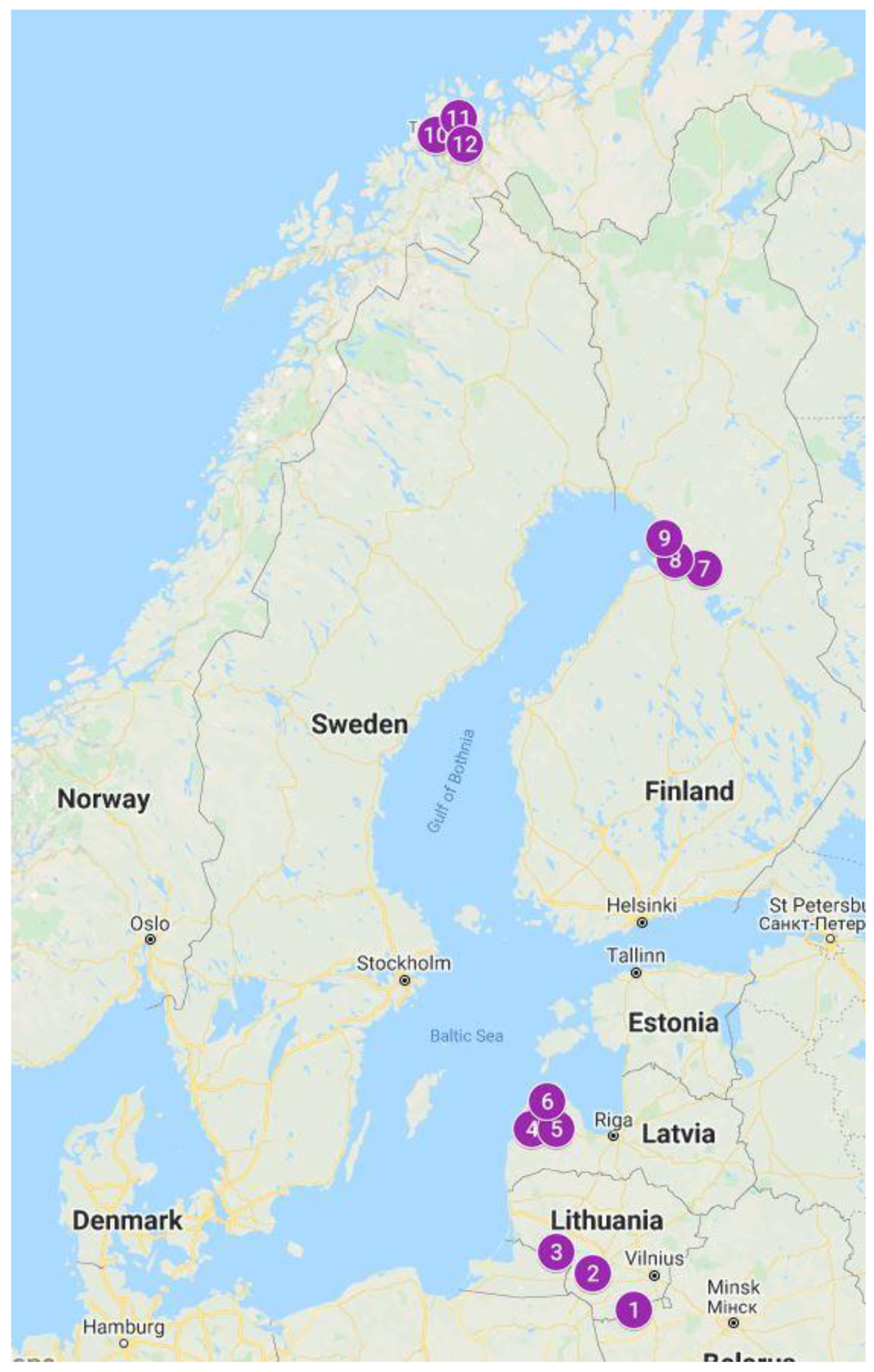
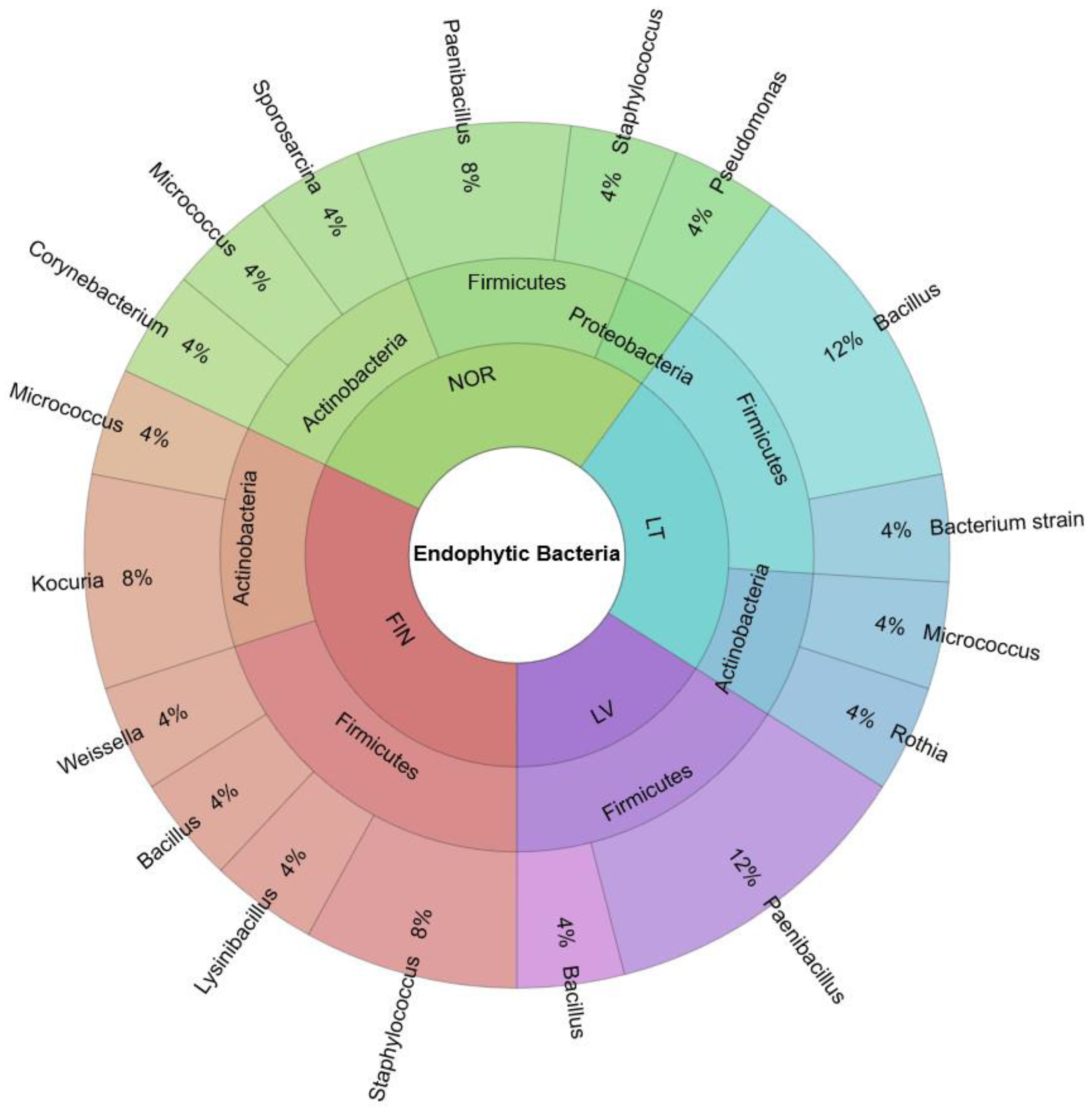
| Isolate | Accession Number in NCBI | Identity Accessions, According NCBI | Sequence Length, bp (Identity, %) |
|---|---|---|---|
| Bil-LT1_1 | MZ469297 | Bacillus halotolerans MK517597.1 B. mojavensis MF040286.1 B. velezensis MT634548.1 B. axarquiensis GU568194.1 B. subtilis AB526464.1 | 1437 (100) |
| Bil-LT1_2 | MZ469298 | Bacillus simplex LK391525.1 Peribacillus butanolivorans CP050509.1 | 1431 (100) |
| Bil-LT4_1 | MZ469299 | Rothia amarae MG905369.1 | 1400 (99.79) |
| Bil-LT4_3 | MZ469300 | Bacterium strain MTL8-4 MH151301.1 | 1439 (99.58) |
| Bil-LT4_7 | MZ469301 | Bacillus zhangzhouensis MN826587.1 B. pumilus CP054310.1 B. safensis KJ542766.1 B. stratosphericus KY203662.1 | 1420 (100) |
| Bil-LT4_8 | MZ469302 | Micrococcus sp. MG132043.1 M. luteus AJ409096.1 | 1398 (100) |
| Bil-LV3_1 | MZ469303 | Bacillus sp. strain MK736127.1 B. aryabhattai MN515130.1 B. megaterium MF988696.1 | 1427 (100) |
| Bil-LV3_3 | MZ469304 | Paenibacillus tundrae HF545335.1 | 1431 (100) |
| Bil-LV3_4 | MZ469305 | Paenibacillus sp. MK290403.1 | 1435 (99.65) |
| Bil-LV3_6 | MZ469306 | Paenibacillus sp. MG758020.1 | 1450 (99.86) |
| Bil-FIN2_3 | MZ469307 | Bacillus cereus MN068934.1 B. thuringiensis CP050183.1 | 1439 (100) |
| Bil-FIN-2_5 | MZ469308 | Staphylococcus warneri CP038242.1 S. pasteuri MW433878.1 | 1437 (100) |
| Bil-FIN2_6 | MZ469309 | Staphylococcus warneri CP038242.1 S. pasteuri MW433878.1 | 1437 (100) |
| Bil-FIN2_7 | MZ469310 | Lysinibacillus macrolides MH542661.1 L. xylanilyticus KP644237.1 L. fusiformis FJ641020.1 | 1427 (100) |
| Bil-FIN2_9 | MZ469311 | Kocuria kristinae KX055834.1 | 1384 (100) |
| Bil-FIN2_10 | MZ469312 | Weissella hellenica CP042399.1 | 1447 (100) |
| Bil-FIN2_12 | MZ469313 | Micrococcus terreus KJ781899.1 | 1385 (100) |
| Bil-FIN2_13 | MZ469314 | Kocuria kristinae KX055834.1 | 1384 (100) |
| Bil-NOR3_11 | MZ469315 | Staphylococcus sp. KM253075.1 | 1431 (100) |
| Bil-NOR3_13 | MZ469316 | Corynebacterium freneyi EF462412.1 | 1393 (99.86) |
| Bil-NOR3_14 | MZ469317 | Micrococcus sp. KX350143.1 M. luteus MN826463.1 | 1385 (100) |
| Bil-NOR3_15 | MZ469318 | Pseudomonas monteilii CP013997.1 | 1422 (100) |
| Bil-NOR3_16 | MZ469319 | Sporosarcina aquimarina MK726086.1 | 1433 (100) |
| Bil-NOR3_17 | MZ469320 | Paenibacillus xylanexedens CP018620.1 | 1436 (99.79) |
| Bil-NOR3_18 | MZ469321 | Paenibacillus sp. KR055031.1 | 1427 (99.86) |
| Endophytic Bacteria Strains in Different Geographic Locations | Amylolytic Activity, mm | Proteolytic Activity, mm | Catalase Reaction | Gene acdS | Gene AcPho |
|---|---|---|---|---|---|
| Bacillus sp. Bil-LT1_1 | 11.9 ± 0.1 | 10.0 ± 0.1 | + | + | - |
| Bacillus sp. Bil-LT1_2 | + | - | - | ||
| Rothia amarae Bil-LT4_1 | 12.0 ± 0.2 | 11.8 ± 0.2 | + | - | - |
| Bacterium strain Bil-LT4_3 | 9.8 ± 0.1 | + | - | - | |
| Bacillus sp. Bil-LT4_7 | 9.0 ± 0.2 | 10.2 ± 01 | + | - | - |
| Micrococcus sp. Bil-LT4_8 | 9.3 ± 0.1 | + | - | - | |
| Bacillus sp. Bil-LV3_1 | 12.5 ± 0.1 | 14.2 ± 0.2 | + | - | - |
| Paenibacillus tundrae Bil-LV3_3 | 10.1 ± 0.2 | 11.9 ± 0.2 | + | - | - |
| Paenibacillus sp. Bil-LV3_4 | 12.1 ± 0.1 | 10.2 ± 0.1 | + | - | - |
| Paenibacillus sp. Bil-LV3_6 | 10.2 ± 0.1 | - | - | - | |
| Bacillus sp. Bil-FIN2_3 | 12.3 ± 0.2 | 14.3 ± 0.3 | - | + | + |
| Staphylococcus sp. Bil-FIN2_5 | + | - | - | ||
| Staphylococcus sp. Bil-FIN2_6 | 11.9 ± 0.2 | + | - | - | |
| Lysinibacillus sp. Bil-FIN2_7 | + | - | - | ||
| Kocuria kristinae Bil-FIN2_9 | + | + | - | ||
| Weissella hellenica Bil-FIN2_10 | - | - | - | ||
| Micrococcus terreus Bil-FIN2_12 | 9.5 ± 0.1 | + | - | + | |
| Kocuria kristinae Bil-FIN2_13 | 8.8 ± 0.1 | + | - | + | |
| Paenibacillus sp. Bil-NOR3_17 | 10.1 ± 0.2 | + | - | - | |
| Paenibacillus sp. Bil-NOR3_18 | + | - | - | ||
| Corynebacterium sp. Bil-NOR3_13 | 12.2 ± 0.1 | + | - | - | |
| Micrococcus sp. Bil-NOR3_14 | 10.3 ± 0.2 | 14.5 ± 0.2 | + | - | - |
| Staphylococcus sp. Bil-NOR3_11 | 10.1 ± 0.1 | + | - | - | |
| Pseudomonas monteilii Bil-NOR3_15 | + | - | - | ||
| Sporosarcina aquimarina Bil-NOR3_16 | + | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mažeikienė, I.; Frercks, B.; Burokienė, D.; Mačionienė, I.; Šalaševičienė, A. Endophytic Community Composition and Genetic-Enzymatic Features of Cultivable Bacteria in Vaccinium myrtillus L. in Forests of the Baltic-Nordic Region. Forests 2021, 12, 1647. https://doi.org/10.3390/f12121647
Mažeikienė I, Frercks B, Burokienė D, Mačionienė I, Šalaševičienė A. Endophytic Community Composition and Genetic-Enzymatic Features of Cultivable Bacteria in Vaccinium myrtillus L. in Forests of the Baltic-Nordic Region. Forests. 2021; 12(12):1647. https://doi.org/10.3390/f12121647
Chicago/Turabian StyleMažeikienė, Ingrida, Birutė Frercks, Daiva Burokienė, Irena Mačionienė, and Alvija Šalaševičienė. 2021. "Endophytic Community Composition and Genetic-Enzymatic Features of Cultivable Bacteria in Vaccinium myrtillus L. in Forests of the Baltic-Nordic Region" Forests 12, no. 12: 1647. https://doi.org/10.3390/f12121647
APA StyleMažeikienė, I., Frercks, B., Burokienė, D., Mačionienė, I., & Šalaševičienė, A. (2021). Endophytic Community Composition and Genetic-Enzymatic Features of Cultivable Bacteria in Vaccinium myrtillus L. in Forests of the Baltic-Nordic Region. Forests, 12(12), 1647. https://doi.org/10.3390/f12121647








