BpTCP3 Transcription Factor Improves Salt Tolerance of Betula platyphylla by Reducing Reactive Oxygen Species Damage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Bioinformatics Analysis
2.3. Gene Cloning and Vector Construction
2.4. Genetic Transformation of B. platyphylla
2.5. Analysis of the Activity of the GUS Reporter Gene
2.6. RT-qPCR
2.7. Growth Measurement and Physiological Traits of BpTCP3 Transgenic B. platyphylla under Salt Stress
2.8. Statistical Analysis
3. Results
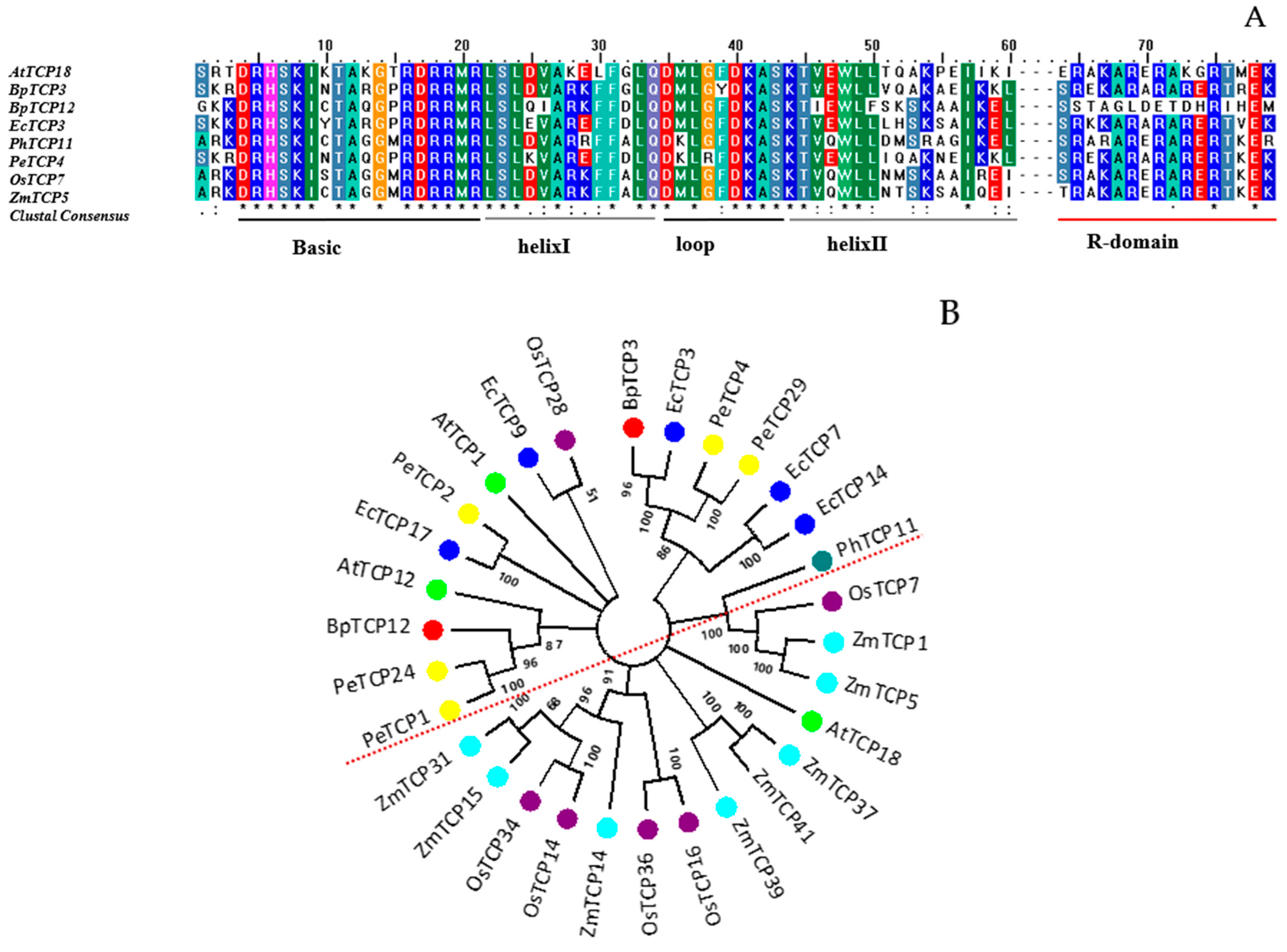
3.1. Structural Identification and Phylogeny of the CYC/TB1 Subclass TCP Genes in B. platyphylla
3.2. The BpTCP3 Promoter Contains Multiple Hormones and Stress-Response Elements
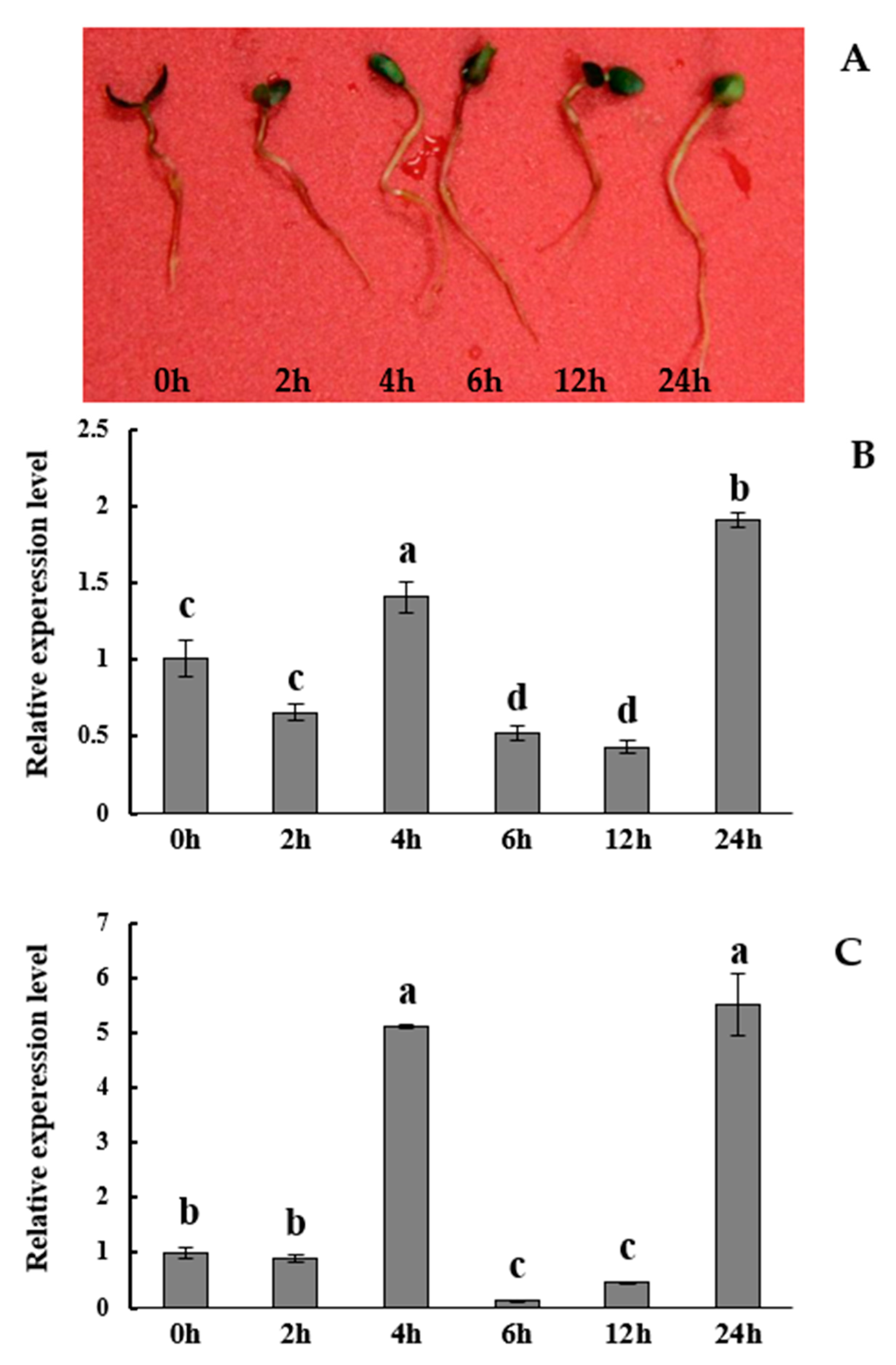
3.3. The BpTCP3 Gene Participates in Salt Treatment Response
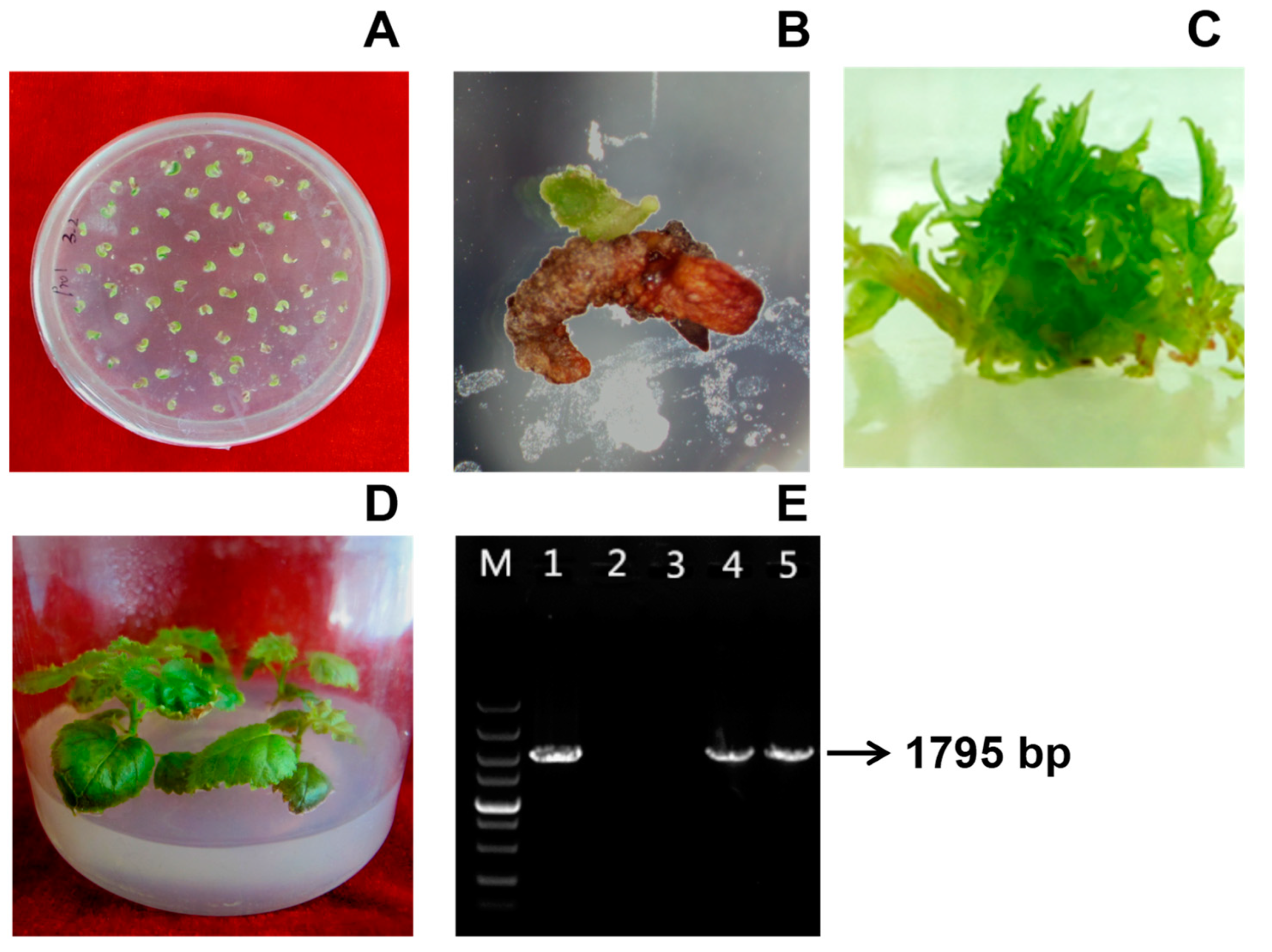
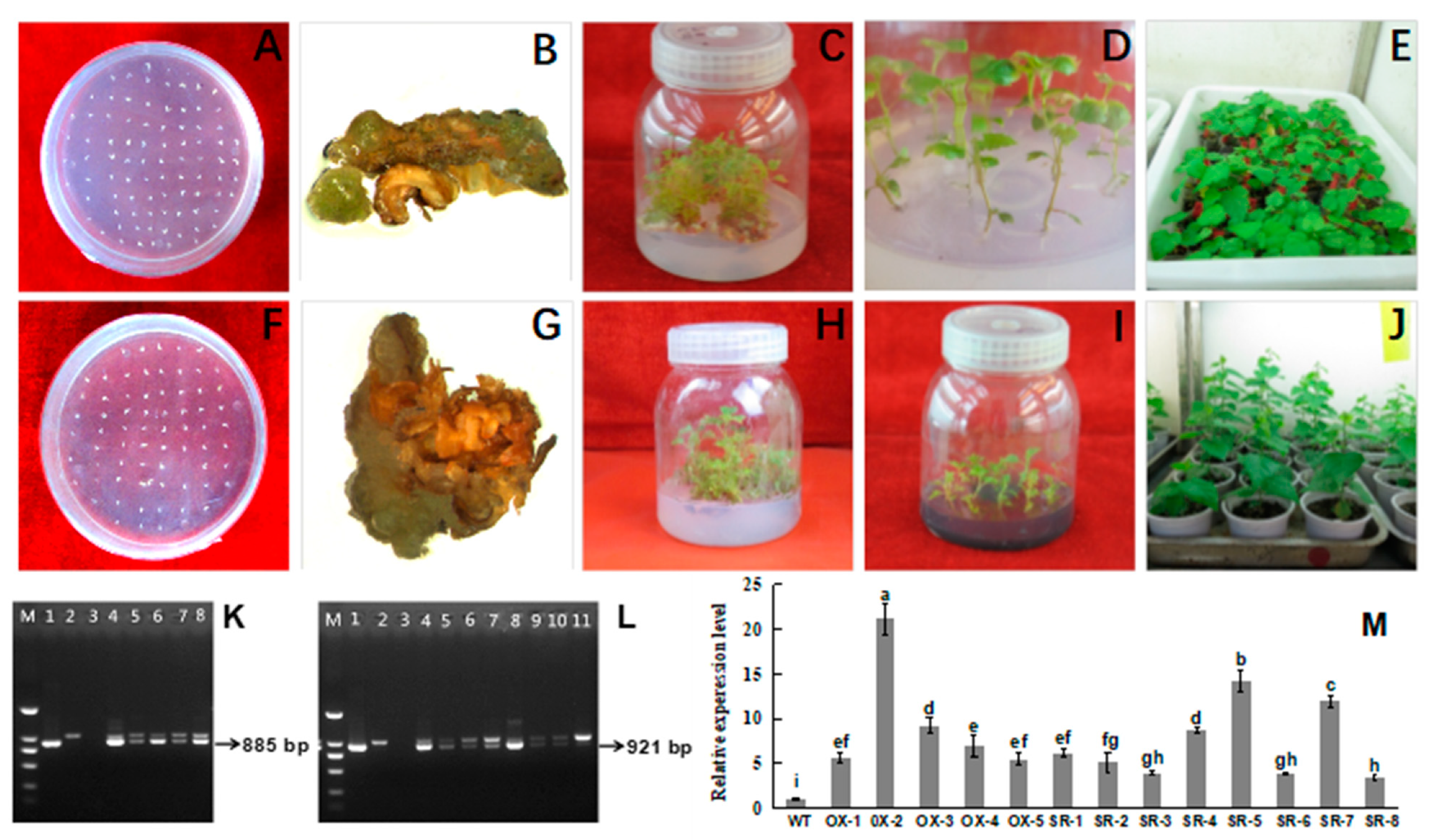
3.4. S::BpTCP3 and 35S::BpTCP3-SRDX Transgenic B. platyphylla Were Obtained
3.5. Salt Treatments Affected the Growth of BpTCP3 Transgenic Lines
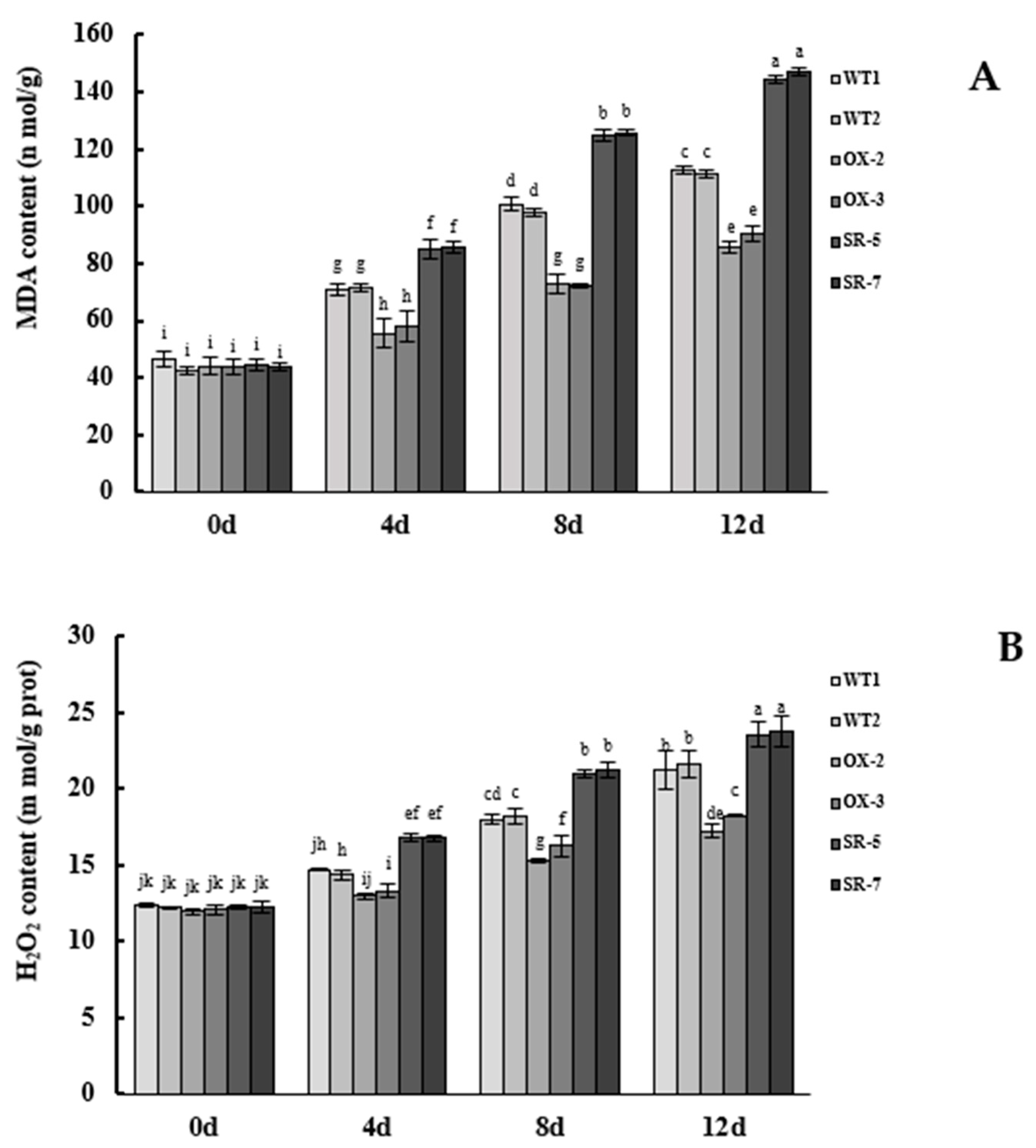
3.6. The BpTCP3 Gene Reduced Reactive Oxygen Species Damage and Improved the Salt Resistance of B. platyphylla
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Gao, W.D.; Bai, S.; Li, Q.M.; Gao, C.Q.; Liu, G.F.; Li, G.D.; Tan, F.L. Overexpression of TaLEA Gene from Tamarix androssowii Improves Salt and Drought Tolerance in Transgenic Poplar (Populus simonii × P. nigra). PLoS ONE 2013, 8, e67462. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.J.; Zhou, B.R.; Zhang, X.M.; Zhao, K.; Cheng, Z.H.; Jiang, T.B. Transcriptome analysis of transcription factor genes under multiple abiotic stresses in Populus simonii × P. nigra. Gene 2019, 707, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.J.; Li, C.Z.; Lin, S.Y.; Wang, J.P.; Zhou, B.R.; Jiang, T.B. Transcriptome analysis of salt-responsive and wood-associated NACs in Populus simonii × Populus nigra. BMC Plant Biol. 2020, 20, 317. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhao, M.Z.; Chen, M.Y.; Li, L.; Jiang, Y.; Liu, S.Z.; Jiang, Y.; Wang, K.Y.; Wang, Y.F.; Sun, C.Y.; et al. The bHLH gene family and its response to saline stress in Jilin ginseng, Panax ginseng C.A. Meyer. Mol. Genet. Genom. 2020, 295, 877–890. [Google Scholar] [CrossRef]
- Chen, J.F.; Zhang, J.; Hu, J.J.; Xiong, W.W.; Du, C.G.; Lu, M.Z. Integrated regulatory network reveals the early salt tolerance mechanism of Populus euphratica. Sci. Rep. 2017, 7, 6769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999, 18, 215–222. [Google Scholar] [CrossRef] [Green Version]
- Navaud, O.; Dabos, P.; Carnus, E.; Tremousaygue, D.; Herve, C. TCP Transcription Factors Predate the Emergence of Land Plants. J. Mol. Evol. 2007, 65, 23–33. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2010, 30, 337–348. [Google Scholar] [CrossRef]
- Martin-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- van Es, S.W.; van der Auweraert, E.B.; Silveira, S.R.; Angenent, G.C.; van Dijk, A.D.J.; Immink, R.G.H. Comprehensive phenotyping reveals interactions and functions of Arabidopsis thaliana TCP genes in yield determination. Plant J. 2019, 99, 316–328. [Google Scholar] [PubMed] [Green Version]
- Tatematsu, K.; Nakabayashi, K.; Kamiya, Y.; Nambara, E. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 2008, 53, 42–52. [Google Scholar] [CrossRef]
- Bresso, E.; Chorostecki, U.; Rodriguez, R.E.; Palatnik, J.F.; Schommera, C. Spatial control of gene expression by miR319-regulated TCP transcription factors in leaf development. Plant Physiol. 2018, 176, 1694–1708. [Google Scholar] [CrossRef]
- Liu, S.R.; Mi, X.Z.; Zhang, R.; An, Y.L.; Zhou, Q.Y.; Yang, T.Y.; Xia, X.B.; Guo, R.; Wang, X.W.; Wei, C.L. Integrated analysis of miRNAs and their targets reveals that miR319c/TCP2 regulates apical bud burst in tea plant (Camellia sinensis). Planta 2019, 250, 1111–1129. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.Z.; Zhang, Y.X.; Wang, W.Y.; Irish, V.F.; Huang, T.B. RABBIT EARS regulates the transcription of TCP4 during petal development in Arabidopsis. J. Exp. Bot. 2016, 67, 6473–6480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Choi, M.H.; Noh, B.; Noh, Y.S. De Novo Shoot Regeneration Controlled by HEN1 and TCP3/4 in Arabidopsis. Plant Cell Physiol. 2020, 61, 1600–1613. [Google Scholar] [CrossRef]
- Sarvepalli, K.; Nath, U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 2011, 67, 595–607. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.J.; Xu, G.X.; Zhao, X.H.; Zhang, Z.J.; Wang, X.X.; Liu, X.; Xiao, W.; Fu, X.L.; Chen, X.D.; Gao, D.S.; et al. TCP transcription factor PpTCP20 is involved in peach bud endodormancy by inhibiting PpDAM5/PpDAM6 and interacting with PpABF2. J. Exp. Bot. 2019, 71, 1585–1597. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Furutani, M.; Tasaka, M.; Ohme-Takagi, M. TCP Transcription Factors Control the Morphology of Shoot Lateral Organs via Negative Regulation of the Expression of Boundary-Specific Genes in Arabidopsis. Plant Cell 2007, 19, 473–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.D.; Wang, C.D.; Xiang, N.; Li, X.; Yang, S.H.; Du, J.C.; Yang, Y.P.; Yang, Y.Q. Activation of secondary cell wall biosynthesis by miR319-targeted TCP4 transcription factor. Plant Biotechnol. J. 2017, 15, 1284–1294. [Google Scholar] [CrossRef] [Green Version]
- Guan, P.Z.; Ripoll, J.J.; Wang, R.H.; Vuong, L.; Bailey-Steinitz, L.J.; Ye, D.N.; Crawford, N.M. Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natls. Acad. Sci. USA 2017, 114, 2419–2424. [Google Scholar] [CrossRef] [Green Version]
- Fan, D.; Ran, L.Y.; Hu, J.; Ye, X.; Xu, D.; Li, J.Q.; Su, H.L.; Wang, X.Q.; Ren, S.; Luo, K.M. The miR319a/TCP module and DELLA protein regulate synergistically trichome initiation and improve insect defenses in Populus tomentosa. New Phytol. 2020, 227, 867–883. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Maurya, J.P.; Azeez, A.; Miskolczi, P.; Tylewicz, S.; Stojkovic, K.; Delhomme, N.; Busov, V.; Bhalerao, R.P. A genetic network mediating the control of bud break in hybrid aspen. Nat. Commun. 2018, 9, 4173. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Son, G.H.; Bhattacharjee, S.; Kim, H.J.; Nam, J.C.; Nguyen, P.D.T.; Hong, J.C.; Gassmann, W. The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J. 2014, 78, 978–989. [Google Scholar] [CrossRef]
- Jiang, J.; Yang, G.P.; Liu, G.F.; Liu, Y.X.; Ren, X.Q. Analysis of genetic variation within and among Betula platyphylla provenaces in Northeast China using RAPD markers. J. Northeast. For. Univ. 2001, 29, 30–34. [Google Scholar]
- Li, Z.X.; Pei, X.N.; Yin, S.P.; Lang, X.B.; Zhao, X.Y.; Qu, G.Z. Plant hormone treatments to alleviate the effects of salt stress on germination of Betula platyphylla seeds. J. For. Res. 2019, 30, 779–787. [Google Scholar] [CrossRef]
- Xing, B.Y.; Gu, C.R.; Zhang, T.X.; Zhang, Q.Z.; Yu, Q.B.; Jiang, J.; Liu, G.F. Functional Study of BpPP2C1 Revealed Its Role in Salt Stress in Betula platyphylla. Front. Plant Sci. 2021, 11, 617635. [Google Scholar] [CrossRef]
- Li, H.Y.; Yuan, H.M.; Liu, F.M.; Luan, J.Y.; Yang, Y.; Ren, L.; An, L.J.; Jiang, J. BpTCP7 gene from Betula platyphylla regulates tolerance to salt and drought stress through multiple hormone pathways. Plant Cell Tissue Organ Cult. 2020, 141, 17–30. [Google Scholar] [CrossRef]
- Dong, J.X.; Ren, L.; Zhang, Y. Bioinformatics and expression analysis of BpTCPs in Betula platyphylla Suk. J. Nanjing For. Univ. 2018, 42, 113–118. [Google Scholar]
- Tao, J.; Zhan, Y.G.; Jiang, J.; Yang, C.P.; Liu, Y.X. Tissue Culture and Regenerative System of Betula platyphylla Suks (Ⅰ). J. Northeast. For. Univ. 1998, 26, 6–9. [Google Scholar]
- Wang, L.Q.; Xu, C.X.; Wang, C.; Wang, Y.C. Characterization of a eukaryotic translation initiation factor 5A homolog from Tamarix androssowii involved in plant abiotic stress tolerance. BMC Plant Biol. 2012, 12, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, K.; Chen, S.; Huang, H.J.; Jiang, J.; Yuan, H.M.; Li, H.Y. Molecular characterization and expression analysis of the SPL gene family with BpSPL9 transgenic lines found to confer tolerance to abiotic stress in Betula platyphylla Suk. Plant Cell Tissue Organ Cult. 2017, 130, 469–481. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiratsu, K.; Matsui, K.; Koyama, T.; Ohme-Takagi, M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003, 34, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Wu, D.Y.; Wang, Z.J.; Liu, F.F.; Liu, G.F.; Jiang, J. BpMADS12 mediates endogenous hormone signaling: Effect on plant development Betula platyphylla. Plant Cell Tissue Organ Cult. 2016, 124, 169–180. [Google Scholar] [CrossRef]
- Liu, C.Y.; Xu, H.W.; Jiang, J.; Wang, S.; Liu, G.F. Analysis of the promoter features of BpCUC2 in Betula platyphylla × Betula pendula. Plant Cell Tissue Organ Cult. 2017, 132, 1–9. [Google Scholar] [CrossRef]
- Jefferson, R.A. Assaying chimeric genes in plants: The GUS Gene Fusion System. Plant Mol. Biol. Report. 1987, 5, 387–405. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef]
- Zhao, H.; Jiang, J.; Li, K.L.; Liu, G.F. Populus simonii x Populus nigra WRKY70 is involved in salt stress and leaf blight disease responses. Tree Physiol. 2017, 37, 827–844. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.M.; Wang, C.; Guo, H.Y.; Wang, Y.C. BplMYB46 from Betula platyphylla Can Form Homodimers and Heterodimers and Is Involved in Salt and Osmotic Stresses. Int. J. Mol. Sci. 2019, 20, 1171. [Google Scholar] [CrossRef] [Green Version]
- Gupta, B.; Huang, B.R. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical, and Molecular Characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef] [PubMed]
- Bose, J.; Rodrigo-Moreno, A.; Shabala, S. ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 2014, 65, 1241–1257. [Google Scholar] [CrossRef]
- Shi, H.T.; Ye, T.T.; Chen, F.F.; Cheng, Z.M.; Wang, Y.P.; Yang, P.F.; Zhang, Y.S.; Chan, Z.L. Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis: Effect on arginine metabolism and ROS accumulation. J. Exp. Bot. 2013, 64, 1367–1379. [Google Scholar] [CrossRef] [Green Version]
- Li, S.T.; Zachgo, S. TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 2013, 76, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Lucero, L.E.; Uberti-Manassero, N.G.; Arce, A.L.; Colombatti, F.; Alemano, S.G.; Gonzalez, D.H. TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J. 2015, 84, 267–282. [Google Scholar] [CrossRef]
- Noh, M.; Shin, J.S.; Hong, J.C.; Kim, S.Y.; Shin, J.S. Arabidopsis TCX8 functions as a senescence modulator by regulating LOX2 expression. Plant Cell Rep. 2021, 40, 677–689. [Google Scholar] [CrossRef]
- Mijiti, M.; Zhang, Y.M.; Zhang, C.R.; Wang, Y.C. Physiological and molecular responses of Betula platyphylla Suk to salt stress. Trees Struct. Funct. 2017, 31, 1653–1665. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.H.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukhopadhyay, P.; Tyagi, A.K. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 2015, 5, 12385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Zhang, D.Z.; An, J.X.; Yin, H.J.; Fang, S.; Chu, J.F.; Zhao, Y.D.; Li, J. TCP transcription factors regulate shade avoidance via directly mediating the expression of both PHYTOCHROME INTERACTING FACTORs and auxin biosynthetic genes. Plant Physiol. 2018, 176, 1850–1861. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.S.; Zhu, M.T.; Cai, M.X.; Zhang, W.P.; Fatima, M.; Jia, H.F.; Li, F.F.; Ming, R. Identification and Expression Analysis of TCP Genes in Saccharum spontaneum L. Trop. Plant Biol. 2019, 12, 206–218. [Google Scholar] [CrossRef]
- Xu, Y.Z.; Liu, H.L.; Gao, Y.M.; Xiong, R.; Wu, M.; Zhang, K.M.; Xiang, Y. The TCP transcription factor PeTCP10 modulates salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 2021, 40, 1971–1987. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Sahu, G.; Shaw, B.P. Insight into the cellular and physiological regulatory modulations of Class-I TCP9 to enhance drought and salinity stress tolerance in cowpea. Physiol. Plant. 2021. [Google Scholar] [CrossRef] [PubMed]



| Primer Name | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| ProBpTCP3 | 5′- CCAAGCTTCCCATCAACACCTGTGAAATGC-3′ (HindIII) | 5′- GCTCTAGAGGGTTGGTCTGAATAAGAGATGG-3′ (XbaI) |
| PBI101 | 5′- ACAGGAAACAGCTATGACCATGATTACG-3′ | 5′- TACAGGACGTAACATAAGGGACTGACC-3′ |
| BpTCP3-PCR | 5′- ATGTATTCCTCATCAAATAGTAAC-3′ | 5′-TCAGGCCTCCCATGCCTTGC-3′ |
| BpTCP3-qPCR | 5′- GGGATCGGAGAATGAGACTTTCG-3′ | 5′- CGTAATAAATTGCAACGTCATCGACG-3′ |
| GUS-qPCR | 5′- CTCTATGAACTGTGCGTCACAGC-3′ | 5′-CGAGCATCTCTTCAGCGTAAGG-3′ |
| BpTubulin | 5′-GCACTGGCCTCCAAGGAT-3′ | 5′-TGGGTCGCTCAATGTCAAGG-3′ |
| Gene | 5′UTR (bp) | 3′UTR (bp) | ORF Length | Protein Length (aa) | Molecular Weight (Mw/kD) | Isoelectric Point (pI) | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| BpTCP3 | 20 | 179 | 885 | 294 | 33.29 | 9.38 | Nucleus |
| BpTCP12 | 210 | 41 | 1200 | 399 | 44.13 | 7.65 | Nucleus |
| Element | Element Sequence | Function | Number |
|---|---|---|---|
| TATA-box | ATTATA; TATAA; TATATA; TATA; ATATAT; TATACA; TACAAAA | Promoter core element | 14 |
| CAAT-box | CAAT; CAAAT; CCCAATTT; CAACCAACTCC | Promoter enhancer conserved elements | 30 |
| MYBCORE | CNGTTR; | MYB binding site, involved in regulation of stress | 3 |
| MBS | CAACTG | MYB binding zone, drought-induced regulatory element | 1 |
| GARE-motif | TCTGTTG | Gibberellin-response element | 1 |
| TGA-element | AACGAC | Auxin-response element | 1 |
| TCA-element | CCATCTTTTT | Salicylic-acid response element | 1 |
| CGTCA-motif | CGTCA | Jasmonic acid-response element | 2 |
| TGACG-motif | TGACG | Jasmonic acid-response element | 2 |
| ABRE | ACGTG; CACGTG | Abscisic acid-response element | 4 |
| CAT-box | GCCACT | Cis-acting regulatory element related to specific expression of meristems | 2 |
| CCGTCC-box | CCGTCC | Cis-acting regulatory element related to specific activation of meristems | 1 |
| Skn-1_motif | GTCAT | Cis-acting regulatory elements related to specific expression of endosperm | 2 |
| MRE | AACCTAA | MYB binding site involved in light responsiveness | 1 |
| ARE | AAACCA | Anaerobic response element | 1 |
| GC-motif | CCCCCG | Anaerobic response element | 1 |
| O2-site | GATGACATGG; GATGATGTGG | Cis-acting regulatory element involved in the regulation of protein metabolism | 2 |
| Box 4 | ATTAAT | Light-responsive element | 2 |
| G-Box | CACGTG | Light-responsive element | 1 |
| Line | Seedling Height before Salt Stress (cm) | Seedling Height after Rehydrated in Water (cm) | Absolute Height Growth (cm) | Relative Height Growth |
|---|---|---|---|---|
| WT1 | 26.60 ± 2.39 a | 33.12 ± 2.31 b | 6.52 ± 0.57 b | 0.24 ± 0.036 b |
| WT2 | 26.42 ± 2.40 a | 33.33 ± 2.80 b | 6.91 ± 0.85 b | 0.26 ± 0.037 b |
| OX-2 | 26.61 ± 1.93 a | 35.91 ± 1.74 a | 9.30 ± 0.85 a | 0.35 ± 0.049 a |
| OX-3 | 26.60 ± 1.54 a | 36.16 ± 2.15 a | 9.56 ± 1.34 a | 0.36 ± 0.053 a |
| SR-5 | 26.43 ± 1.62 a | 31.45 ± 1.90 c | 5.02 ± 0.64 c | 0.19 ± 0.024 c |
| SR-7 | 26.54 ± 1.86 a | 31.00 ± 1.75 c | 4.47 ± 0.65 c | 0.17 ± 0.031 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, L.; Li, F.; Jiang, J.; Li, H. BpTCP3 Transcription Factor Improves Salt Tolerance of Betula platyphylla by Reducing Reactive Oxygen Species Damage. Forests 2021, 12, 1633. https://doi.org/10.3390/f12121633
Ren L, Li F, Jiang J, Li H. BpTCP3 Transcription Factor Improves Salt Tolerance of Betula platyphylla by Reducing Reactive Oxygen Species Damage. Forests. 2021; 12(12):1633. https://doi.org/10.3390/f12121633
Chicago/Turabian StyleRen, Li, Fangrui Li, Jing Jiang, and Huiyu Li. 2021. "BpTCP3 Transcription Factor Improves Salt Tolerance of Betula platyphylla by Reducing Reactive Oxygen Species Damage" Forests 12, no. 12: 1633. https://doi.org/10.3390/f12121633
APA StyleRen, L., Li, F., Jiang, J., & Li, H. (2021). BpTCP3 Transcription Factor Improves Salt Tolerance of Betula platyphylla by Reducing Reactive Oxygen Species Damage. Forests, 12(12), 1633. https://doi.org/10.3390/f12121633





