Morpho-Histology, Endogenous Hormone Dynamics, and Transcriptome Profiling in Dacrydium Pectinatum during Male Cone Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
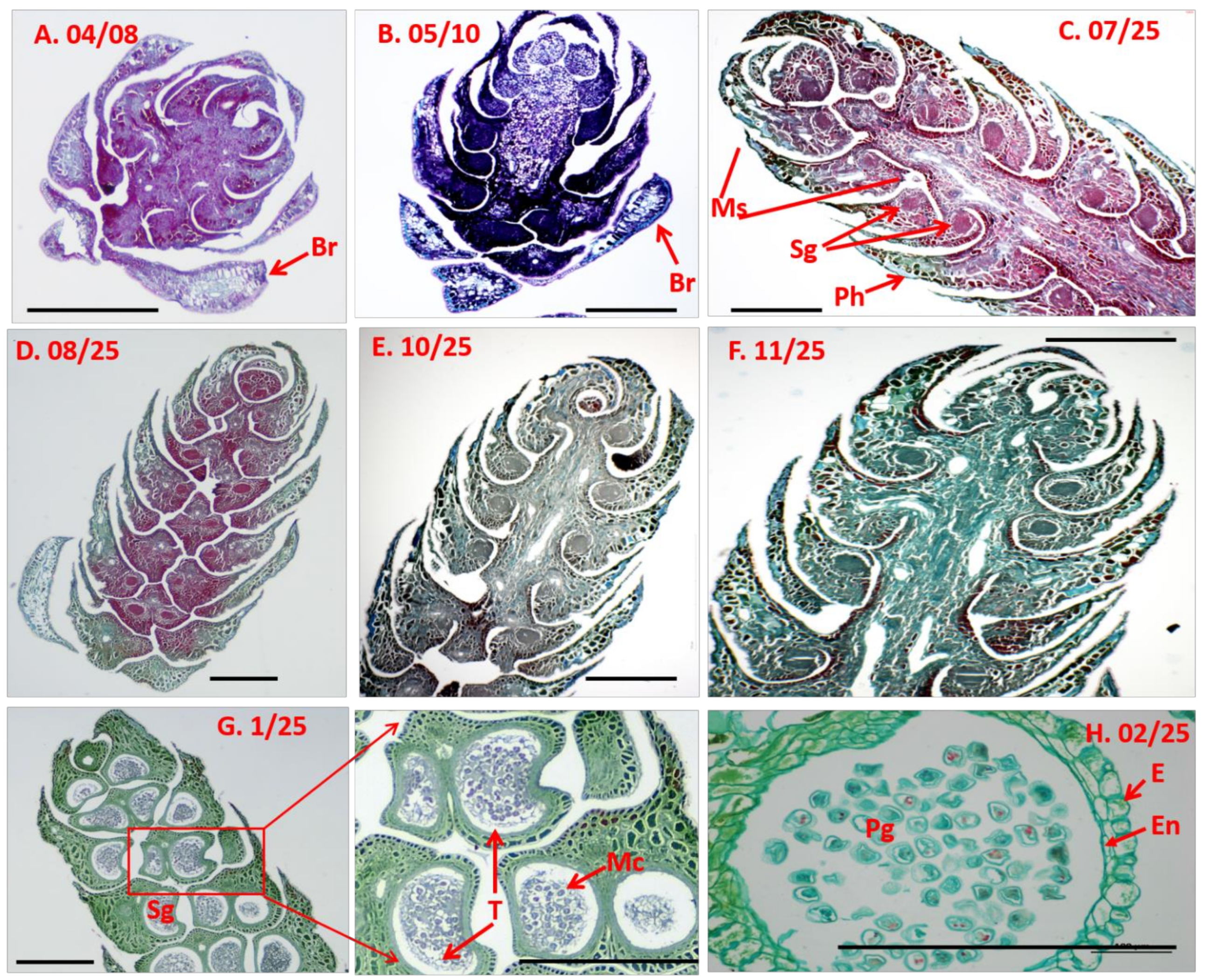
2.2. Semi-Thin Sectioning
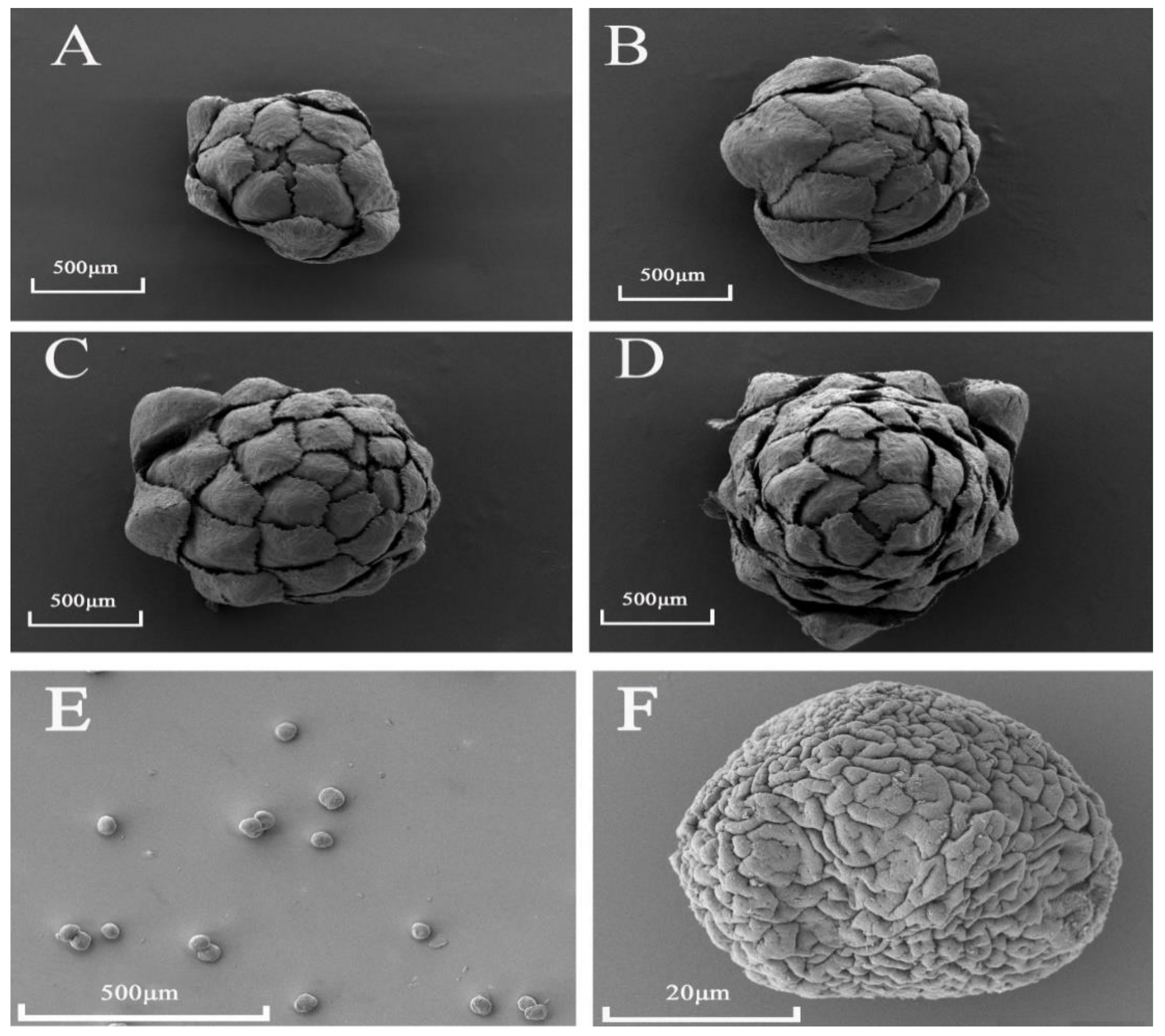
2.3. Observation of Male Cone Morphology by Scanning Electron Microscopy (SEM)
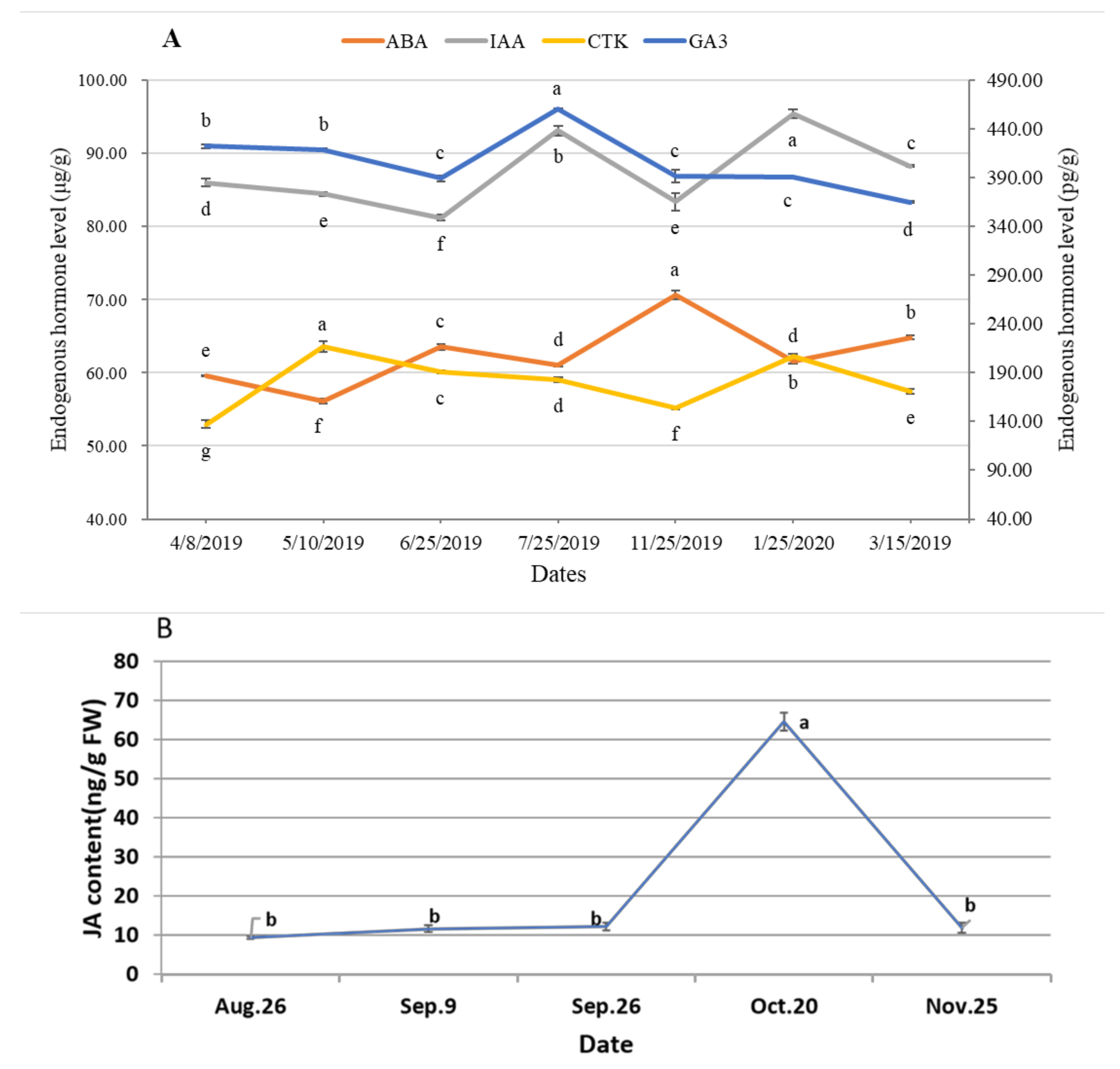
2.4. Detection of Endogenous Hormones
2.5. RNA Sequencing and De Novo Transcriptome Assembly
2.6. Differentially Expressed Gene (DEG) Selection
2.7. Statistical Analysis
3. Results
3.1. Arrangement and Phenology of Male Cones
3.2. Microscopic Anatomy of D. pectinatum Male Cones
3.3. Dynamic of Endogenous Hormones during Male Cone Development
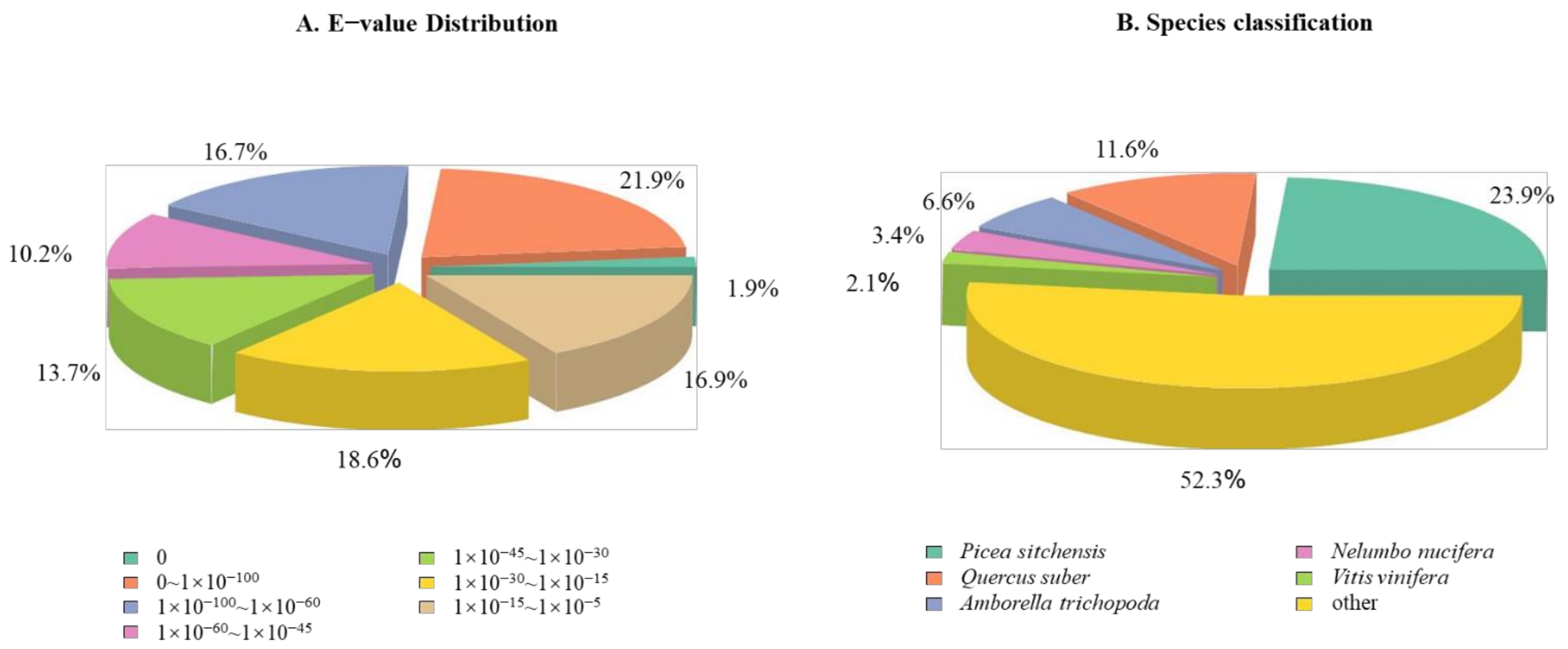
3.4. De Novo Assembly of Dacrydium Transcriptome
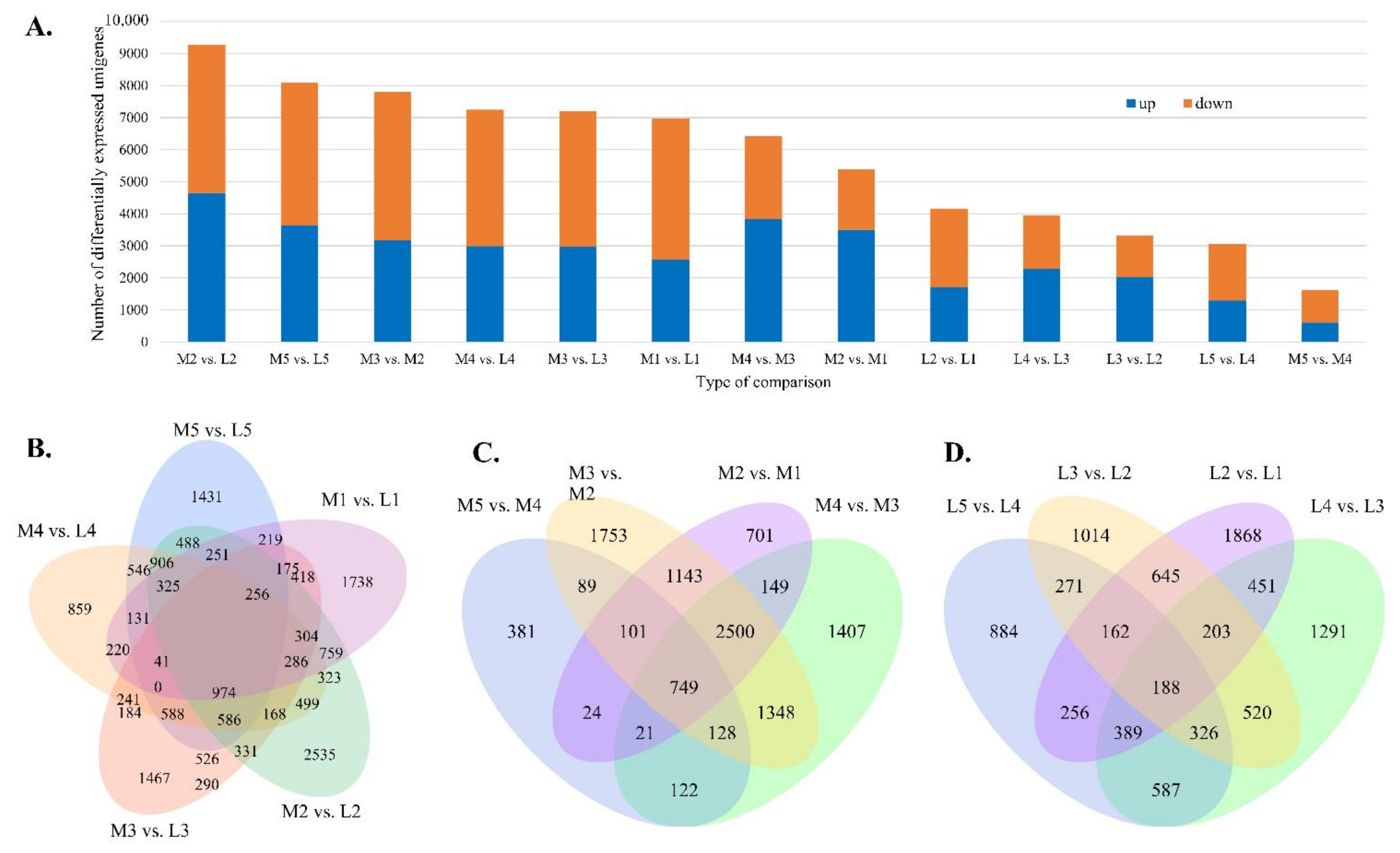
3.5. Differentially Expressed (DE) Dacrydium Unigenes
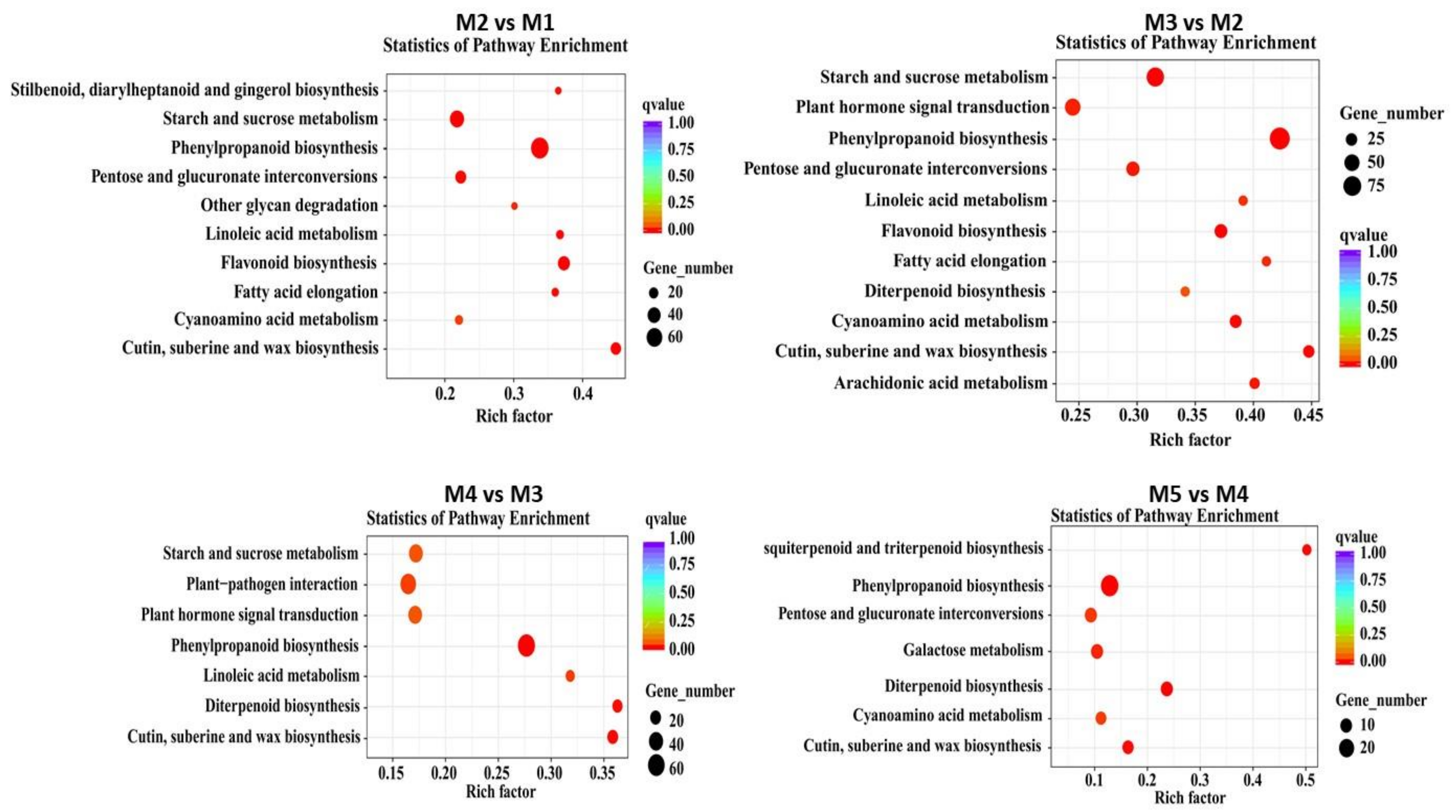
3.6. DE Dacrydium Unigenes Involved in Hormone Metabolism and Hormone Signal Transduction Pathway during Male Cone Development
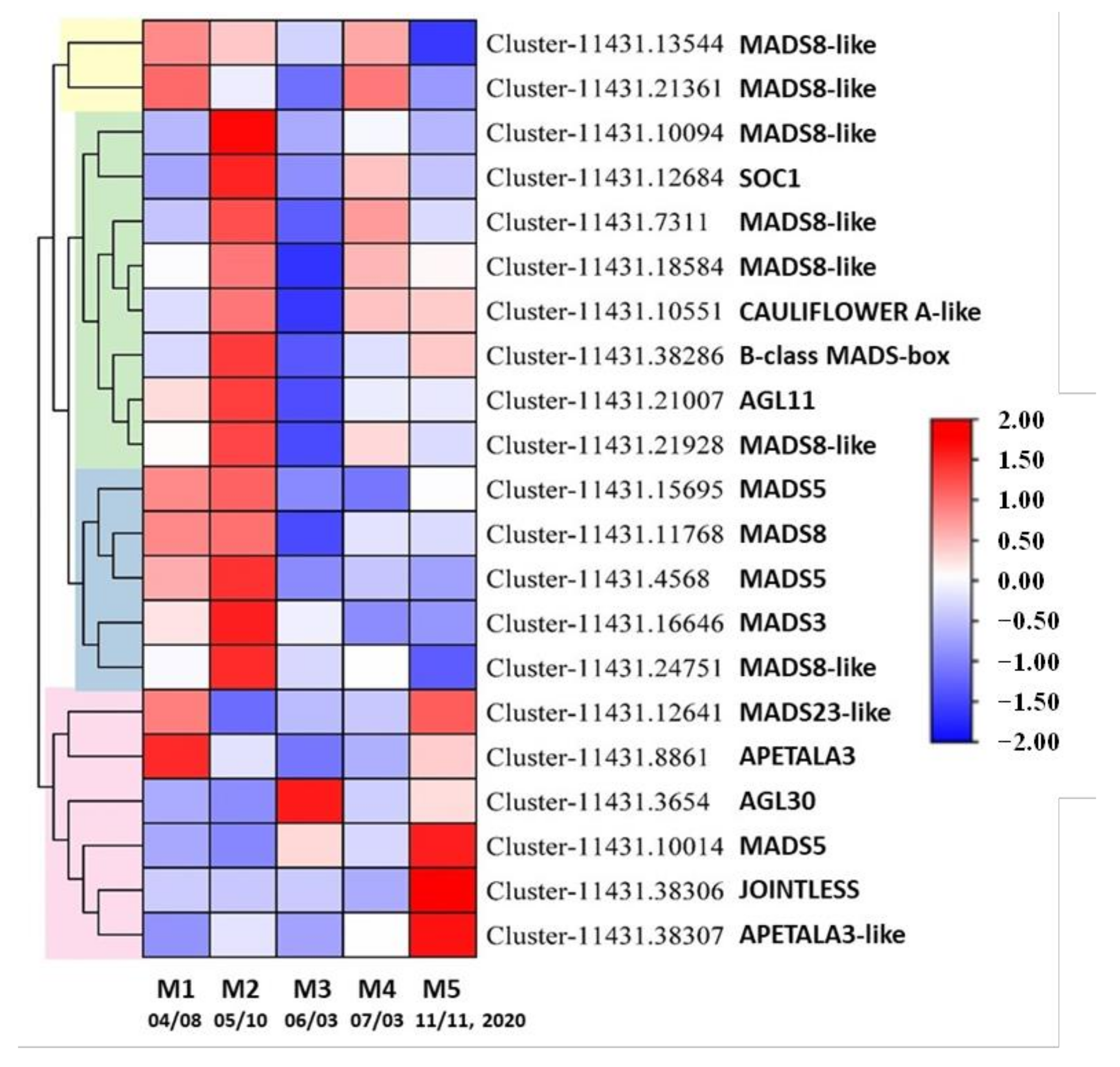
3.7. DE Unigenes Involved in the Development of Reproductive Structures
4. Discussion
4.1. Development of D. pectinatum Male Cone Lasts for a Year and Its Microsporophylls Are Spirally Arranged
4.2. Endogenous Hormones Fluctuate during the Process of Male Cone Development
4.3. RNA Sequencing Reveals Many Dacrydium Genes and SSR Makers
4.4. Gene Expression Is Modulated during D. pectinatum Male Cone Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De, L.; David, J. A revision of the melanesia and pacific rainforest conifers, I. Podocarpaceae, in part. J. Arnold Arboretum. 1969, 50, 274–314. [Google Scholar] [CrossRef]
- Ding, Y.; Zang, R.; Liu, S.; He, F.; Letcher, S.G. Recovery of woody plant diversity in tropical rain forests in southern China after logging and shifting cultivation. Biol. Conserv. 2012, 145, 225–233. [Google Scholar] [CrossRef]
- Wu, C.Y.; Chen, Y.F.; Chen, Q.; Hong, X.J.; Han, W.T.; Li, X.C. Characteristics of seed rain and soil seed bank of Dacrydium pierrei in Bawangling, Hainan. J. Trop. Subtrop. Bot. 2018, 26, 13–23. (In Chinese) [Google Scholar] [CrossRef]
- Chen, Q.; Chen, Y.F.; Wang, W.Q.; Li, Z.C.; Li, X.L.; Han, W.T. An initial report on Dacrydium pierrei seed quantity and quality in Bawangling, Hainan. Forest Sci. Technol. 2016, 10, 29–34. (In Chinese) [Google Scholar] [CrossRef]
- Huang, L.; Deng, Q.; Li, N.; Su, Y.; Wang, T. A set of microsatellite markers developed for Dacrydium pectinatum (Podocarpaceae), a vulnerable conifer in China. Conserv. Genet. Resour. 2014, 6, 167–168. [Google Scholar] [CrossRef]
- Keppel, G.; Prentis, P.; Biffin, E.; Hodgskiss, P.; Tuisese, S.; Tuiwawa, M.V.; Lowe, A.J. Diversification history and hybridisation of Dacrydium (Podocarpaceae) in remote Oceania. Aust. J. Bot. 2011, 59, 262–273. [Google Scholar] [CrossRef]
- Su, Y.J.; Wang, T.; Deng, F. Population genetic variation, differentiation and bottlenecks of Dacrydium pectinatum (Podocarpaceae) in Hainan Island, China: Implications for its conservation. Aust. J. Bot. 2010, 58, 318–326. [Google Scholar] [CrossRef]
- Yang, Y.C.; Zhang, W.Y.; Lin, R.C.; Yang, X.S. Study on structure and species diversity in post harvested tropical montane Rain forest dominated by Dacridium pierrii in Bawangling, Hainan Island. For. Res. 2018, 21, 37–43. [Google Scholar] [CrossRef]
- Liang, H.Y.; Yin, W.L. Flower induction in Metasequoia glyptostroboides Hu et Cheng by fowering regulator. In Growth and Development Control and Biotechnology in Woody Plants; Wang, S.S., Jiang, X.N., Eds.; China Forestry Publishing House: Beijing, China, 1994; pp. 215–219. [Google Scholar]
- Kurita, M.; Mishima, K.; Tsubomura, M.; Takashima, Y.; Nose, M.; Hirao, T.; Takahashi, M. Transcriptome analysis in male strobilus induction by gibberellin treatment in Cryptomeria japonica D. Don. Forests 2020, 11, 633. [Google Scholar] [CrossRef]
- Kang, J.H.; Lee, Y.J.; Oh, B.K.; Lee, S.K.; Hyun, B.R.; Lee, B.W. Microstructure of the water spider (Argyroneta aquatica) using the scanning electron microscope. J. Asia-Pacific Biodivers. 2014, 7, 484–488. [Google Scholar] [CrossRef][Green Version]
- Ma, Z.; Ge, L.; Lee, A.S.Y.; Yong, J.W.H.; Tan, S.N.; Ong, E.S. Simultaneous analysis of different classes of phytohormones in coconut (Cocos nucifera L.) water using high-performance liquid chromatography and liquid chromatography-tandem mass spectrometry after solid-phase extraction. Anal. Chim. Acta 2008, 610, 274–281. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Chen, Y.; Gao, M.; Zhao, Y.; Xu, Z.; Cao, P. Transcriptome analysis of Litsea cubeba floral buds reveals the role of hormones and transcription factors in the differentiation process. G3 Genes Genomes Genet. 2018, 8, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2013, 29, 644–652. [Google Scholar] [CrossRef]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014, 15, 410. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- De Laubenfels, D.J. Coniferales Dordrecht: Kluwer academic. Flora Males 1988, 10, 337–453. [Google Scholar]
- Quinn, C.J. Taxonomy of dacrydium Sol. ex lamb. emend. de laub. (Podocarpaceae). Aust. J. Bot. 1982, 30, 311–320. [Google Scholar] [CrossRef]
- Norton, D.A.; Herbert, J.W.; Beveridge, A.E. The ecology of Dacrydium cupressinam: A review. N. Z. J. Bot. 1988, 26, 37–62. [Google Scholar] [CrossRef]
- Jin, B.; Tang, L.; Lu, Y.; Wang, D.; Zhang, M. Temporal and spatial characteristics of male cone development in Metasequoia glyptostroboides Hu et Cheng. Plant Signal. Behav. 2012, 7, 1687–1694. [Google Scholar] [CrossRef]
- Leslie, A.B. Predation and protection in the macroevolutionary history of conifer cones. Proc. R. Soc. B Biol. Sci. 2011, 278, 3003–3008. [Google Scholar] [CrossRef]
- Leslie, A.B. Shifting functional roles and the evolution of conifer pollen-producing and seed-producing cones. Paleobiology 2011, 37, 587–602. [Google Scholar] [CrossRef]
- Davis, S.J. Integrating hormones into the floral-transition, pathway of Arabidopsis thaliana. Plant Cell Environ. 2009, 32, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Lyons, R. The link between flowering timeand stress tolerance. J. Exp. Bot. 2016, 67, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Lan, Q.; Liu, J.F.; Shi, S.Q.; Deng, N.; Jiang, Z.P.; Chang, E.M. Anatomy, microstructure, and endogenous hormone changes in Gnetum parvifolium during anthesis. J. Syst. Evol. 2018, 56, 14–24. [Google Scholar] [CrossRef]
- Niu, S.; Yuan, L.; Zhang, Y.; Chen, X.; Li, W. Isolation and expression profiles of gibberellin metabolism genes in developing male and female cones of Pinus tabuliformis. Funct. Integr. Genom. 2014, 14, 697–705. [Google Scholar] [CrossRef]
- Kong, L.; Abrams, S.R.; Owen, S.J.; Graham, H.; Von, A.P. Phytohormones and their metabolites during long shoot development in Douglas-fir following cone induction by gibberellin injection. Tree Physiol. 2008, 28, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, M.S.; Adams, G.W.; Gillespie, M.V. Stimulation of flowering by grafted black spruce and white spruce: A comparative study of the effects of gibberellin A4/7, cultural treatments, and environment. Can. J. For. Res. 2011, 21, 395–400. [Google Scholar] [CrossRef]
- Kong, L.; Von, A.P.; Zaharia, I. Analysis of phytohormone profiles during male and female cone initiation and early differentiation in long-shoot buds of lodgepole pine. J. Plant Growth Regul. 2012, 31, 478–489. [Google Scholar] [CrossRef][Green Version]
- Piotrowska, A.; Bajguz, A.; Godlewska, Z.B.; Czerpak, R.; Kaminska, M. Jasmonic acid as modulator of lead toxicity in aquatic plant Wolffia arrhiza (Lemnaceae). Environ. Exp. Bot. 2009, 66, 507–513. [Google Scholar] [CrossRef]
- Wasternack, C. Action of jasmonates in plant stress responses and development—Applied aspects. Biotechnol. Adv. 2014, 32, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Liu, B.; Liu, L.Y.; Song, S.S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef]
- Yuan, Z.; Zhang, D. Roles of jasmonate signalling in plant inflorescence and flower development. Curr. Opin. Plant Biol. 2015, 27, 44–51. [Google Scholar] [CrossRef]
- Ruan, X.; Wang, Z.; Wang, T.; Su, Y. Characterization and application of EST-SSR markers developed from the transcriptome of Amentotaxus argotaenia (Taxaceae), a relict vulnerable Conifer. Front. Genet. 2019, 10, 1014. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Zhang, Z.; Wang, Z.; Zhang, X.; Wu, G. Transcriptome sequencing and development of EST-SSR markers in Pinus dabeshanensis, an endangered conifer endemic to China. Mol. Breeding 2015, 35, 158. [Google Scholar] [CrossRef]
- Wakushima, S.; Yoshioka, H.; Sakurai, N. Promotion of lateral female strobili production in Pinus densiflora by cytokinin application at a specific stage. J. For. Res. 1997, 2, 51–57. [Google Scholar] [CrossRef]
- Wakushima, S.; Yoshioka, H.; Sakurai, N. Lateral female strobili production in a Japanese red pine (Pinus densiflora Sieb. Et Zucc.) clone by exogenous cytokinin application. J. For. Res. 1996, 1, 143–148. [Google Scholar] [CrossRef]
- Fei, Y.; Liu, Z.; Qi, R.; Tang, W. Cloning and Functional Analysis of TCAP3 Gene in Taxus Chinensis var. mairei. Int. J. Environ. 2017, 3, 25–35. [Google Scholar]
- Verelst, W.; Twell, D.; de Folter, S.; Immink, R.; Saedler, H.; Münster, T. MADS-complexes regulate transcriptome dynamics during pollen maturation. Genome Biol. 2007, 8, R249. [Google Scholar] [CrossRef]
- Niu, S.; Yuan, H.; Sun, X.; Porth, I.; Li, Y.; El-Kassaby, Y.A.; Li, W. A transcriptomics investigation into pine reproductive organ development. New Phytol. 2016, 209, 1278–1289. [Google Scholar] [CrossRef]
- Rudall, P.J.; Hilton, J.; Vergara-Silva, F.; Bateman, R.M. Recurrent abnormalities in conifer cones and the evolutionary origins of flower-like structures. Trends Plant Sci. 2011, 16, 151–159. [Google Scholar] [CrossRef]
- Zheng, Q.; Zheng, Y.; Ji, H.; Burnie, W.; Perry, S.E. Gene Regulation by the AGL15 transcription factor reveals hormone interactions in somatic embryogenesis. Plant Physiol. 2016, 172, 2374–2387. [Google Scholar] [CrossRef]
- Pineda, B.; Giménez-Caminero, E.; García-Sogo, B.; Antón, M.T.; Atarés, A.; Capel, J.; Lozano, R.; Angosto, T.; Moreno, V. Genetic and physiological characterization of the arlequin insertional mutant reveals a key regulator of reproductive development in tomato. Plant Cell Physiol. 2010, 51, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Begum, D.; Chuang, H.W.; Budiman, M.A.; Szymkowiak, E.J.; Irish, E.E.; Wing, R.A. JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 2000, 406, 910–913. [Google Scholar] [CrossRef] [PubMed]
- Ireland, H.S.; Yao, J.L.; Tomes, S.; Sutherland, P.W.; Nieuwenhuizen, N.; Gunaseelan, K.; Winz, R.A.; David, K.M.; Schaffer, R.J. Apple SEPALLATA1/2-like genes control fruit flesh development and ripening. Plant J. 2013, 73, 1044–1056. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Wu, S.; Liang, X.; Wang, K.; Li, Y.; Li, B.; Ma, J.; Liang, H. Paclobutrazol modulates endogenous level of phytohormones in inducing early flowering in Camellia tamdaoensis Hakoda et Ninh, a golden Camellia species. HortScience 2021, 56, 1258–1262. [Google Scholar] [CrossRef]
- Wei, X.J.; Ma, J.; Wang, K.; Liang, X.J.; Lan, J.X.; Li, Y.J.; Li, K.X.; Liang, H. Early flowering induction in golden Camellia seedlings and effects of paclobutrazol. HortScience 2018, 53, 1849–1854. [Google Scholar] [CrossRef]
- Wei, X.J.; Ma, J.; Li, K.X.; Liang, X.J.; Liang, H. Flowering induction in Camellia chrysantha, a golden Camellia species, with paclobutrazol and urea. HortScience 2017, 52, 1537–1543. [Google Scholar] [CrossRef]
- Wongsrisakulkaew, Y.; Boonprakob, U.; Sethpakdee, R.; Juntawong, N. Effect of paclobutrazol concentrations and time of foliar application on flowering of ‘Namdokmai-sitong’mango. Int. J. Geomate 2017, 12, 41–45. [Google Scholar] [CrossRef]
- Sha, J.; Ge, S.; Zhu, Z.; Du, X.; Zhang, X.; Xu, X.; Wang, F.; Chen, Q.; Tian, G.; Jiang, Y. Paclobutrazol regulates hormone and carbon-nitrogen nutrition of autumn branches, improves fruit quality and enhances storage nutrition in ‘Fuji’apple. Sci. Hortic. 2021, 282, 110022. [Google Scholar] [CrossRef]


| Hormone Name | M2 vs. M1 | M3 vs. M2 | M4 vs. M3 | M5 vs. M4 | Total DE Unigenes | Total Annotated Genes | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | |||
| IAA | 6 | 0 | 1 | 6 | 2 | 1 | 2 | 0 | 18 | 3 (Auxin transporter-like protein 1, Auxin transporter-like protein 2, Methylesterase 1) |
| CTK | 3 | 0 | 2 | 3 | 1 | 0 | 3 | 0 | 12 | 4 (Cytokinin trans-hydroxylase, Cytokinin riboside 5′-monophosphate phosphoribohydrolase, Cytokinin dehydrogenase, Adenylate kinase) |
| GA | 4 | 1 | 3 | 7 | 7 | 3 | 7 | 0 | 32 | 2 (ent-Copalyl diphosphate synthase, ent-Kaurene oxidase) |
| ABA | 1 | 5 | 3 | 2 | 1 | 1 | 0 | 0 | 13 | 4 (9-cis-epoxycarotenoid dioxygenase, Short-chain alcohol dehydrogenase/reductase, Molybdenum cofactor sulfurase, Violaxanthin de-epoxidase) |
| Ethelene | 3 | 2 | 3 | 6 | 2 | 2 | 1 | 0 | 19 | 2 (1-aminocyclopropane-1-carboxylic acid synthase, 1-aminocyclopropane-1-carboxylic acid oxidase) |
| JA | 14 | 11 | 12 | 15 | 13 | 13 | 2 | 9 | 89 | 4 (Phospholipase A1, Lipoxygenase, Allene oxide synthase, Allene oxide cyclase) |
| Hormone Name | M2 vs. M1 | M3 vs. M2 | M4 vs. M3 | M5 vs. M4 | TotalDE Unigenes | TotalAnnotation Categories | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | |||
| IAA | 14 | 2 | 19 | 7 | 13 | 4 | 1 | 4 | 64 | 4 |
| CTK | 3 | 2 | 4 | 5 | 2 | 1 | 1 | 0 | 18 | 4 |
| GA | 1 | 1 | 3 | 4 | 2 | 1 | 0 | 1 | 13 | 3 |
| ABA | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 4 | 1 |
| Ethylene | 0 | 2 | 1 | 0 | 0 | 1 | 0 | 1 | 5 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, W.; Wang, E.; Zhou, W.; Li, Y.; Li, Z.; Song, X.; Wang, J.; Ren, M.; Yang, D.; Huo, S.; et al. Morpho-Histology, Endogenous Hormone Dynamics, and Transcriptome Profiling in Dacrydium Pectinatum during Male Cone Development. Forests 2021, 12, 1598. https://doi.org/10.3390/f12111598
Lu W, Wang E, Zhou W, Li Y, Li Z, Song X, Wang J, Ren M, Yang D, Huo S, et al. Morpho-Histology, Endogenous Hormone Dynamics, and Transcriptome Profiling in Dacrydium Pectinatum during Male Cone Development. Forests. 2021; 12(11):1598. https://doi.org/10.3390/f12111598
Chicago/Turabian StyleLu, Wenju, Enbo Wang, Weijuan Zhou, Yifan Li, Zhaoji Li, Xiqiang Song, Jian Wang, Mingxun Ren, Donghua Yang, Shaojie Huo, and et al. 2021. "Morpho-Histology, Endogenous Hormone Dynamics, and Transcriptome Profiling in Dacrydium Pectinatum during Male Cone Development" Forests 12, no. 11: 1598. https://doi.org/10.3390/f12111598
APA StyleLu, W., Wang, E., Zhou, W., Li, Y., Li, Z., Song, X., Wang, J., Ren, M., Yang, D., Huo, S., Zhao, Y., & Liang, H. (2021). Morpho-Histology, Endogenous Hormone Dynamics, and Transcriptome Profiling in Dacrydium Pectinatum during Male Cone Development. Forests, 12(11), 1598. https://doi.org/10.3390/f12111598









