Characteristics and Driving Factors of Rhizosphere Bacterial Communities of Chinese Fir Provenances
Abstract
:1. Introduction
2. Method and Materials
2.1. Study Site
2.2. Sample Collection
2.3. Sequencing and Analysis of Soil Bacterial Community
2.4. BioinformaticsAnalysis
2.5. Data Analysis
3. Results
3.1. Quality Analysis of Sample Sequencing
3.2. Analysis of the Rhizosphere Soil Bacterial Community Compositionunder Different Chinese Fir Provenances
3.3. Analysis of Biomarkers in the Rhizosphere of Chinese Fir Provenances
3.4. α-Diversity Analysis of Bacterial Community in Rhizosphere Soil
3.5. Correlation Analysis of Rhizosphere Bacterial Community Composition and α-Diversity with Environmental Factors
4. Discussion
4.1. Geographical Distribution of Rhizosphere Soil Bacteria
4.2. Driving Factors of the Geographical Distribution of Rhizosphere Bacteria
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, J.; Guo, X.; Xiao, F.; Xiong, C.; Yin, Y. Influences of provenance and rotation age on heartwood ratio, stem diameter and radial variation in tracheid dimension of Cunninghamia lanceolata. Eur. J. Wood Wood Prod. 2018, 76, 669–677. [Google Scholar] [CrossRef]
- Xu, Y.; Du, C.; Huang, G.; Li, Z.; Xu, X.; Zheng, J.; Wu, C. Morphological characteristics of tree crowns of Cunninghamia lanceolata var. Luotian. J. For. Res. 2020, 31, 837–856. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Nguyen, T.H.N.; Qin, J.; Jiao, Y.; Li, Z.; Ding, S.; Lu, Y.; Liu, Q.; Luo, Z.-B. Phosphorus assimilation of Chinese fir from two provenances during acclimation to changing phosphorus availability. Environ. Exp. Botany. 2018, 153, 21–34. [Google Scholar] [CrossRef]
- Li, K.P.; Wei, Z.C.; Huang, K.Y.; Dong, L.J.; Huang, H.F.; Chen, Q.; Dai, J.; Tan, W.J. Research on Variation Pattern of Wood Properties of Red-heart Chinese Fir Plus Trees, a Featured Provenance from Rongshui of Guangxi. For. Res. 2017, 30, 424–429. [Google Scholar] [CrossRef]
- Wu, P.F.; Tigabu, M.; Ma, X.Q.; Oden, P.C.; He, Y.L.; Yu, X.T.; He, Z.Y. Variations in biomass, nutrient contents and nutrient use efficiency among Chinese fir provenances. Silvae Genet. 2011, 60, 95–105. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.B.; Duan, A.G.; Zhang, J.G.; Sun, J.J. Effect of Long-term Selection of Chinese fir (Cunninghamia lanceolata (Lamb.)Hook) Provenances. For. Res. 2019, 32, 9–17. [Google Scholar] [CrossRef]
- Zheng, H.; Duan, H.; Hu, D.; Li, Y.; Hao, Y. Genotypic variation of Cunninghamia lanceolata revealed by phenotypic traits and SRAP markers. Dendrobiology 2015, 74, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Liu, Y.-X.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X.; et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef]
- Bakker, P.A.H.M.; Pieterse, C.M.J.; de Jonge, R.; Berendsen, R.L. The Soil-Borne Legacy. Cell 2018, 172, 1178–1180. [Google Scholar] [CrossRef] [Green Version]
- Gaiero, J.R.; McCall, C.A.; Thompson, K.A.; Day, N.J.; Best, A.S.; Dunfield, K.E. Inside the root microbiome: Bacterial root endophytes and plant growth promotion. Am. J. Bot. 2013, 100, 1738–1750. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Haig, T.; Pratley, J.; Lemerle, D.; An, M. Allelochemicals in Wheat (Triticum aestivum L.): Cultivar Difference in the Exudation of Phenolic Acids. J. Agric. Food Chem. 2001, 49, 3742–3745. [Google Scholar] [CrossRef]
- Singh, G.; Mukerji, K.G. Root Exudates as Determinant of Rhizospheric Microbial Biodiversity. In Microbial Activity in the Rhizoshere; Springer: Berlin/Heidelberg, Germany, 2006; pp. 39–53. [Google Scholar] [CrossRef]
- Rolfe, S.A.; Griffiths, J.; Ton, J. Crying out for help with root exudates: Adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr. Opin. Microbiol. 2019, 49, 73–82. [Google Scholar] [CrossRef]
- Pham, V.H.T.; Kim, J. Cultivation of unculturable soil bacteria. Trends Biotechnol. 2012, 30, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ke, X.; Hernández, M.; Wang, B.; Dumont, M.G.; Jia, Z.; Conrad, R. Autotrophic Growth of Bacterial and Archaeal Ammonia Oxidizers in Freshwater Sediment Microcosms Incubated at Different Temperatures. Appl. Environ. Microbiol. 2013, 79, 3076–3084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Połka, J.; Rebecchi, A.; Pisacane, V.; Morelli, L.; Puglisi, E. Bacterial diversity in typical Italian salami at different ripening stages as revealed by high-throughput sequencing of 16S rRNA amplicons. Food Microbiol. 2015, 46, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; Bodegom, P.M.; Bengtsson-Palme, J.; Anslan, S.; Coelho, L.P.; Harend, H.; et al. Structure and function of the global topsoil microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Trivedi, P.; Osanai, Y.; Liu, Y.R.; Hamonts, K.; Jeffries, T.C.; Singh, B.K. Carbon content and climate variability drive global soil bacterial diversity patterns. Ecol. Monogr. 2016, 86, 373–390. [Google Scholar] [CrossRef]
- Wan, X.; Huang, Z.; He, Z.; Yu, Z.; Wang, M.; Davis, M.R.; Yang, Y. Soil C:N ratio is the major determinant of soil microbial community structure in subtropical coniferous and broadleaf forest plantations. Plant Soil 2015, 387, 103–116. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-Oter, R.; Munch, P.C.; Weiman, A.; Droge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, S.K.; Mohammed, M.; Dakora, F.D. Microbial community structure in the rhizosphere of the orphan legume Kersting’s groundnut [Macrotyloma geocarpum (Harms) Marechal & Baudet]. Mol. Biol. Rep. 2019, 46, 4471–4481. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The Diversity and Biogeography of Soil Bacterial Communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, D.; Lee-Cruz, L.; Kim, W.-S.; Kerfahi, D.; Chun, J.-H.; Adams, J.M. Strong elevational trends in soil bacterial community composition on Mt. Halla, South Korea. Soil Biol. Biochem. 2014, 68, 140–149. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M. Obscure soil microbes and where to find them. ISME J. 2019, 13, 2120–2124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fei, Y.-C.; Wu, Q.-Z.; Lu, J.; Ji, C.-S.; Zheng, H.; Cao, S.-J.; Lin, K.-M.; Cao, G.-Q. Effects of Undergrowth Vegetation Management Measures on the Soil Bacterial Community Structure of Large Diameter Timber Plantation of Cunninghamia Lanceolata. Chin. J. Appl. Ecol. 2020, 31, 407–416. [Google Scholar] [CrossRef]
- Cheng, X.; Xing, W.; Yuan, H.; Yu, M. Long-Term Thinning Does not Significantly Affect Soil Water-Stable Aggregates and Diversity of Bacteria and Fungi in Chinese Fir (Cunninghamia lanceolata) Plantations in Eastern China. Forests 2018, 9, 687. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Li, J.; Zheng, J.; Liu, J.; Liu, S.; Lin, W.; Wu, C. Soil microbial community structure and catabolic activity are significantly degenerated in successive rotations of Chinese fir plantations. Sci. Rep. 2017, 7, 6691–6697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, J.; Zheng, Y.; Yang, Y. Phylogenetic Structure of Soil Bacterial Communities along Age Sequence of Subtropical Cunninghamia Lanceolata Plantations. Sustainability 2020, 12, 1864. [Google Scholar] [CrossRef] [Green Version]
- Lei, X.; Shen, F.; Lei, X.; Liu, W.; Duan, H.; Fan, H.; Wu, J. Assessing influence of simulated canopy nitrogen deposition and under-story removal on soil microbial community structure in a Cunninghamia lanceolata plantation. Biodivers. Sci. 2018, 26, 962–971. [Google Scholar] [CrossRef]
- Wu, Z.-Y.; Lin, W.-X.; Li, J.-J.; Liu, J.-F.; Li, B.-L.; Wu, L.-K.; Fang, C.-X.; Zhang, Z.-X. Effects of seasonal variations on soil microbial community composition of two typical zonal vegetation types in the Wuyi Mountains. J. Mt. Sci. 2016, 13, 1056–1065. [Google Scholar] [CrossRef]
- Xie, L.; Zhang, Q.; Cao, J.; Liu, X.; Xiong, D.; Kong, Q.; Yang, Y. Effects of Warming and Nitrogen Addition on the Soil Bacterial Community in a Subtropical Chinese Fir Plantation. Forests 2019, 10, 861. [Google Scholar] [CrossRef] [Green Version]
- Riley, D.; Barber, S.A. Bicarbonate Accumulation and pH Changes at the Soybean (Glycine max (L.) Merr.) Root-Soil Interface. Soil Sci. Soc. Am. J. 1969, 63, 905–908. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, C.; Yu, W.; Turak, A.; Chen, D.; Huang, Y.; Ao, J.; Jiang, Y.; Huang, Z. Effects of Nitrogen and Phosphorus Inputs on Soil Bacterial Abundance, Diversity, and Community Composition in Chinese Fir Plantations. Front. Microbiol. 2018, 9, 1543. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef] [Green Version]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an Ecological Classification of Soil Bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- van Niftrik, L.; Geerts, W.J.C.; van Donselaar, E.G.; Humbel, B.M.; Webb, R.I.; Harhangi, H.R.; den Camp, H.J.M.O.; Fuerst, J.A.; Verkleij, A.J.; Jetten, M.S.M.; et al. Cell division ring, a new cell division protein and vertical inheritance of a bacterial organelle in anammox planctomycetes. Mol. Microbiol. 2009, 73, 1009–1019. [Google Scholar] [CrossRef]
- Brown, M.E. Seed and Root Bacterization. Annu. Rev. Phytopathol. 1974, 12, 181–197. [Google Scholar] [CrossRef]
- Mayak, S.; Tirosh, T.; Glick, B.R. Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol. Biochem. 2004, 42, 565–572. [Google Scholar] [CrossRef]
- Getzke, F.; Thiergart, T.; Hacquard, S. Contribution of bacterial-fungal balance to plant and animal health. Curr. Opin. Microbiol. 2019, 49, 66–72. [Google Scholar] [CrossRef]
- Hu, L.; Robert, C.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; Van Der Heijden, M.G.A.; et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018, 9, 2713–2738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordovez, V.; Dini-Andreote, F.; Carrión, V.J.; Raaijmakers, J.M. Ecology and Evolution of Plant Microbiomes. Annu. Rev. Microbiol. 2019, 73, 69–88. [Google Scholar] [CrossRef] [PubMed]
- Dick, R.P.; Rasmussen, P.E.; Kerle, E.A. Influence of long-term residue management on soil enzyme activities in relation to soil chemical properties of a wheat-fallow system. Biol. Fertil. Soils 1988, 6, 159–164. [Google Scholar] [CrossRef]
- Xia, Z.; Bai, E.; Wang, Q.; Gao, D.; Zhou, J.; Jiang, P.; Wu, J. Biogeographic Distribution Patterns of Bacteria in Typical Chinese Forest Soils. Front. Microbiol. 2016, 7, 1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Axelsson, E.P.; Girona, M.M.; Senior, J.K. Continuous-cover forestry maintains soil fungal communities in Norway spruce dominated boreal forests. For. Ecol. Manag. 2021, 480, 118659. [Google Scholar] [CrossRef]
- Bach, L.H.; Grytnes, J.-A.; Halvorsen, R.; Ohlson, M. Tree influence on soil microbial community structure. Soil Biol. Biochem. 2010, 42, 1934–1943. [Google Scholar] [CrossRef]
- Yang, T.; Tedersoo, L.; Soltis, P.S.; Soltis, D.E.; Gilbert, J.A.; Sun, M.; Shi, Y.; Wang, H.; Li, Y.; Zhang, J.; et al. Phylogenetic imprint of woody plants on the soil mycobiome in natural mountain forests of eastern China. ISME J. 2019, 13, 686–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez, K.S.; Snoek, L.B.; Koorem, K.; Geisen, S.; Bloem, L.J.; ten Hooven, F.; Kostenko, O.; Krigas, N.; Manrubia, M.; Caković, D.; et al. Range-expansion effects on the belowground plant microbiome. Nat. Ecol. Evol. 2019, 3, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.-S.; Yang, J.W.; Ghim, S.-Y.; Ryu, C.-M. A cry for help from leaf to root: Aboveground insect feeding leads to the recruitment of rhizosphere microbes for plant self-protection against subsequent diverse attacks. Plant Signal. Behav. 2011, 6, 1192–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.H.M.; et al. Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
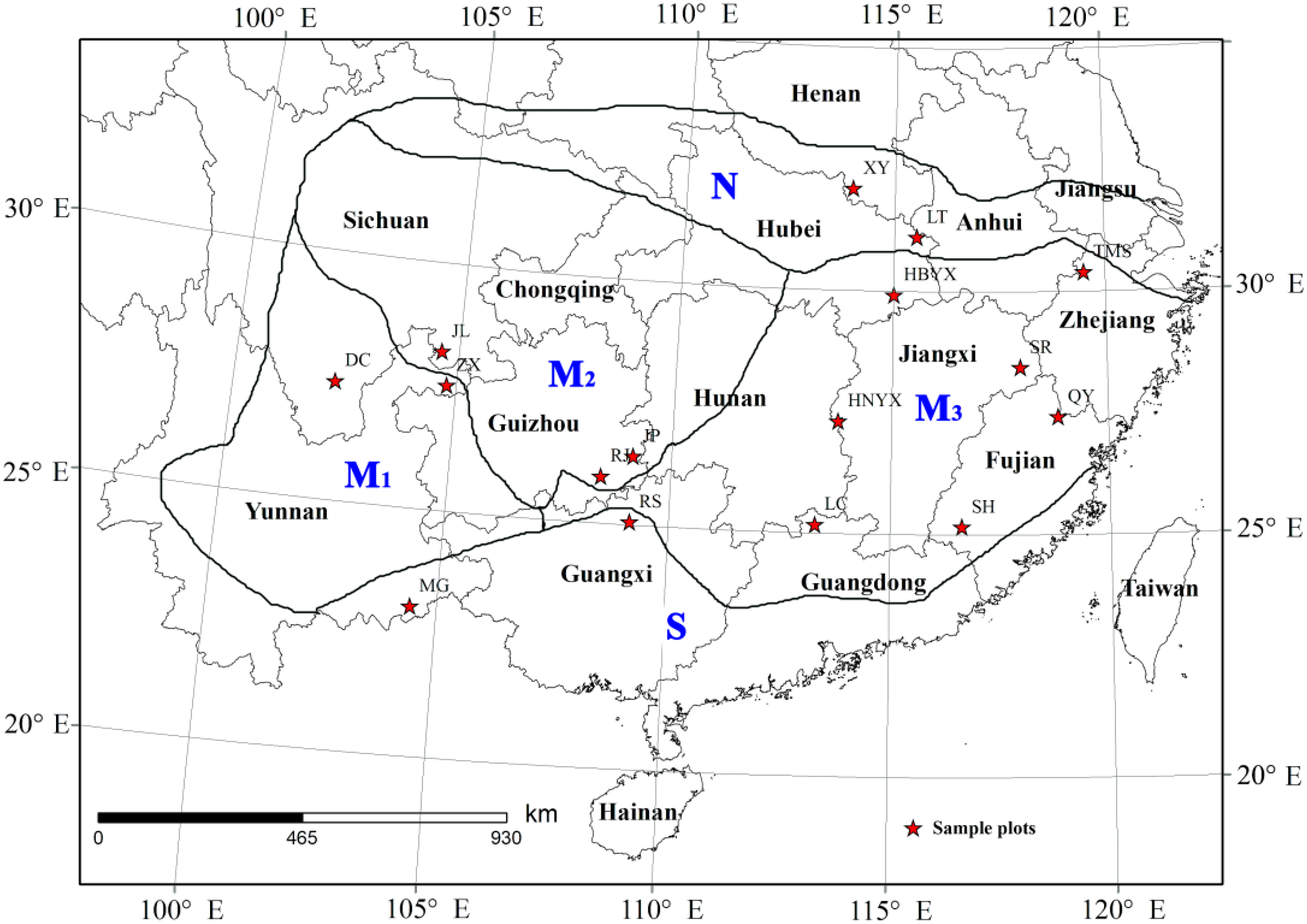







| Producing Regions | Provenances | Longitude | Latitude | Altitude (m) | AAP (mm) | AAT (°C) | Stand Age (year) | Average Tree Height (m) | Average DBH (cm) | Average Crown (m) |
|---|---|---|---|---|---|---|---|---|---|---|
| Southern belt (S) | MG | 104°25′58″ | 23°2′5″ | 1594 | 1345 | 16.9 | 45 | 28.7 | 39.4 | 4.7 |
| RS | 109°8′37″ | 25°3′54″ | 550 | 1813 | 19.6 | 42 | 25.4 | 37.9 | 3.9 | |
| Eastern middle belt (M3) | SH | 116°38′7″ | 25°9′47″ | 571 | 1518.2 | 19.8 | 40 | 29.1 | 36.7 | 3.4 |
| LC | 113°18′44″ | 25°10′34″ | 501 | 1464 | 19.8 | 39 | 21.3 | 35.6 | 3.5 | |
| HNYX | 113°46′55″ | 27°18′58″ | 582 | 1410 | 17.8 | 37 | 29.3 | 34.5 | 4.5 | |
| QY | 118°50′51″ | 27°25′38″ | 930 | 1760 | 17.4 | 37 | 34.4 | 43.1 | 2.6 | |
| SR | 117°59′13″ | 28°26′47″ | 150 | 1780 | 18.3 | 42 | 30.3 | 35.9 | 3.3 | |
| HBYX | 115°1′6″ | 29°55′53″ | 436 | 1389.6 | 16.8 | 40 | 29.3 | 32.1 | 4.3 | |
| TMS | 119°30′7″ | 30°23′57″ | 708 | 1613.9 | 11.7 | 40 | 36.2 | 42.3 | 4.1 | |
| Central middle belt (M2) | RJ | 108°25′45″ | 25°57′48″ | 560 | 1250 | 18.1 | 43 | 37.1 | 44.1 | 2.9 |
| JP | 109°8′9″ | 26°24′32″ | 531 | 1300 | 16.4 | 43 | 38.7 | 43.7 | 3.8 | |
| JL | 104°36′46″ | 28°12′50″ | 990 | 1100 | 17.6 | 32 | 31.4 | 40.6 | 3.1 | |
| Western middle belt (M1) | DC | 102°17′2″ | 27°23′30″ | 735 | 1074.4 | 17.7 | 39 | 28.1 | 37.5 | 2.5 |
| ZX | 104°47′32″ | 27°32′27″ | 1722 | 1334.6 | 11.3 | 40 | 33.6 | 41.5 | 4.2 | |
| Northern belt (N) | LT | 115°32′25″ | 31°7′15″ | 443 | 1330 | 16.4 | 38 | 27.6 | 38.7 | 3.7 |
| XY | 113°59′52″ | 32°5′58″ | 137 | 1050 | 15.5 | 39 | 26.8 | 30.8 | 4.3 |
| Factor | Explains/% | F | P |
|---|---|---|---|
| Ca/Mg | 8.1 | 4.0 | 0.032 * |
| AAP | 6.9 | 3.7 | 0.044 * |
| K | 9.0 | 5.2 | 0.014 * |
| Mg | 10.9 | 7.2 | 0.006 ** |
| K/Ca | 5.3 | 3.7 | 0.032 * |
| N | 4.2 | 3.1 | 0.090 |
| P | 9.8 | 8.5 | 0.006 ** |
| AAT | 2.4 | 2.1 | 0.132 |
| LO | 2.5 | 2.3 | 0.118 |
| LA | 2.3 | 2.2 | 0.114 |
| N/P | 3.9 | 4.0 | 0.022 * |
| AL | 1.8 | 1.9 | 0.128 |
| K/Mg | 0.9 | 0.9 | 0.362 |
| C/N | 1.0 | 1.1 | 0.312 |
| C/P | 0.5 | 0.6 | 0.516 |
| pH | 0.4 | 0.4 | 0.624 |
| C | 0.2 | 0.2 | 0.812 |
| Ca | 0.1 | 0.1 | 0.874 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Y.; Li, B.; Huang, Z.; Zhang, H.; Wu, X.; Farooq, T.H.; Wu, P.; Li, M.; Ma, X. Characteristics and Driving Factors of Rhizosphere Bacterial Communities of Chinese Fir Provenances. Forests 2021, 12, 1362. https://doi.org/10.3390/f12101362
Yan Y, Li B, Huang Z, Zhang H, Wu X, Farooq TH, Wu P, Li M, Ma X. Characteristics and Driving Factors of Rhizosphere Bacterial Communities of Chinese Fir Provenances. Forests. 2021; 12(10):1362. https://doi.org/10.3390/f12101362
Chicago/Turabian StyleYan, Yao, Bingjun Li, Zhijun Huang, Hui Zhang, Xiaojian Wu, Taimoor Hassan Farooq, Pengfei Wu, Ming Li, and Xiangqing Ma. 2021. "Characteristics and Driving Factors of Rhizosphere Bacterial Communities of Chinese Fir Provenances" Forests 12, no. 10: 1362. https://doi.org/10.3390/f12101362
APA StyleYan, Y., Li, B., Huang, Z., Zhang, H., Wu, X., Farooq, T. H., Wu, P., Li, M., & Ma, X. (2021). Characteristics and Driving Factors of Rhizosphere Bacterial Communities of Chinese Fir Provenances. Forests, 12(10), 1362. https://doi.org/10.3390/f12101362







