Identification of a Natural Hybrid between Castanopsis sclerophylla and Castanopsis tibetana (Fagaceae) Based on Chloroplast and Nuclear DNA Sequences
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
2.2. Chloroplast DNA Sequencing and Nuclear Microsatellite Genotyping
2.3. Data Analyses
3. Results
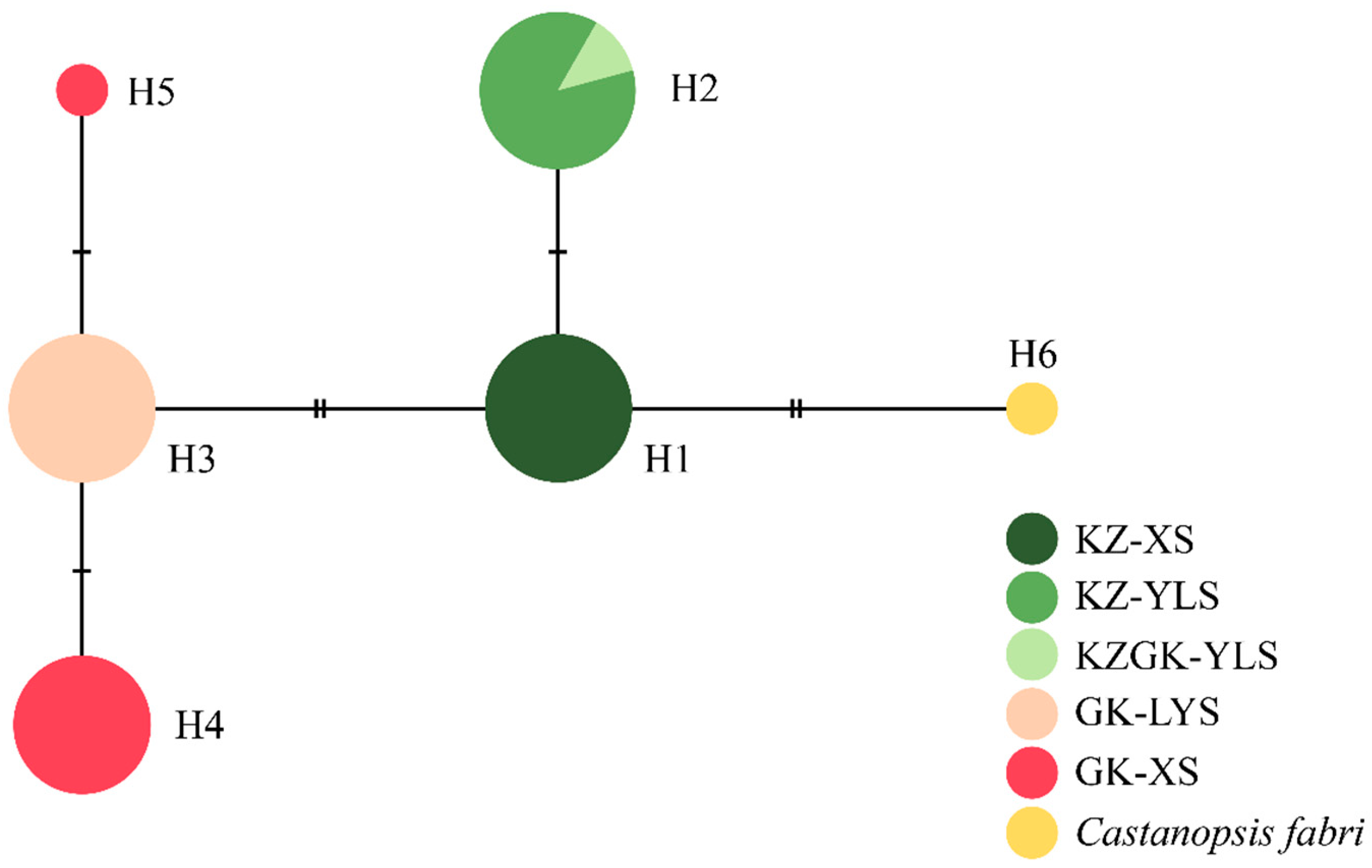
3.1. Chloroplatst DNA Variation
3.2. Genetic Diversity at Nuclear Microsatellite Loci
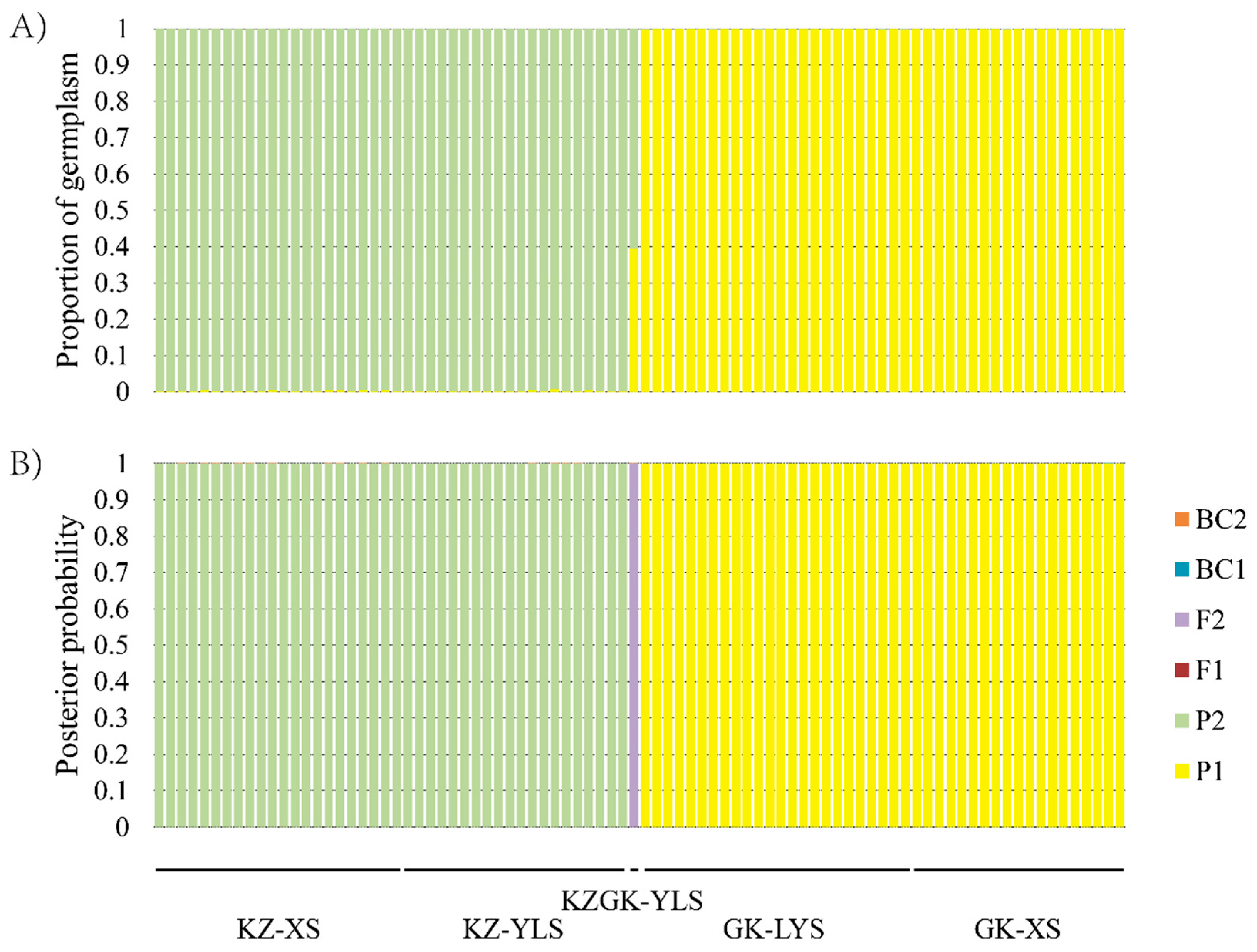
3.3. STRUCTURE and NewHybrids Analyses Based on Nuclear Microsatellite Markers
4. Discussion
4.1. SSR Transferability among Castanopsis Species
4.2. Genetic Diversity of C. sclerophylla and C. tibetana
4.3. Molecular Evidence of Natural Hybrid and Taxonomic Status for C. × kuchugouzhui
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Harrison, R.G.; Larson, E.L. Hybridization, Introgression, and the Nature of Species Boundaries. J. Hered. 2014, 105, 795–809. [Google Scholar] [CrossRef] [PubMed]
- Mallet, J. Hybrid speciation. Nature 2007, 446, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Ungerer, M.C.; Baird, S.J.E.; Pan, J.; Rieseberg, L.H. Rapid hybrid speciation in wild sunflowers. Proc. Natl. Acad. Sci. USA 1998, 95, 11757–11762. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.L.; Zhang, C.Q.; Gao, L.M.; Yang, J.B.; Li, H.T. Natural hybridization origin of Rhododendron agastum (Ericaceae) in Yunnan, China: Inferred from morphological and molecular evidence. J. Plant Res. 2007, 120, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Wei, W.; Zhang, R.; Liu, T.; Chen, Y.; Shi, S.; Zhou, R. Molecular evidence for hybrid origin of Melastoma intermedium. Biochem. Syst. Ecol. 2012, 41, 136–141. [Google Scholar] [CrossRef]
- Liu, T.; Chen, Y.; Chao, L.; Wang, S.; Wu, W.; Dai, S.; Wang, F.; Fan, Q.; Zhou, R. Extensive Hybridization and Introgression between Melastoma candidum and M. sanguineum. PLoS ONE 2014, 9, e96680. [Google Scholar] [CrossRef]
- Rieseberg, L.H. Hybrid Origins of Plant Species. Annu. Rev. Ecol. Syst. 1997, 28, 359–389. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Carney, S.E. Plant hybridization. New Phytol. 1998, 140, 599–624. [Google Scholar] [CrossRef]
- Barton, N.H. The role of hybridization in evolution. Mol. Ecol. 2001, 10, 551–568. [Google Scholar] [CrossRef]
- Zalapa, J.E.; Brunet, J.; Guries, R.P. The extent of hybridization and its impact on the genetic diversity and population structure of an invasive tree, Ulmus pumila (Ulmaceae). Evol. Appl. 2010, 3, 157–168. [Google Scholar] [CrossRef]
- Ellstrand, N.C.; Whitkus, R.; Rieseberg, L.H. Distribution of Spontaneous Plant Hybrids. Proc. Natl. Acad. Sci. USA 1996, 93, 5090–5093. [Google Scholar] [CrossRef] [PubMed]
- Rieseberg, L.H.; Raymond, O.; Rosenthal, D.M.; Lai, Z.; Livingstone, K.; Nakazato, T.; Durphy, J.L.; Schwarzbach, A.E.; Donovan, L.A.; Lexer, C. Major Ecological Transitions in Wild Sunflowers Facilitated by Hybridization. Science 2003, 301, 1211–1216. [Google Scholar] [CrossRef] [PubMed]
- Stukenbrock, E.H. The Role of Hybridization in the Evolution and Emergence of New Fungal Plant Pathogens. Phytopathology 2016, 106, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Stebbins, G.L. The Role of Hybridization in Evolution. Proc. Am. Philos. Soc. 1959, 103, 231–251. [Google Scholar]
- Isoda, K.; Shiraishi, S.; Watanabe, S.; Kitamura, K. Molecular evidence of natural hybridization between Abies veitchii and A. homolepis (Pinaceae) revealed by chloroplast, mitochondrial and nuclear DNA markers. Mol. Ecol. 2000, 9, 1965–1974. [Google Scholar] [CrossRef]
- Ning, H.; Yu, J.; Gong, X. Bidirectional natural hybridization between sympatric Ligularia vellerea and L. subspicata. Plant Divers. 2017, 39, 214–220. [Google Scholar] [CrossRef]
- Wu, R.; Zou, P.; Tan, G.; Hu, Z.; Wang, Y.; Ning, Z.; Wu, W.; Liu, Y.; He, S.; Zhou, R. Molecular identification of natural hybridization between Melastoma malabathricum and Melastoama beccarianum in Sarawak, Malaysia. Ecol. Evol. 2019, 9, 5766–5776. [Google Scholar] [CrossRef]
- Huang, C.J.; Chang, Y.T. Flora Reipublicae Popularis Sinicae. In Castanopsis; Chun, W.Y., Huang, C.J., Eds.; Science Press: Beijing, China, 1998; pp. 13–80. [Google Scholar]
- Huang, C.J.; Zhang, Y.T.; Bruce, B. Flora of China. In Fagaceae; Wu, Z.Y., Raven, P.H., Hong, D.Y., Eds.; Science Press and Missouri Botanical Garden Press: Beijing, China, 1999; pp. 315–333. [Google Scholar]
- Kress, W.J.; Wurdack, K.J.; Zimmer, E.A.; Weigt, L.A.; Janzen, D.H. Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. USA 2005, 102, 8369–8374. [Google Scholar] [CrossRef]
- Cheng, Y.P.; Hwang, S.Y.; Lin, T.P. Potential refugia in Taiwan revealed by the phylogeographic study of Castanopsis carlesii Hayata (Fagaceae). Mol. Ecol. 2005, 14, 2075–2085. [Google Scholar] [CrossRef]
- Shi, Y.S.; Zhang, J.; Jiang, K.; Cui, M.Y.; Li, Y.Y. Development and characterization of polymorphic microsatellite markers in Castanopsis sclerophylla (Fagaceae). Am. J. Bot. 2011, 98, e19–e21. [Google Scholar] [CrossRef]
- Ye, L.J.; Wang, J.; Sun, P.; Dong, S.P.; Zhang, Z.Y. The Transferability of Nuclear Microsatellite Markers in Four Castanopsis Species to Castanopsis tibetana (Fagaceae). Plant Divers. Resour. 2014, 36, 443–448. [Google Scholar]
- Fu, D.D.; Wang, J.; Liu, Y.F.; Huang, H.W. Isolation of Microsatellite Markers for Castanopsis fargesii (Fagaceae). J. Trop. Subtrop. Botany 2010, 18, 541–546. [Google Scholar]
- Ueno, S.; Aoki, K.; Tsumura, Y. Generation of Expressed Sequence Tags and development of microsatellite markers for Castanopsis sieboldii var. sieboldii (Fagaceae). Ann. For. Sci. 2009, 66, 509. [Google Scholar] [CrossRef]
- Waikham, P.; Thongkumkoon, P.; Chomdej, S.; Liu, A.; Wangpakapattanawong, P. Development of 13 microsatellite markers for Castanopsis tribuloides (Fagaceae) using next-generation sequencing. Mol. Biol. Rep. 2018, 45, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Marinoni, D.; Akkak, A.; Bounous, G.; Edwards, K.J.; Botta, R. Development and characterization of microsatellite markers in Castanea sativa (Mill.). Mol. Breed. 2003, 11, 127–136. [Google Scholar] [CrossRef]
- Li, C.; Sun, Y.; Huang, H.W.; Cannon, C.H. Footprints of divergent selection in natural populations of Castanopsis fargesii (Fagaceae). Heredity 2014, 113, 533–541. [Google Scholar] [CrossRef]
- Holland, M.M.; Parson, W. GeneMarker® HID: A Reliable Software Tool for the Analysis of Forensic STR Data. J. Forensic Sci. 2011, 56, 29–35. [Google Scholar] [CrossRef]
- Alzohairy, A.M. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. FSTAT (Version 2.9.4), A Program (for Windows 95 and Above) to Estimate and Test Population Genetics Parameters; University of Lausanne: Lausanne, Switzerland, 2003. [Google Scholar]
- Pritchard, J.K.; Wen, W. Documentation for STRUCTURE Software: Version 2; University of Chicago: Chicago, IL, USA, 2003. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.C.; Thompson, E.A. A Model-Based Method for Identifying Species Hybrids Using Multilocus Genetic Data. Genetics 2002, 160, 1217–1229. [Google Scholar]
- Milne, R.I.; Abbott, R.J. Reproductive isolation among two interfertile Rhododendron species: Low frequency of post-F1 hybrid genotypes in alpine hybrid zones. Mol. Ecol. 2008, 17, 1108–1121. [Google Scholar] [CrossRef]
- Peakall, R.; Gilmore, S.; Keys, W.; Morgante, M.; Rafalski, A. Cross-Species Amplification of Soybean (Glycine max) Simple Sequence Repeats (SSRs) Within the Genus and Other Legume Genera: Implications for the Transferability of SSRs in Plants. Mol. Biol. Evol. 1998, 15, 1275–1287. [Google Scholar] [CrossRef]
- Li, C.; Sun, Y. Transferability analysis of EST-SSR markers of Castanea mollissima to Castanopsis fargesii. Guihaia 2012, 32, 293–297. [Google Scholar]
- Young, A.; Boyle, T.; Brown, T. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol. (Amst.) 1996, 11, 413–418. [Google Scholar] [CrossRef]
- Blakesley, D.; Pakkad, G.; James, C.; Torre, F.; Elliott, S. Genetic diversity of Castanopsis acuminatissima (Bl.) A. DC. in northern Thailand and the selection of seed trees for forest restoration. New For. (Dordr.) 2004, 27, 89–100. [Google Scholar]
- Aoki, K.; Tamaki, I.; Nakao, K.; Ueno, S.; Kamijo, T.; Setoguchi, H.; Murakami, N.; Kato, M.; Tsumura, Y. Approximate Bayesian computation analysis of Est-associated microsatellites indicates that the broadleaved evergreen tree Castanopsis sieboldii survived the Last Glacial Maximum in multiple refugia in Japan. Heredity 2019, 122, 326–340. [Google Scholar] [CrossRef]
- Liu, B.; Wang, R.; Liu, Y.L.; Xu, G.F.; Chen, X.Y. Seed dispersal and predation of Castanopsis sclerophylla by small rodents in habits with different disturbance intensity. Chin. J. Ecol. 2011, 30, 1668–1673. [Google Scholar]
- Xu, Y.; Shen, Z.; Li, D.; Guo, Q. Pre-dispersal seed predation in a species-rich forest community: Patterns and the interplay with determinants. PLoS ONE 2015, 10, e0143040. [Google Scholar] [CrossRef] [PubMed]
- Nakanish, A.; Yoshimaru, H.; Tomaru, N.; Miura, M.; Manabe, T.; Yamamoto, S. Patterns of pollen flow in a dense population of the insect-pollinated canopy tree species Castanopsis sieboldii. J. Hered. 2012, 103, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002; pp. 366–394. [Google Scholar]
| Species | Location/Population Code | Latitude (N) | Longitude (E) | Sample Size | A | AR | H | FIS |
|---|---|---|---|---|---|---|---|---|
| Castanopsis sclerophylla | Xiushui/KZ-XS | 28°55′23″ | 114°43′12″ | 22 | 5.6 | 5.426 | 0.629 | 0.103 * |
| Yuelushan/KZ-YLS | 28°10′49″ | 112°56′28″ | 20 | 5.5 | 5.430 | 0.580 | 0.052 | |
| Average | 21 | 5.6 | 5.428 | 0.605 | 0.078 | |||
| Castanopsis × kuchugouzhui | Yuelushan/KZGK-YLS | 28°10′49″ | 112°56′28″ | 1 | ||||
| Castanopsis tibetana | Liangyeshan/GK-LYS | 25°12′35″ | 116°10′48″ | 24 | 3.3 | 3.219 | 0.481 | −0.057 |
| Xiushui/GK-XS | 28°55′23″ | 114°43′12″ | 19 | 2.9 | 2.862 | 0.427 | −0.068 | |
| Average | 22 | 3.1 | 3.041 | 0.454 | −0.063 |
| Species | Population Code | Haplotype | Frequency | Variable Site | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| psbA-trnH | trnM-trnV | |||||||||
| 100 | 258 | 284 | 478 | 491 | 672 | 892 | ||||
| Castanopsis sclerophylla | KZ-XS | H1 | 8 | C | T | A | C | C | G | C |
| KZ-YLS | H2 | 8 | C | T | A | C | A | G | C | |
| Castanopsis × kuchugouzhui | KZGK-YLS | H2 | 1 | C | T | A | C | A | G | C |
| Castanopsis tibetana | GK-LYS | H3 | 8 | C | T | A | C | C | T | A |
| GK-XS | H4 | 7 | C | T | A | A | C | T | A | |
| H5 | 1 | C | C | A | C | C | T | A | ||
| Castanopsis fabri | outgroup | H6 | 1 | T | T | T | C | C | G | C |
| Locus | Allele Size | A | HO | HS | HT | FIS | FST |
|---|---|---|---|---|---|---|---|
| CC-30080 | 151–163 | 5 | 0.473 | 0.549 | 0.545 | 0.132 | −0.013 |
| CC-33079-1 | 189–209 | 7 | 0.259 | 0.468 | 0.546 | 0.455 * | 0.244 |
| CC-935 | 131–149 | 6 | 0.668 | 0.662 | 0.669 | −0.011 | 0.022 |
| CsCAT34 | 149–161 | 5 | 0.475 | 0.486 | 0.521 | 0.026 | 0.128 |
| CC-14826 | 228–244 | 9 | 0.423 | 0.461 | 0.462 | 0.079 | 0.006 |
| CC-2994 | 269–272 | 2 | 0.150 | 0.498 | 0.499 | 0.714 * | 0.004 |
| CC-3722 | 131–145 | 5 | 0.548 | 0.570 | 0.598 | 0.042 | 0.087 |
| CC-6538 | 169–185 | 6 | 0.734 | 0.713 | 0.727 | −0.029 | 0.037 |
| CC-4950 | 247–283 | 15 | 0.693 | 0.900 | 0.907 | 0.233 * | 0.016 |
| CcC02022 | 351–375 | 11 | 0.764 | 0.795 | 0.807 | 0.040 | 0.029 |
| CFA22 | 156–162 | 3 | 0.186 | 0.286 | 0.312 | 0.354 | 0.150 |
| CC-25032 | 176–194 | 5 | 0.143 | 0.454 | 0.455 | 0.688 * | 0.002 |
| CC-41284 | 271–289 | 10 | 0.759 | 0.825 | 0.862 | 0.073 | 0.081 |
| CS05 | 156–178 | 7 | 0.764 | 0.741 | 0.744 | −0.025 | 0.008 |
| CC-43042 | 186–210 | 6 | 0.464 | 0.582 | 0.622 | 0.215 | 0.120 |
| CFA71 | 174–204 | 9 | 0.809 | 0.840 | 0.845 | 0.037 | 0.011 |
| CC-42621 | 288–297 | 3 | 0.366 | 0.308 | 0.319 | −0.183 | 0.064 |
| CsCAT14 | 124–154 | 15 | 0.932 | 0.856 | 0.865 | −0.087 | 0.020 |
| CC-4562 | 144–156 | 3 | 0.782 | 0.657 | 0.661 | −0.194 | 0.011 |
| CC-26213 | 122–152 | 4 | 0.505 | 0.619 | 0.637 | 0.190 | 0.055 |
| CFA61 | 184–212 | 11 | 0.761 | 0.601 | 0.601 | −0.268 | 0.001 |
| CS24 | 123–144 | 7 | 0.784 | 0.730 | 0.725 | −0.077 | −0.015 |
| CS43 | 148–168 | 7 | 0.755 | 0.702 | 0.718 | −0.079 | 0.042 |
| CT161 | 148–157 | 4 | 0.395 | 0.362 | 0.388 | −0.094 | 0.124 |
| CT128 | 163–202 | 4 | 0.455 | 0.456 | 0.480 | −0.003 | 0.099 |
| CS20 | 249–273 | 8 | 0.741 | 0.728 | 0.780 | −0.024 | 0.128 |
| CS44 | 135–147 | 7 | 0.748 | 0.797 | 0.807 | 0.050 | 0.025 |
| CC-33079 | 243–259 | 6 | 0.310 | 0.370 | 0.387 | 0.171 | 0.081 |
| CC-4323 | 231–249 | 5 | 0.309 | 0.521 | 0.543 | 0.411 * | 0.077 |
| Mean | 6.7 | 0.557 | 0.605 | 0.622 | 0.080 | 0.053 |
| Locus | Allele Size | A | HO | HS | HT | FIS | FST |
|---|---|---|---|---|---|---|---|
| CC-30080 | 148–166 | 7 | 0.465 | 0.680 | 0.722 | 0.268 * | 0.113 |
| CC-33079-1 | 197–205 | 3 | 0.644 | 0.495 | 0.547 | −0.293 | 0.173 |
| CC-935 | 131–133 | 2 | 0.372 | 0.356 | 0.356 | −0.030 | −0.002 |
| CsCAT34 | 147–153 | 3 | 0.530 | 0.417 | 0.419 | −0.246 | 0.008 |
| CC-14826 | 226–244 | 6 | 0.702 | 0.614 | 0.734 | −0.135 | 0.281 |
| CC-2994 | 266–275 | 2 | 0.404 | 0.353 | 0.350 | −0.126 | −0.019 |
| CC-3722 | 137–141 | 3 | 0.721 | 0.573 | 0.598 | −0.275 | 0.079 |
| CC-6538 | 161–187 | 7 | 0.627 | 0.547 | 0.711 | −0.150 | 0.366 |
| CC-4950 | 247–257 | 2 | 0.503 | 0.469 | 0.470 | −0.049 | 0.003 |
| CcC02022 | 353–361 | 5 | 0.753 | 0.697 | 0.731 | −0.088 | 0.086 |
| CFA22 | 156–158 | 2 | 0.342 | 0.228 | 0.285 | −0.502 | 0.363 |
| CC-25032 | 182–194 | 2 | 0.921 | 0.495 | 0.498 | −0.876 | 0.010 |
| CC-41284 | 271–273 | 2 | 0.419 | 0.361 | 0.503 | −0.152 | 0.439 |
| CS05 | 156–160 | 3 | 0.021 | 0.163 | 0.175 | 0.872 * | 0.115 |
| CC-43042 | 204–240 | 3 | 0.219 | 0.196 | 0.199 | −0.125 | 0.032 |
| CFA71 | 184–214 | 8 | 0.702 | 0.663 | 0.786 | −0.045 | 0.269 |
| CC-42621 | 291–294 | 2 | 0.354 | 0.231 | 0.439 | −0.532 | 0.616 |
| CsCAT14 | 130–138 | 2 | 0.167 | 0.194 | 0.198 | 0.089 | 0.037 |
| CC-4562 | 144–168 | 6 | 0.659 | 0.575 | 0.772 | −0.132 * | 0.398 |
| CC-26213 | 137–157 | 3 | 0.346 | 0.680 | 0.673 | 0.521 * | −0.021 |
| CFA61 | 184–196 | 3 | 0.246 | 0.221 | 0.222 | −0.113 | 0.004 |
| CS24 | 117–144 | 9 | 0.602 | 0.640 | 0.683 | 0.042 | 0.119 |
| CS43 | 144–164 | 7 | 0.791 | 0.746 | 0.765 | −0.066 | 0.049 |
| CT161 | 145–148 | 2 | 0.414 | 0.320 | 0.331 | −0.279 | 0.066 |
| CT128 | 181–199 | 6 | 0.451 | 0.468 | 0.722 | 0.027 * | 0.528 |
| CS20 | 239–253 | 7 | 0.555 | 0.672 | 0.707 | 0.171 | 0.095 |
| CS44 | 137–141 | 3 | 0.325 | 0.508 | 0.513 | 0.363 | 0.020 |
| CC-33079 | 247–257 | 3 | 0.539 | 0.448 | 0.591 | −0.183 | 0.387 |
| CC-4323 | 237–240 | 2 | 0.188 | 0.155 | 0.171 | −0.208 | 0.156 |
| Mean | 4.0 | 0.482 | 0.454 | 0.513 | −0.061 | 0.204 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, X.; Chen, R.; Bian, Y.; Qin, X.; Zhang, Z.; Sun, Y. Identification of a Natural Hybrid between Castanopsis sclerophylla and Castanopsis tibetana (Fagaceae) Based on Chloroplast and Nuclear DNA Sequences. Forests 2020, 11, 873. https://doi.org/10.3390/f11080873
Zeng X, Chen R, Bian Y, Qin X, Zhang Z, Sun Y. Identification of a Natural Hybrid between Castanopsis sclerophylla and Castanopsis tibetana (Fagaceae) Based on Chloroplast and Nuclear DNA Sequences. Forests. 2020; 11(8):873. https://doi.org/10.3390/f11080873
Chicago/Turabian StyleZeng, Xiaorong, Risheng Chen, Yunxin Bian, Xinsheng Qin, Zhuoxin Zhang, and Ye Sun. 2020. "Identification of a Natural Hybrid between Castanopsis sclerophylla and Castanopsis tibetana (Fagaceae) Based on Chloroplast and Nuclear DNA Sequences" Forests 11, no. 8: 873. https://doi.org/10.3390/f11080873
APA StyleZeng, X., Chen, R., Bian, Y., Qin, X., Zhang, Z., & Sun, Y. (2020). Identification of a Natural Hybrid between Castanopsis sclerophylla and Castanopsis tibetana (Fagaceae) Based on Chloroplast and Nuclear DNA Sequences. Forests, 11(8), 873. https://doi.org/10.3390/f11080873





