Mutation Mechanism of Leaf Color in Plants: A Review
Abstract
1. Introduction
2. Genetic Model of Plant Leaf Color Mutants
3. Molecular Mechanisms of Plant Leaf Color Mutations
3.1. Abnormal Chlorophyll Metabolism Pathway
3.1.1. Mutations of Genes Related to the Chlorophyll Synthesis Pathway
3.1.2. Mutations of Genes Related to the Chlorophyll Degradation Pathway
3.1.3. Mutations of Genes Related to the Heme Metabolism Pathway
3.2. Abnormal Chloroplast Development and Differentiation
3.3. Abnormal Carotenoid Metabolism Pathway
3.4. Abnormal Anthocyanin Metabolism Pathway
3.4.1. Mutations of Structural Genes Related to the Anthocyanin Synthesis Pathway
3.4.2. Abnormal Regulatory Factors Related to the Anthocyanin Synthesis Pathway
4. Mapping and Cloning of Leaf Color Mutational Genes
4.1. Using Map-Based Cloning to Discover Mutant Genes
4.2. Using RNA-Seq to Discover Mutant Genes
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Awan, M.A.; Konzak, C.F.; Rutger, J.N.; Nilan, R.A. Mutagenic effects of sodium azide in rice. Crop Sci. 1980, 20, 663–668. [Google Scholar] [CrossRef]
- Wang, L.; Yue, C.; Cao, H.; Zhou, Y.; Zeng, J.; Yang, Y.; Wang, X. Biochemical and transcriptome analyses of a novel chlorophyll-deficient chlorina tea plant cultivar. BMC Plant Biol. 2014, 14. [Google Scholar] [CrossRef] [PubMed]
- Beale, S.I.; Appleman, D. Chlorophyll synthesis in chlorella: Regulation by degree of light limitation of growth. Plant Physiol. 1971, 47, 230–235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brestic, M.; Zivcak, M.; Kunderlikova, K.; Allakhverdiev, S.I. High temperature specifically affects the photoprotective responses of chlorophyll b-deficient wheat mutant lines. Photosynth. Res. 2016, 130, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.M.; Zhang, X.; Wang, J.L.; Wan, J.M. Leaf chloroplast ultrastructure and photosynthetic properties of a chlorophyll-deficient mutant of rice. Photosynthetica 2014, 52, 217–222. [Google Scholar] [CrossRef]
- Seetharami Reddi, T.V.; Reddi, V.R. Frequency and spectrum of chlorophyll mutants induced in rice by chemical mutagens. Theor. Appl. Genet. 1984, 67, 231–233. [Google Scholar] [CrossRef]
- Falbel, T.G.; Staehelin, L.A. Partial blocks in the early steps of the chlorophyll synthesis pathway: A common feature of chlorophyll b-deficient mutants. Physiol. Plant. 1996, 97, 311–320. [Google Scholar] [CrossRef]
- Tanaka, A.; Tanaka, R. Chlorophyll metabolism. Curr. Opin. Plant Biol. 2006, 9, 248–255. [Google Scholar] [CrossRef]
- Chen, G.; Bi, Y.R.; Li, N. EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J. 2005, 41, 364–375. [Google Scholar] [CrossRef]
- Lonosky, P.M.; Zhang, X.S.; Honavar, V.G.; Dobbs, D.L.; Fu, A.; Rodermel, S.R. A proteomic analysis of maize chloroplast biogenesis. Plant Physiol. 2004, 134, 560–574. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, B. Arbuscular mycorrhiza induced putrescine degradation into γ-aminobutyric acid, malic acid accumulation, and improvement of nitrogen assimilation in roots of water-stressed maize plants. Mycorrhiza 2020, 30, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.-S.; Van Phap, N.; Jeon, H.-W.; Kim, M.-H.; Eom, S.H.; Lim, Y.J.; Kim, W.-C.; Park, E.-J.; Choi, Y.-I.; Ko, J.-H. Overexpression of PtrMYB119, a R2R3-MYB transcription factor from Populus trichocarpa, promotes anthocyanin production in hybrid poplar. Tree Physiol. 2016, 36, 1162–1176. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Peuke, A.D.; Zhao, X.; Yan, J.; Li, C. Effects of simulated atmospheric nitrogen deposition on foliar chemistry and physiology of hybrid poplar seedlings. Plant Physiol. Biochem. 2019, 143, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Guo, Y.; Adil, M.F.; Sehar, S.; Cai, B.; Xiang, Z.; Tu, Y.; Zhao, D.; Shamsi, I.H. Comparative proteomic analysis by iTRAQ reveals that plastid pigment metabolism contributes to leaf color changes in tobacco (Nicotiana tabacum) during curing. Int. J. Mol. Sci. 2020, 21, 2394. [Google Scholar] [CrossRef] [PubMed]
- Zhou, h.; Wang, K.; Wang, F.; Espley, R.V.; Ren, F.; Zhao, J.; Ogutu, C.; He, H.; Jiang, Q.; Allan, A.C.; et al. Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 2019, 221, 1919–1934. [Google Scholar] [CrossRef] [PubMed]
- Bae, C.H.; Abe, T.; Matsuyama, T.; Fukunishi, N.; Nagata, N.; Nakano, T.; Kaneko, Y.; Miyoshi, K.; Matsushima, H.; Yoshida, S. Regulation of chloroplast gene expression is affected in ali, a novel tobacco albino mutant. Ann. Bot. 2001, 88, 545–553. [Google Scholar] [CrossRef]
- Tominaga-Wada, R.; Nukumizu, Y.; Wada, T. Flowering is delayed by mutations in homologous genes CAPRICE and TRYPTICHON in the early flowering Arabidopsis cpl3 mutant. J. Plant Physiol. 2013, 170, 1466–1468. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Zhu, L.; Shen, H.; Huq, E. PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 9433–9438. [Google Scholar] [CrossRef]
- Aluru, M.R.; Yu, F.; Fu, A.; Rodermel, S. Arabidopsis variegation mutants: New insights into chloroplast biogenesis. J. Exp. Bot. 2006, 57, 1871–1881. [Google Scholar] [CrossRef]
- Jarvis, P.; Chen, L.J.; Li, H.; Peto, C.A.; Fankhauser, C.; Chory, J. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 1998, 282, 100–103. [Google Scholar] [CrossRef]
- Zheng, J.; Wu, H.; Zhu, H.; Huang, C.; Liu, C.; Chang, Y.; Kong, Z.; Zhou, Z.; Wang, G.; Lin, Y.; et al. Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytol. 2019, 223, 705–721. [Google Scholar] [CrossRef]
- Deng, L.; Qin, P.; Liu, Z.; Wang, G.; Chen, W.; Tong, J.; Xiao, L.; Tu, B.; Sun, Y.; Yan, W.; et al. Characterization and fine-mapping of a novel premature leaf senescence mutant yellow leaf and dwarf 1 in rice. Plant Physiol. Biochem. 2017, 111, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Guo, S.; Wang, Z.; Du, Q.; Xing, Y.; Zhang, T.; Shen, W.; Sang, X.; Ling, Y.; He, G. Map-based cloning and functional analysis of YGL8, which controls leaf colour in rice (Oryza sativa). BMC Plant Biol. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.H.; Hur, J.; Ryu, C.H.; Choi, Y.; Chung, Y.Y.; Miyao, A.; Hirochika, H.; An, G.H. Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant Cell Physiol. 2003, 44, 463–472. [Google Scholar] [CrossRef]
- Wang, S.; Wang, P.; Gao, L.; Yang, R.; Li, L.; Zhang, E.; Wang, Q.; Li, Y.; Yin, Z. Characterization and complementation of a chlorophyll-less dominant mutant GL1 in Lagerstroemia indica. DNA Cell Biol. 2017, 36, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Z.; Wang, P.; Wang, S.; Ma, L.; Li, L.; Yang, R.; Ma, Y.; Wang, Q. Comprehensive transcriptome analysis discovers novel candidate genes related to leaf color in a Lagerstroemia indica yellow leaf mutant. Genes Genom. 2015, 37, 851–863. [Google Scholar] [CrossRef]
- Liu, L.; Sun, T.; Liu, X.; Guo, Y.; Huang, X.; Gao, P.; Wang, X. Genetic analysis and mapping of a striped rind gene (st3) in melon (Cucumis melo L.). Euphytica 2019, 215. [Google Scholar] [CrossRef]
- Kusumi, K.; Sakata, C.; Nakamura, T.; Kawasaki, S.; Yoshimura, A.; Iba, K. A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions. Plant J. 2011, 68, 1039–1050. [Google Scholar] [CrossRef]
- Su, N.; Hu, M.-L.; Wu, D.-X.; Wu, F.-Q.; Fei, G.-L.; Lan, Y.; Chen, X.-L.; Shu, X.-L.; Zhang, X.; Guo, X.-P.; et al. Disruption of a rice pentatricopeptide repeat protein causes a seedling-specific albino phenotype and its utilization to enhance seed purity in hybrid rice production. Plant Physiol. 2012, 159, 227–238. [Google Scholar] [CrossRef]
- Yoo, S.-C.; Cho, S.-H.; Sugimoto, H.; Li, J.; Kusumi, K.; Koh, H.-J.; Iba, K.; Paek, N.-C. Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol. 2009, 150, 388–401. [Google Scholar] [CrossRef]
- Cheng, G.-X.; Zhang, R.-X.; Liu, S.; He, Y.-M.; Feng, X.-H.; Ul Haq, S.; Luo, D.-X.; Gong, Z.-H. Leaf-color mutation induced by ethyl methane sulfonate and genetic and physio-biochemical characterization of leaf-color mutants in pepper (Capsicum annuum L.). Sci. Hortic. 2019, 257. [Google Scholar] [CrossRef]
- Cao, W.; Du, Y.; Wang, C.; Xu, L.; Wu, T. Cscs encoding chorismate synthase is a candidate gene for leaf variegation mutation in cucumber. Breed. Sci. 2018, 68, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.-M.; Wei, S.-L.; Chen, J.; Wu, Y.; Li, F.; Wei, L.-B.; Li, C.; Zeng, Y.-J.; Tian, Y.; Wang, D.-Y.; et al. Cytological, genetic, and proteomic analysis of a sesame (Sesamum indicum L.) mutant Siyl-1 with yellow-green leaf color. Genes Genom. 2020, 42, 25–39. [Google Scholar] [CrossRef] [PubMed]
- La Rocca, N.; Rascio, N.; Oster, U.; Rudiger, W. Amitrole treatment of etiolated barley seedlings leads to deregulation of tetrapyrrole synthesis and to reduced expression of Lhc and RbcS genes. Planta 2001, 213, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Monde, R.A.; Zito, F.; Olive, J.; Wollman, F.A.; Stern, D.B. Post-transcriptional defects in tobacco chloroplast mutants lacking the cytochrome b6/f complex. Plant J. 2000, 21, 61–72. [Google Scholar] [CrossRef]
- Hosticka, L.P.; Hanson, M.R. Induction of plastid mutations in tomatoes by nitrosomethylurea. J. Hered. 1984, 75, 242–246. [Google Scholar] [CrossRef]
- Li, J.; Yu, X.; Cai, Z.; Wu, F.; Luo, J.; Zheng, L.; Chu, W. An overview of chlorophyll biosynthesis in higher plants. Mol. Plant Breed. 2019, 17, 6013–6019. [Google Scholar] [CrossRef]
- Beale, S.I. Green genes gleaned. Trends Plant Sci. 2005, 10, 309–312. [Google Scholar] [CrossRef]
- Matringe, M.; Camadro, J.M.; Block, M.A.; Joyard, J.; Scalla, R.; Labbe, P.; Douce, R. Localization within chloroplasts of protoporphyrinogen oxidase, the target enzyme for diphenylether-like herbicides. J. Biol. Chem. 1992, 267, 4646–4651. [Google Scholar]
- Tanaka, R.; Kobayashi, K.; Masuda, T. Tetrapyrrole Metabolism in Arabidopsis thaliana. Arab. Book 2011, 9, e0145. [Google Scholar] [CrossRef]
- Sampath, P.; Mazumder, B.; Seshadri, V.; Gerber, C.A.; Chavatte, L.; Kinter, M.; Ting, S.M.; Dignam, J.D.; Kim, S.; Driscoll, D.M.; et al. Noncanonical function of glutamyl-prolyi-tRNA synthetase: Gene-specific silencing of translation. Cell 2004, 119, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, J.H.; Yoo, E.; Lee, C.H.; Hirochika, H.; An, G.H. Differential regulation of chlorophyll a oxygenase genes in rice. Plant Mol. Biol. 2005, 57, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Schelbert, S.; Park, S.-Y.; Han, S.-H.; Lee, B.-D.; Andres, C.B.; Kessler, F.; Hoertensteiner, S.; Paek, N.-C. Stay-green and chlorophyll catabolic enzymes interact at light-harvesting complex ii for chlorophyll detoxification during leaf senescence in arabidopsis. Plant Cell 2012, 24, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.-R.; Zhang, F.-T.; Gao, J.-X.; Sun, X.-Q.; Deng, X.-J. An overview of chlorophyll biosynthesis in higher plants. Acta Bot. Boreali Occident. Sin. 2009, 29, 629–636. [Google Scholar] [CrossRef]
- Cornah, J.E.; Terry, M.J.; Smith, A.G. Green or red: What stops the traffic in the tetrapyrrole pathway? Trends Plant Sci. 2003, 8, 224–230. [Google Scholar] [CrossRef]
- Schmied, J.; Hedtke, B.; Grimm, B. Overexpression of HEMA1 encoding glutamyl-tRNA reductase. J. Plant Physiol. 2011, 168, 1372–1379. [Google Scholar] [CrossRef]
- Kumar, A.M.; Csankovszki, G.; Soll, D. A second and differentially expressed glutamyl-tRNA reductase gene from Arabidopsis thaliana. Plant Mol. Biol. 1996, 30, 419–426. [Google Scholar] [CrossRef]
- Ilag, L.L.; Kumar, A.M.; Soll, D. Light regulation of chlorophyll biosynthesis at the level of 5-aminolevulinate formation in Arabidopsis. Plant Cell 1994, 6, 265–275. [Google Scholar] [CrossRef]
- Kumar, A.M.; Soll, D. Antisense HEMA1 RNA expression inhibits heme and chlorophyll biosynthesis in Arabidopsis. Plant Physiol. 2000, 122, 49–56. [Google Scholar] [CrossRef]
- Goslings, D.; Meskauskiene, R.; Kim, C.H.; Lee, K.P.; Nater, M.; Apel, K. Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J. 2004, 40, 957–967. [Google Scholar] [CrossRef]
- Walker, C.J.; Willows, R.D. Mechanism and regulation of Mg-chelatase. Biochem. J. 1997, 327 Pt 2, 321–333. [Google Scholar] [CrossRef]
- Strand, A.; Asami, T.; Alonso, J.; Ecker, J.R.; Chory, J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 2003, 421, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Rissler, H.M.; Collakova, E.; DellaPenna, D.; Whelan, J.; Pogson, B.J. Chlorophyll biosynthesis. Expression of a second chl I gene of magnesium chelatase in Arabidopsis supports only limited chlorophyll synthesis. Plant Physiol. 2002, 128, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Papenbrock, J.; Mock, H.P.; Tanaka, R.; Kruse, E.; Grimm, B. Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol. 2000, 122, 1161–1169. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, N.; Brusslan, J.A.; Larkin, R.; Nagatani, A.; Chory, J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 2001, 98, 2053–2058. [Google Scholar] [CrossRef]
- Shi, Y.; He, Y.; Guo, D.; Lv, X.; Huang, Q.; Wu, J. Genetic analysis and gene mapping of a pale green leaf mutant HM133 in rice. Chin. J. Rice Sci. 2016, 30, 603–610. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, Z.; Shan, X.; Li, C.; Tang, X.; Chi, M.; Feng, H. Physiological properties and chlorophyll biosynthesis in a Pak-choi (Brassica rapa L. ssp chinensis) yellow leaf mutant, pylm. Acta Physiol. Plant. 2017, 39. [Google Scholar] [CrossRef]
- Wenkai, D.; Yuan, S.; Hu, F. Research progress on molecular mechanisms of the leaf color mutation. Mol. Plant Breed. 2019, 17, 1888–1897. [Google Scholar] [CrossRef]
- Hoertensteiner, S. Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 2006, 57, 55–77. [Google Scholar] [CrossRef]
- You, S.-C.; Cho, S.-H.; Zhang, H.; Paik, H.-C.; Lee, C.-H.; Li, J.; Yoo, J.-H.; Lee, B.-W.; Koh, H.-J.; Seo, H.S.; et al. Quantitative trait loci associated with functional stay-green SNU-SG1 in rice. Mol. Cells 2007, 24, 83–94. [Google Scholar]
- Zhu, G.; Yang, F.; Shi, S.; Li, D.; Wang, Z.; Liu, H.; Huang, D.; Wang, C. Transcriptome characterization of cymbidium sinense ‘dharma’ using 454 pyrosequencing and its application in the identification of genes associated with leaf color variation. PLoS ONE 2015, 10, e0128592. [Google Scholar] [CrossRef] [PubMed]
- Wilks, A. Heme oxygenase: Evolution, structure, and mechanism. Antioxid. Redox Signal. 2002, 4, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Terry, K. Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol. 1999, 119, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Linley, P.J.; Landsberger, M.; Kohchi, T.; Cooper, J.B.; Terry, M.J. The molecular basis of heme oxygenase deficiency in the pcd1 mutant of pea. FEBS J. 2006, 273, 2594–2606. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Cheng, Z.; Ma, X.; Wu, H.; Liu, Y.; Zhou, K.; Chen, Y.; Ma, W.; Bi, J.; Zhang, X.; et al. A knockdown mutation of YELLOW-GREEN LEAF2 blocks chlorophyll biosynthesis in rice. Plant Cell Rep. 2013, 32, 1855–1867. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Chai, J.; Wang, L.; Wang, C.; Long, W.; Wang, D.; Wang, Y.; Zheng, M.; Peng, C.; et al. Green-revertible Chlorina 1 (grc1) is required for the biosynthesis of chlorophyll and the early development of chloroplasts in rice. J. Plant Biol. 2013, 56, 326–335. [Google Scholar] [CrossRef]
- Fankhauser, C.; Chory, J. Light control of plant development. Annu. Rev. Cell Dev. Biol. 1997, 13, 203–229. [Google Scholar] [CrossRef]
- Chan, K.X.; Crisp, P.A.; Estavillo, G.M.; Pogson, B.J. Chloroplast-to-nucleus communication: Current knowledge, experimental strategies and relationship to drought stress signaling. Plant Signal. Behav. 2010, 5, 1575–1582. [Google Scholar] [CrossRef]
- Pogson, B.J.; Albrecht, V. Genetic dissection of chloroplast biogenesis and development: An overview. Plant Physiol. 2011, 155, 1545–1551. [Google Scholar] [CrossRef]
- Koussevitzky, S.; Nott, A.; Mockler, T.C.; Hong, F.; Sachetto-Martins, G.; Surpin, M.; Lim, I.J.; Mittler, R.; Chory, J. Signals from chloroplasts converge to regulate nuclear gene expression. Science 2007, 316, 715–719. [Google Scholar] [CrossRef]
- Lopez-Juez, E.; Jarvis, R.P.; Takeuchi, A.; Page, A.M.; Chory, J. New Arabidopsis cue mutants suggest a close connection between plastid- and phytochrome regulation of nuclear gene expression. Plant Physiol. 1998, 118, 803–815. [Google Scholar] [CrossRef]
- Miura, E.; Kato, Y.; Matsushima, R.; Albrecht, V.; Laalami, S.; Sakamoto, W. The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell 2007, 19, 1313–1328. [Google Scholar] [CrossRef]
- Mei, J.; Li, F.; Liu, X.; Hu, G.; Fu, Y.; Liu, W. Newly identified CSP41b gene localized in chloroplasts affects leaf color in rice. Plant Sci. 2017, 256, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, V.; Ingenfeld, A.; Apel, K. Characterization of the Snowy cotyledon 1 mutant of Arabidopsis thaliana: The impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol. Biol. 2006, 60, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Li, M.; Xu, F.; Xu, Q.; Ge, S.; Dong, J. Analysis of agronomic characters and preliminary mapping of regulatory genes of a barley yellow-green leaf mutant ygl. J. Triticeae Crop. 2019, 39, 653–658. [Google Scholar]
- Ren, Y.; Zhu, Y.; Sun, D.; Deng, Y.; An, G.; Li, W.; Liu, J. Physiological characteristic analysis of a leaf-yellowing mutant in watermelon. J. Fruit Sci. 2020, 37, 565–573. [Google Scholar] [CrossRef]
- Polivka, T.; Frank, H.A. Molecular factors controlling photosynthetic light harvesting by carotenoids. Acc. Chem. Res. 2010, 43, 1125–1134. [Google Scholar] [CrossRef]
- Giorio, G.; Stigliani, A.L.; D’Ambrosio, C. Phytoene synthase genes in tomato (Solanum lycopersicum L.)—New data on the structures, the deduced amino acid sequences and the expression patterns. FEBS J. 2008, 275, 527–535. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Rodriguez-Concepcion, M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 2010, 107, 11626–11631. [Google Scholar] [CrossRef]
- Busch, M.; Seuter, A.; Hain, R. Functional analysis of the early steps of carotenoid biosynthesis in tobacco. Plant Physiol. 2002, 128, 439–453. [Google Scholar] [CrossRef] [PubMed]
- Aluru, M.R.; Bae, H.; Wu, D.; Rodermel, S.R. The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiol. 2001, 127, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Aluru, M.R.; Zola, J.; Foudree, A.; Rodermel, S.R. Chloroplast photooxidation-induced transcriptome reprogramming in Arabidopsis immutans white leaf sectors. Plant Physiol. 2009, 150, 904–923. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Chai, C.; Qian, Q.; Li, C.; Tang, J.; Sun, L.; Huang, Z.; Guo, X.; Sun, C.; Liu, M.; et al. Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J. 2008, 54, 177–189. [Google Scholar] [CrossRef]
- Park, H.; Kreunen, S.S.; Cuttriss, A.J.; Della, P.D.; Pogson, B.J. Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 2002, 14, 321–332. [Google Scholar] [CrossRef]
- Seeram, N.P.; Nair, M.G. Inhibition of lipid peroxidation and structure-activity-related studies of the dietary constituents anthocyanins, anthocyanidins, and catechins. J. Agric. Food Chem. 2002, 50, 5308–5312. [Google Scholar] [CrossRef]
- Zhao, D.; Tao, J. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, H.; Wang, Y.; Guan, S.; Wang, F.; Tang, J.; Zhang, R.; Xie, L.; Lu, Y. Characterization of the cis elements in the proximal promoter regions of the anthocyanin pathway genes reveals a common regulatory logic that governs pathway regulation. J. Exp. Bot. 2015, 66, 3775–3789. [Google Scholar] [CrossRef]
- Davies, K.M.; Albert, N.W.; Schwinn, K.E. From landing lights to mimicry: The molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 2012, 39, 619–638. [Google Scholar] [CrossRef]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef]
- Holton, T.A.; Cornish, E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Liao, L.; Zhou, H.; Wang, L.; Deng, X.; Han, Y. Constitutive activation of an anthocyanin regulatory gene PcMYB10.6 is related to red coloration in Purple-Foliage Plum. PLoS ONE 2015, 10, e0135159. [Google Scholar] [CrossRef] [PubMed]
- Fukusaki, E.; Kawasaki, K.; Kajiyama, S.; An, C.I.; Suzuki, K.; Tanaka, Y.; Kobayashi, A. Flower color modulations of Torenia hybrida by downregulation of chalcone synthase genes with RNA interference. J. Biotechnol. 2004, 111, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Meng, X.; Liang, L.; Jiang, W.; Huang, Y.; He, J.; Hu, H.; Almqvist, J.; Gao, X.; Wang, L. Molecular and biochemical analysis of chalcone synthase from freesia hybrid in flavonoid biosynthetic pathway. PLoS ONE 2015, 10, e0119154. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, M.; Nakatsuka, T.; Yamamura, S. Flavonoid components and flower color change in transgenic tobacco plants by suppression of chalcone isomerase gene. FEBS Lett. 2005, 579, 6074–6078. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Higeta, D.; Suzuki, A.; Yoshida, H.; Ozeki, Y. Excision of transposable elements from the chalcone isomerase and dihydroflavonol 4-reductase genes may contribute to the variegation of the yellow-flowered carnation (Dianthus caryophyllus). Plant Cell Physiol. 2002, 43, 578–585. [Google Scholar] [CrossRef]
- Kim, E.-Y.; Kim, C.-W.; Kim, S. Identification of two novel mutant ANS alleles responsible for inactivation of anthocyanidin synthase and failure of anthocyanin production in onion (Allium cepa L.). Euphytica 2016, 212, 427–437. [Google Scholar] [CrossRef]
- Brugliera, F.; Tao, G.-Q.; Tems, U.; Kalc, G.; Mouradova, E.; Price, K.; Stevenson, K.; Nakamura, N.; Stacey, I.; Katsumoto, Y.; et al. Violet/blue chrysanthemums-metabolic engineering of the anthocyanin biosynthetic pathway results in novel petal colors. Plant Cell Physiol. 2013, 54, 1696–1710. [Google Scholar] [CrossRef]
- Katsumoto, Y.; Fukuchi-Mizutani, M.; Fukui, Y.; Brugliera, F.; Holton, T.A.; Karan, M.; Nakamura, N.; Yonekura-Sakakibara, K.; Togami, J.; Pigeaire, A.; et al. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 2007, 48, 1589–1600. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB transcription factors as regulators of secondary metabolism in plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, M.; Toda, S.; Tasaki, K. The novel allele of the LhMYB12 gene is involved in splatter-type spot formation on the flower tepals of Asiatic hybrid lilies (Lilium spp.). New Phytol. 2014, 201, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Borevitz, J.O.; Xia, Y.; Blount, J.; DIXon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383–2394. [Google Scholar] [CrossRef]
- Zhou, H.; Kui, L.-W.; Wang, H.; Gu, C.; Dare, A.P.; Espley, R.V.; He, H.; Allan, A.C.; Han, Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015, 82, 105–121. [Google Scholar] [CrossRef]
- Yang, P.; Zhou, B.; Li, Y. The bHLH transcription factors involved in anthocyanin biosynthesis in plants. Plant Physiol. J. 2012, 48, 747–758. [Google Scholar] [CrossRef]
- Bai, H.; Song, Z.; Zhang, Y.; Li, Z.; Wang, Y.; Liu, X.; Ma, J.; Quan, J.; Wu, X.; Liu, M.; et al. The bHLH transcription factor PPLS1 regulates the color of pulvinus and leaf sheath in foxtail millet (Setaria italica). Theor. Appl. Genet. 2020, 133, 1911–1926. [Google Scholar] [CrossRef]
- Li, P.; Chen, B.; Zhang, G.; Chen, L.; Dong, Q.; Wen, J.; Mysore, K.S.; Zhao, J. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 2016, 210, 905–921. [Google Scholar] [CrossRef]
- Sharma, S.; Holme, I.B.; Dionisio, G.; Kodama, M.; Dzhanfezova, T.; Joernsgaard, B.; Brinch-Pedersen, H. Cyanidin based anthocyanin biosynthesis in orange carrot is restored by expression of AmRosea1 and AmDelila, MYB and bHLH transcription factors. Plant Mol. Biol. 2020, 103, 443–456. [Google Scholar] [CrossRef]
- Xiang, L.-L.; Liu, X.-F.; Li, X.; Yin, X.-R.; Grierson, D.; Li, F.; Chen, K.-S. A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in Chrysanthemums (Chrysanthemum morifolium Ramat.). PLoS ONE 2015, 10, e0143892. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, P.; Li, J.; Zhang, C.; Wang, L.; Ren, Z. Genome-wide analysis of the WD-repeat protein family in cucumber and Arabidopsis. Mol. Genet. Genom. 2014, 289, 103–124. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Grain, D.; Le Gourrierec, J.; Harscoet, E.; Berger, A.; Jauvion, V.; Scagnelli, A.; Berger, N.; Bidzinski, P.; Kelemen, Z.; et al. Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol. 2013, 198, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Broun, P. Transcriptional control of flavonoid biosynthesis: A complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 2005, 8, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Jun, J.H.; Dixon, R.A. MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula. Plant Physiol. 2014, 165, 1424–1439. [Google Scholar] [CrossRef]
- Neale, D.B.; Kremer, A. Forest tree genomics: Growing resources and applications. Nat. Rev. Genet. 2011, 12, 111–122. [Google Scholar] [CrossRef]
- Chen, S.; Lin, X.; Zhang, D.; Li, Q.; Zhao, X.; Chen, S. Genome-wide analysis of NAC gene family in Betula pendula. Forests 2019, 10, 741. [Google Scholar] [CrossRef]
- Wang, S.; Huang, H.; Han, R.; Liu, C.; Qiu, Z.; Liu, G.; Chen, S.; Jiang, J. Negative feedback loop between BpAP1 and BpPI/BpDEF heterodimer in Betula platyphylla x B. pendula. Plant Sci. 2019, 289. [Google Scholar] [CrossRef]
- Voichek, Y.; Weigel, D. Identifying genetic variants underlying phenotypic variation in plants without complete genomes. Nat. Genet. 2020, 52, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.-A.; Zhang, H.; Liu, Z.; Shi, M.; et al. Pan-genome of wild and cultivated soybeans. Cell 2020, 182, 162. [Google Scholar] [CrossRef]
- Tanksley, S.D.; Ganal, M.W.; Martin, G.B. Chromosome landing: A paradigm for map-based gene cloning in plants with large genomes. Trends Genet. 1995, 11, 63–68. [Google Scholar] [CrossRef]
- Lin, T.; Sun, L.; Jing, D.; Qian, H.; Yu, B.; Zeng, S.; Li, C.; Gong, H. Characterization and gene mapping of a Yellow Green Leaf Mutant ygl14(t) in Rice. J. Nucl. Agric. Sci. 2018, 32, 216–226. [Google Scholar]
- Gao, S.; Gao, W.; Liao, X.; Xiong, C.; Yu, G.; Yang, Q.; Yang, C.; Ye, Z. The tomato WV gene encoding a thioredoxin protein is essential for chloroplast development at low temperature and high light intensity. BMC Plant Biol. 2019, 19. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Cui, H.; Zhang, B.; Liu, X.; Yang, L.; Zhuang, M.; Lv, H.; Li, Z.; Wang, Y.; Fang, Z.; et al. Map-based cloning and characterization of BoCCD4, a gene responsible for white/yellow petal color in B-oleracea. BMC Genom. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Ming, M.; Allan, A.C.; Gu, C.; Li, L.; Wu, X.; Wang, R.; Chang, Y.; Qi, K.; Zhang, S.; et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017, 92, 437–451. [Google Scholar] [CrossRef]
- Gao, M.; Hu, L.; Li, Y.; Weng, Y. The chlorophyll-deficient golden leaf mutation in cucumber is due to a single nucleotide substitution in CsChlI for magnesium chelatase I subunit. Theor. Appl. Genet. 2016, 129, 1961–1973. [Google Scholar] [CrossRef]
- Mueller, A.H.; Dockter, C.; Gough, S.P.; Lundqvist, U.; von Wettstein, D.; Hansson, M. Characterization of mutations in barley fch2 encoding chlorophyllide a oxygenase. Plant Cell Physiol. 2012, 53, 1232–1246. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, X.; Xu, B.; Li, Y.; Ma, Y.; Wang, G. Phenotype and transcriptome analysis reveals chloroplast development and pigment biosynthesis together influenced the leaf color formation in mutants of Anthurium andraeanum ‘Sonate’. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Cao, H.; Li, H.; Miao, Z.; Fu, G.; Yang, C.; Wu, L.; Zhao, P.; Shan, Q.; Ruan, J.; Wang, G.; et al. The preliminary study of leaf-color mutant in dendrobium officinale. J. Nucl. Agric. Sci. 2017, 31, 461–471. [Google Scholar]
- Sun, C.; Deng, L.; Du, M.; Zhao, J.; Chen, Q.; Huang, T.; Jiang, H.; Li, C.-B.; Li, C. A Transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol. Plant 2020, 13, 42–58. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hensel, G.; Mascher, M.; Melzer, M.; Budhagatapalli, N.; Rutten, T.; Himmelbach, A.; Beier, S.; Korzun, V.; Kumlehn, J.; et al. Leaf variegation and impaired chloroplast development caused by a truncated CCT domain gene in albostrians barley. Plant Cell 2019, 31, 1430–1445. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Shi, N.; An, X.; Liu, C.; Fu, H.; Cao, L.; Feng, Y.; Sun, D.; Zhang, L. Candidate genes for yellow leaf color in common wheat (Triticum aestivum L.) and major related metabolic pathways according to transcriptome profiling. Int. J. Mol. Sci. 2018, 19, 1594. [Google Scholar] [CrossRef] [PubMed]
- Gang, H.; Li, R.; Zhao, Y.; Liu, G.; Chen, S.; Jiang, J. Loss of GLK1 transcription factor function reveals new insights in chlorophyll biosynthesis and chloroplast development. J. Exp. Bot. 2019, 70, 3125–3138. [Google Scholar] [CrossRef]
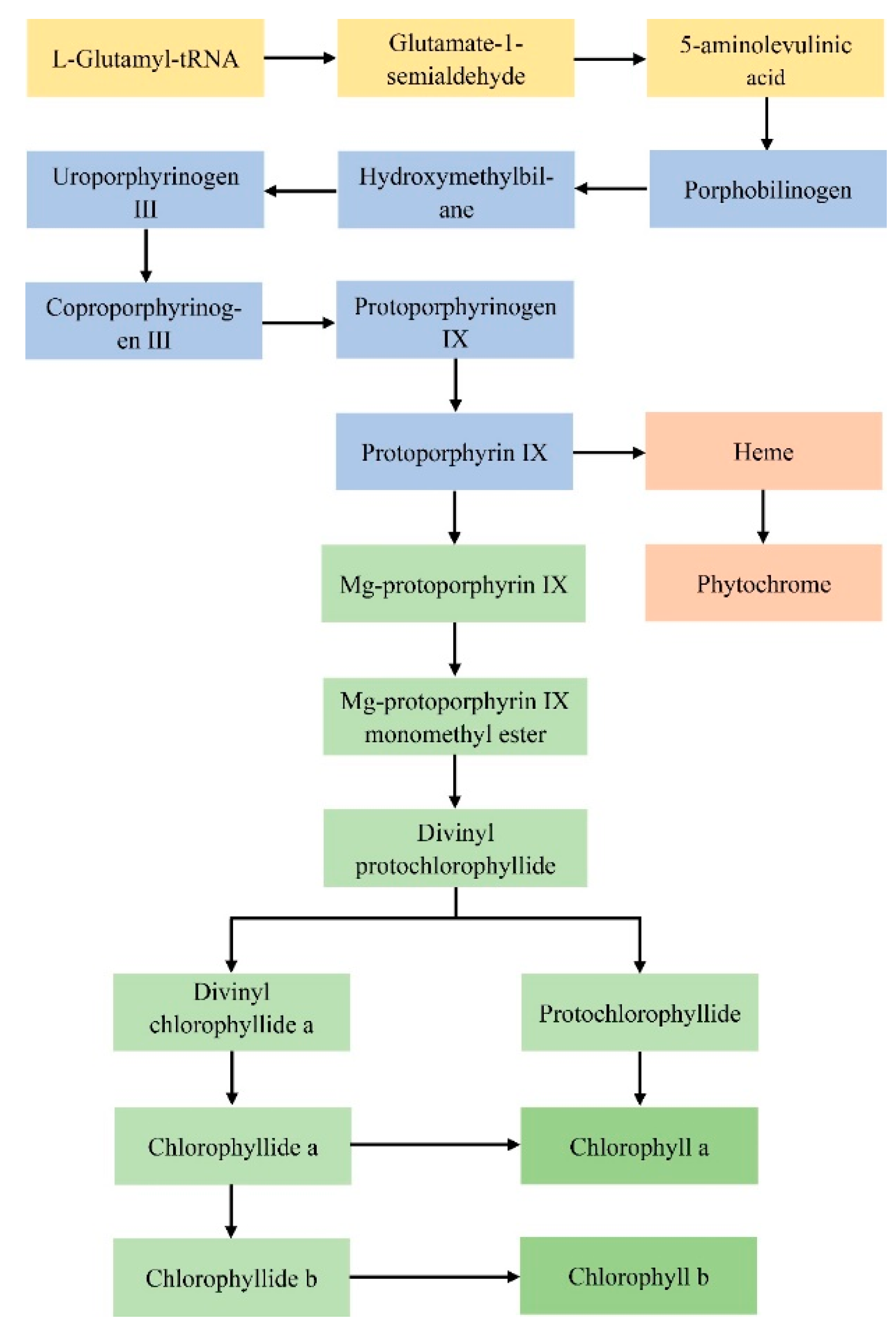
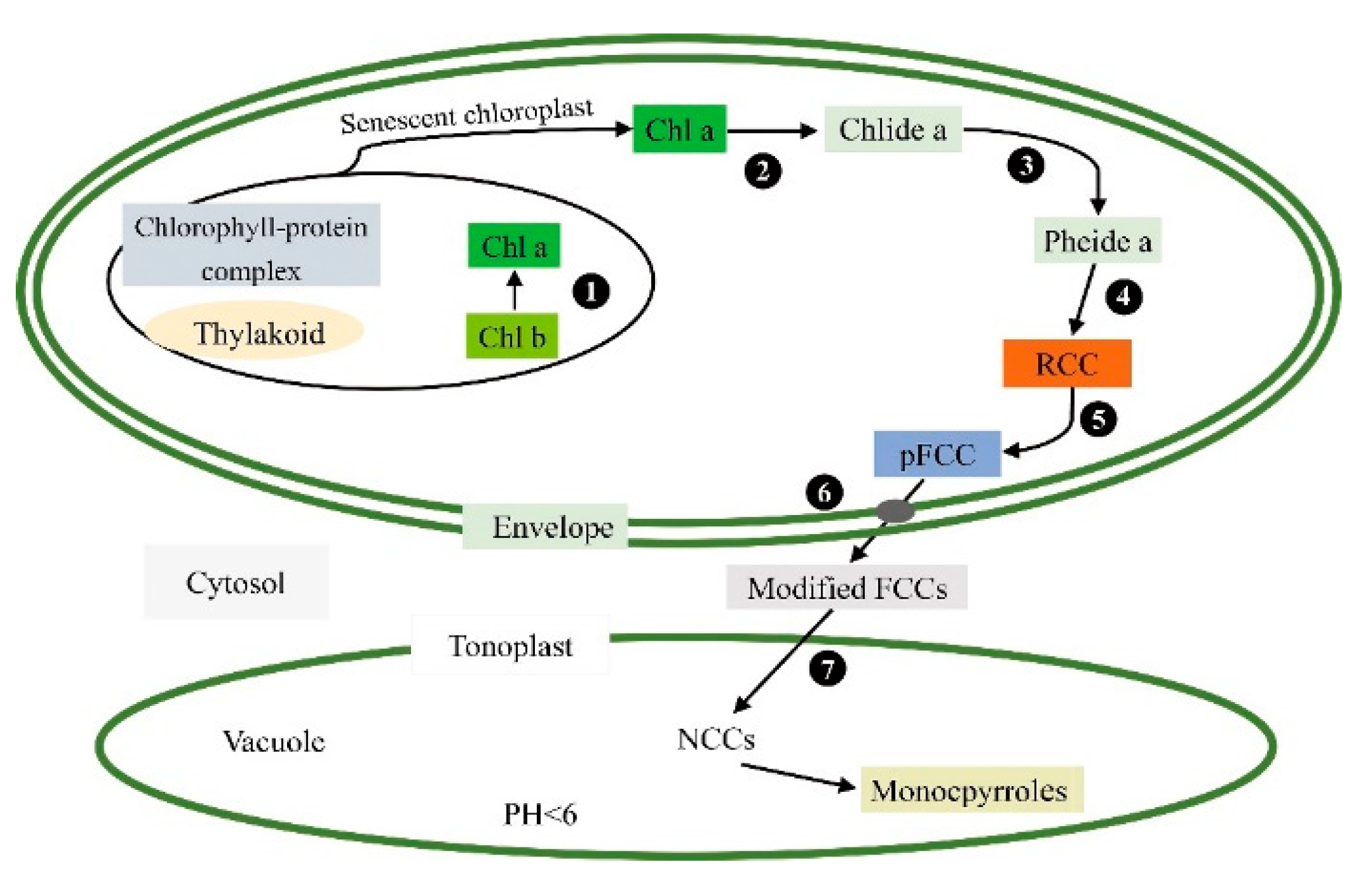
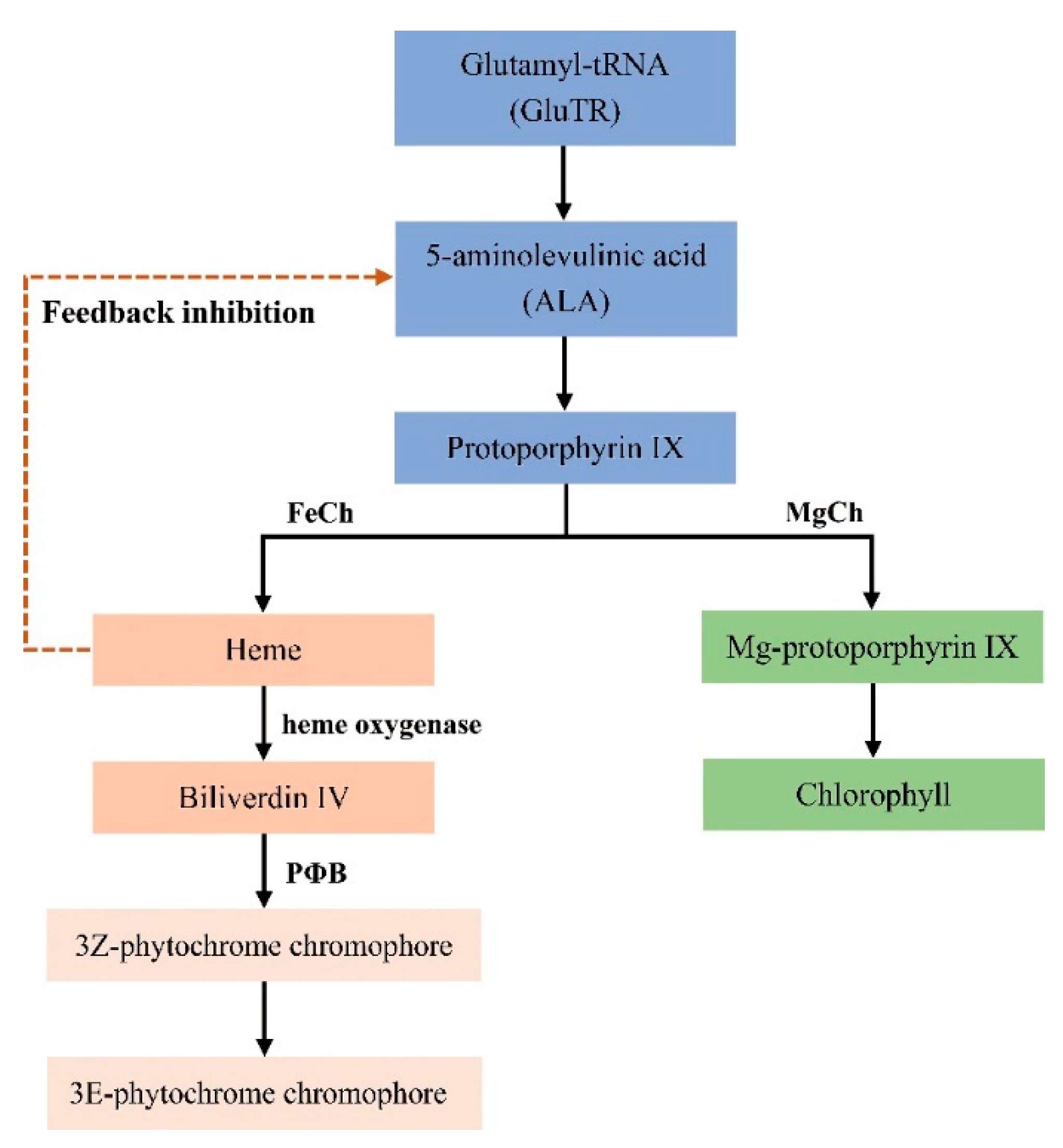
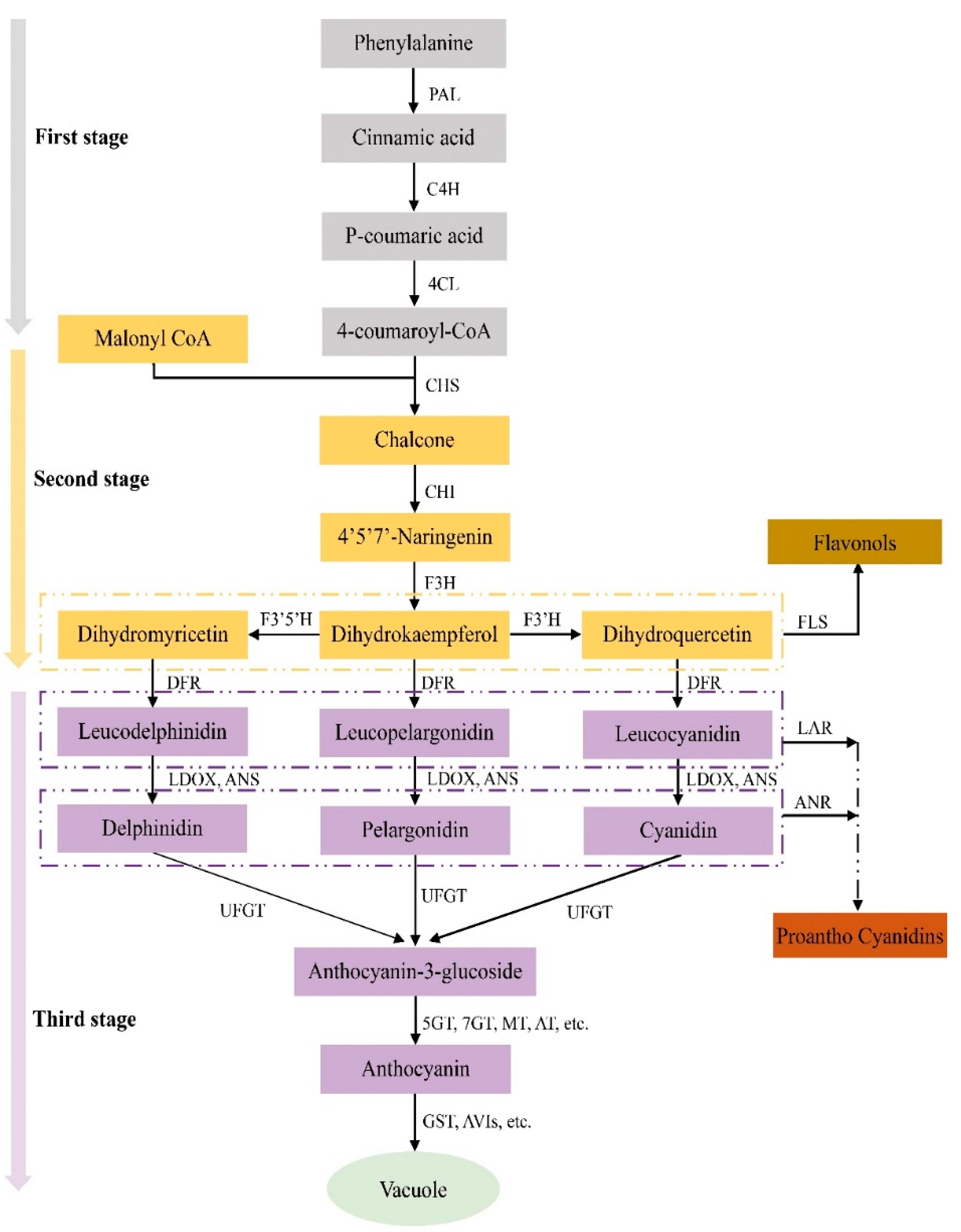
| Step | Enzyme Name | Abbreviated Name of Enzyme | Gene Name | Locus Name in Arabidopsis |
|---|---|---|---|---|
| 1 | Glutamyl-tRNA reductase | GluTR | HEMA1 | AT1G58290 |
| HEMA2 | AT1G09940 | |||
| HEMA3 | AT2G31250 | |||
| 2 | Glutamate-1-semialdehyde 2, 1-aminomutase | GSA-AM | GSA1 (HEML1) | AT5G63570 |
| GSA2 (HEML2) | AT3G48730 | |||
| 3 | 5-Aminolevulinate dehydratase | PBGS (ALAD) | HEMB1 | AT1G69740 |
| HEMB2 | AT1G44318 | |||
| 4 | Porphobilinogen deaminase | PBGD | HEMC | AT5G08280 |
| 5 | Uroporphyrinogen III synthase | UROS | HEMD | AT2G26540 |
| 6 | Uroporphyrinogen III decarboxylase | UROD | HEME1 | AT3G14930 |
| HEME2 | AT2G40490 | |||
| 7 | Coproporphyrinogen III oxidase | CPOX | HEMF1 | AT1G03475 |
| HEMF2 | AT4G03205 | |||
| 8 | Protoporphyrinogen oxidase | PPOX | HEMG1 | AT4G01690 |
| HEMG2 | AT5G14220 | |||
| 9 | Magnesium chelatase H subunit | MgCh | CHLH | AT5G13630 |
| Magnesium chelatase I subunit | CHL11 | AT4G18480 | ||
| CHL12 | AT5G45930 | |||
| Magnesium chelatase D subunit | CHLD | AT1G08520 | ||
| 10 | Mg-protoporphyrin IX methyltransferase | MgPMT | CHLM | AT4G25080 |
| 11 | Mg-protoporphyrin IX monomethyl ester | MgPME | CRD1 (ACSF) | AT3G56940 |
| 12 | 3,8-Divinyl protochlorophyllide a 8-vinyl reductase | DVR | DVR | AT5G18660 |
| 13 | Protochlorophyllide oxidoreductase | POR | PORA | AT5G54190 |
| PORB | AT4G27440 | |||
| PORC | AT1G03630 | |||
| 14 | Chlorophyll synthase | CHLG | CHLG | AT3G51820 |
| 15 | Chlorophyllide a oxygenase | CAO | CAO (CHL) | AT1G44446 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, M.-H.; Li, X.; Zhang, X.-X.; Zhang, H.; Zhao, X.-Y. Mutation Mechanism of Leaf Color in Plants: A Review. Forests 2020, 11, 851. https://doi.org/10.3390/f11080851
Zhao M-H, Li X, Zhang X-X, Zhang H, Zhao X-Y. Mutation Mechanism of Leaf Color in Plants: A Review. Forests. 2020; 11(8):851. https://doi.org/10.3390/f11080851
Chicago/Turabian StyleZhao, Ming-Hui, Xiang Li, Xin-Xin Zhang, Heng Zhang, and Xi-Yang Zhao. 2020. "Mutation Mechanism of Leaf Color in Plants: A Review" Forests 11, no. 8: 851. https://doi.org/10.3390/f11080851
APA StyleZhao, M.-H., Li, X., Zhang, X.-X., Zhang, H., & Zhao, X.-Y. (2020). Mutation Mechanism of Leaf Color in Plants: A Review. Forests, 11(8), 851. https://doi.org/10.3390/f11080851






