Abstract
Phlebiopsis gigantea (Fr.) Jülich has been successfully used as a biological control fungus for Heterobasidion annosum (Fr.) Bref., an important pathogen of pine and spruce trees. The P. gigantea species has been known for many years, but our understanding of the relationship between various isolates of this fungus has been substantially improved through the application of DNA sequence comparisons. In this study, relationships between P. gigantea and selected Basidiomycota species was determined, based on elongation factor 1-alpha (EF1α) partial DNA sequence and in silico data. A total of 12 isolates, representing the most representatives of P. gigantea, with diverse geographic distributions and hosts, were included in this study. Phylogenetic trees generated for sequences obtained in this research, grouped the European taxa of P. gigantea and partial sequence of the genome deposed in NCBI database, in a strongly supported clade, basal to the rest of the strains included in the study. P. gigantea isolates originating from Poland, Finland, Sweden, Great Britain and partial sequence of genome formed a monophyletic group. Within this group, isolates of P. gigantea constituted two subclades, showing their partial difference like the two SNPs (single nucleotide polymorphisms) between one and the rest of isolates. The intron and exon relationships among P. gigantea isolates were moreover resolved. The results obtained using the EF1α region should be useful in the selection of more efficient P. gigantea isolates for limiting forest tree root pathogens.
1. Introduction
Phlebiopsis gigantea (Fr.) Jülich has widely been used as a biological control of the fungus Heterobasidion annosum (Fr.) Bref., the causative factor of conifer root rot infections [1,2,3,4,5,6,7,8,9,10,11,12]. In the literature this species is described with multiple synonymous names, e.g., Corticium giganteum (Fr.) Fr., Peniophora gigantea (Fr.) Massee, Peniophora gigantea f. pruinosa Pilát, Phanerochaete gigantea (Fr.) S.S. Rattan, Phlebia gigantea (Fr.) Donk. The fungus, after the taxonomic revision made by Jülich (1978) In: Parmasto and Hallenberg [13], was finally placed in the new genus Phlebiopsis, which is now commonly accepted. The fungus is a typical saprotroph, regularly colonizing dead parts of coniferous woods, occurring mostly in the Northern Hemisphere, but it has also has a worldwide distribution; for example, few strains have been retrieved from South Africa, New Zealand and Canada [9,14].
According to the Index Fungorum database (www.indexfungorum.org) the taxonomic status of P. gigantea is well recognized and there are at least 23 species included in the genus Phlebiopsis, based on morphological characteristics. A number of techniques have been employed to identify P. gigantea and traditionally, the morphological characters of the spores were used for this purpose. Although the classical methods are reasonably easy and fast to apply [15], molecular techniques confirm identification of this fungus and are very useful for identification of species [16,17].
Various molecular markers used in barcoding, like ITS 1/2, β-tubulin, histone H3 and elongation factor α (syn. EF1α), are the most commonly applied in fungal taxonomy. Their application allows determination of fungal genotypes at a species level [18,19,20]. Comparisons of DNA sequence data are increasingly being used in order to gain knowledge concerning the phylogenetic relationships among P. gigantea isolates [7,10,11,21,22]. Many studies have utilised DNA sequence data of the EF1α gene for phylogenetic analyses including a wide range species of fungi [23,24].
The objectives of this study were to obtain DNA partial sequences for EF1α for P. gigantea strains, and to compare them with other some Basidiomycota species from the NCBI database. This gene is a highly conserved ubiquitous protein involved in translation that has been suggested to have desirable properties for phylogenetic inference [25]. It has been successfully used in phylogenetic studies as a phylogenetic marker for Eukaryotes, Acomycetes and Basidiomycota [23,24,25] Additionally, the partial sequences (intron and exon partial regions) obtained in the study of EF1α gene isolates of P. gigantea from Poland, Finland, Sweden and Great Britain, and partial genome sequence (gi:752829739) deposited in GenBank by Hori et al. [26] were investigated. To date, in studies on the differentiation of chosen P. gigantea isolates on the known activity of linear growth and wood decay [3,4,11,27] the EF1α gene has not been studied.
Our study provides an additional gene region useful for testing taxonomic groupings and phylogenetic relationships, previously identified based on the other gene regions like ITS [2,3]. Several factors can affect biological activity of the fungus. Grossbard [28] reported, that the presence of some fungi in soil can modify biological traits of co-occurring taxons. Schardl and Craven [29] described that the variation in enzyme and decay activity of fungal isolates in time may suggested risks in lost or change the molecular and biochemical characteristics. The cause is showed in possible hybridisation from the mating of clearly homozygous individuals. Żółciak et al. [3] and Sierota et al. [27] suggested the changes in the activity of different P. gigantea isolates with time, its origin, and wood density. For the effective use of competitive fungi used in biopreparations against pathogens in biological control (e.g., P. gigantea), there is a need for periodic exchange of strains for more effective ones [4]. Checking the utility of the EF1α region can be a valuable clue and can help in making decision regarding the selection of the most effective P. gigantea isolates as a competitor of Heterobasidion spp.
2. Materials and Methods
2.1. Cultivation of Isolates
Twelve previously identified and tested isolates of P. gigantea [11] were used in the experiment: six from Poland (not registered as biocontrol agent) and one from Finland, one from Sweden and four from Great Britain (registered as biocontrol agent) (Table 1). The number of Polish isolates was limited to six due to difficulties in obtaining homogeneous single-spore cultures, while the Finnish and British isolates were accepted as previously tested and approved. Isolates were grown on potato dextrose agar (PDA) medium (DifcoTM, Sparks, MD, USA) in Petri dishes for ten days at 20 °C according to Kwaśna et al. [30].

Table 1.
List of P. gigantea isolates used in the study.
2.2. DNA Extraction
Total fungal DNA of P. gigantea was extracted from mycelium grown on PDA by using DNeasy Plant Mini Kit (Qiagen, Hilden, Germany), according to the protocol. Quality of the DNA was checked with an Infinite 200 PRO multimode plate reader (Tecan, Group Ltd., Männedorf, Switzerland).
2.3. Primers and PCR Conditions
The PCR reactions were done in 25 μL volumes using a Veriti Thermal Cycler (Applied Biosystems, Foster City, CA, USA). Each reaction contained 1 unit (0.25 μL) of Taq DNA polymerase (recombinant) (Thermo Scientific, Life Technologies Inc., Carlsbad, CA, USA), 2.5 μL of 10 × Taq buffer, 2 mM of each dNTP (0.5 μL), 1.5 mM of MgCl2 (1.5 μL), 12.5 pmol of forward/reverse primers (0.125 μL), and 20 ng (1 μL) of DNA.
The primers and the touchdown PCR reaction conditions were used according to modified procedure of Rehner [18]. Amplicons of the partial region of the EF1& gene were generated using two overlapping primer combinations, 526F (5′ GTC GTY GTY ATY GGH CAY GT 3′) × 1567R (5′ AC HGT RCC RAT ACC ACC RAT CTT 3′) and EFdF (5′ AAG GAY GGN CAR ACY CGN GAR CAY GC 3′) × 2218R (5′ AT GAC ACC RAC RGC RAC RGT YTG 3′). For 526F × 1567R primers the touchdown PCR was as follows: 5 min at 94 °C (an initial denaturation), 30 s at 94 °C (denaturation) and 45 s at 60 °C (annealing) in the first cycle, successively reducing the Tm by 1 °C per cycle over the next 9 cycles to a final Tm 50 °C, which was used in the remaining 36 cycles. An extension step per cycle was 1 min 30 s at 72 °C. The final extension step was 1 cycle of 7 min at 72 °C.
PCR parameters for EFdF × 2218R primers were as follows: 5 min at 94 °C, 45 cycles of (30 s at 94 °C, 30 s at 63 °C, 1 min 30 s at 72 °C) and 7 min at 72 °C. Amplicons were run in 1.5% (w/v) ethidium bromide-stained agarose gels (Sigma-Aldrich, St. Louis, MO, USA) and the bands were visualized under UV illumination. PCR products were sequenced using Sanger’s sequencing method at the Polish Academy of Sciences Institute of Biochemistry and Biophysics (Warsaw, Poland).
2.4. DNA Sequence Analysis
Sequencing results were analyzed using the BLAST algorithm on The National Center for Biotechnology Information (NCBI, https://blast.ncbi.nlm.nih.gov/) database and MEGA version 5 [31]. The DNA sequences were aligned using Clustal W version 2.0. [32]. Genetic variability was estimated for the sequences of P. gigantea and realigned separately for intron and exon regions using FGENESH 2.6 [33,34].
The obtained sequences were analyzed using Tajima’s Neutrality Test. The number of sequences (m), number of segregating sites (S), mean number of segregating sites (Ps), nucleotide diversity (π) and estimates of Theta (Θ) per site as well as the Tajima test statistic (D) were estimated using MEGA5 program [35,36].
EF1α partial gene sequences of P. gigantea and selected Basidiomycota species from the NCBI database were analyzed using the maximum likelihood approach for phylogeny reconstruction and tested by bootstrapping with 1,000 replicates. Missing and ambiguous characters were excluded from the analysis. Phylogenetic trees were generated based on maximum likelihood method [37]. Two sequences of P. gigantea were deposited in GenBank NCBI (Accession numbers: KU886024 and KU886025, www.ncbi.nlm.nih.gov.genbank/).
3. Results
EF1α Partial Gene of P. gigantea Analysis
The region of the EF1α gene was successfully amplified for all P. gigantea isolates described in this study. All amplifications yielded a single band approximately 1,500 bp long. The final analyzed length partial sequence of the EF1α gene was 1,411 bp, the mRNA length was 1,403 bp. The structure of EF1α partial gene from the P. gigantea (Accession numbers: KU886024 and KU886025) is shown in Figure 1.
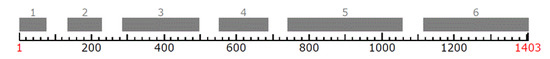
Figure 1.
The structure of EF1α partial gene from the Phlebiopsis gigantea. The positions for exons are presented respectively: exon 1 (1–75), exon 2 (134–228), exon 3 (284–496), exon 4 (550–686), exon 5 (739–1056), exon 6 (1113–1403). (mRNA) 1–6 exon (s): 1–1403. The numbers indicate the positions of the bases identified using FGENESH software.
Genetic diversity was calculated for all obtained sequences including exons and introns of 12 partial sequences of the EF1α gene and partial genome sequence (gi:752829739) of P. gigantea (Figure 1) and separately for exons and introns (Table 2) The number of segregating sites (S) and mean number of segregating sites (Ps) were 32 and 0.022679 respectively. The value of the Θ, expressing the total variability was 0.007308 for analyzed 13 partial sequences mentioned above. The nucleotide diversity (π) was 0.005761. The value of Tajima’s D was below zero (−0.931820).

Table 2.
Genetic polymorphism and neutrality tests of EF1α partial gene sequences of 12 Polish isolates and partial genome sequence (gi:752829739) of P. gigantea, including introns and exons.
In the I1 and I2 intron region the number of segregating sites was one. The remaining introns I3, I4 and I5 showed three and six segregating sites, respectively. The value of the Θ, expressing the total variability ranged from 0.005556 to 0.034526 for all intron part sequences. The nucleotide diversity (π) ranged from 0.004863 to 0.025641. The analysis of exons of the coding regions showed no segregating sites for partial E1, however the number of segregating sites ranged from one for E2 to seven for E6. The value of the Θ was zero for partial E1 and the highest 0.007544 for E6. Among exons the nucleotide diversity (π) ranged from zero to 0.006176. The value of Tajima’s D was below zero for all intron and E2–E6 exon regions (Table 2).
Bootstrap values supported the separation of species into distinct clades (Figure 2). Among partial 12 sequences of P. gigantea the closest one to the partial genome sequence of P. gigantea (gi: 752829739) was GB Pg 16 (KU886025) from Great Britain. The couple of single mutations differentiated GB Pg 16 sequence from the rest of identical isolates of this fungus.
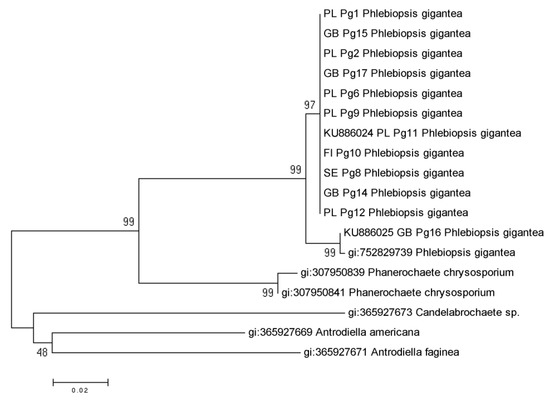
Figure 2.
Phylogenetic tree for Phlebiopsis gigantea isolates and other species from the Phanerochaetaceae, based on EF1α gene partial sequences. Branch length values were shown, the tree was reconstructed using the maximum likelihood approach and tested by bootstrapping (1,000 replicates).
For Phanerochaetaceae family, two major and well supported groups were identified. One of the group represented all P. gigantea and Phanerochaete chrysosporium Burds. partial sequences (97–99% bootstrap support). The second group included: Candelabrochaete sp., Antrodiella Americana Ryvarden & Gilb. and Antrodiella faginea Vampola & Pouzar (48% bootstrap support). Phylogenetic analysis of partial sequences of EF1α gene for P. gigantea and selected Basidiomycota species, showed separate clades for following orders: Polyporales, Agaricales, Boletales, Atheliales, Russulales and Sebacinales (Figure 3).
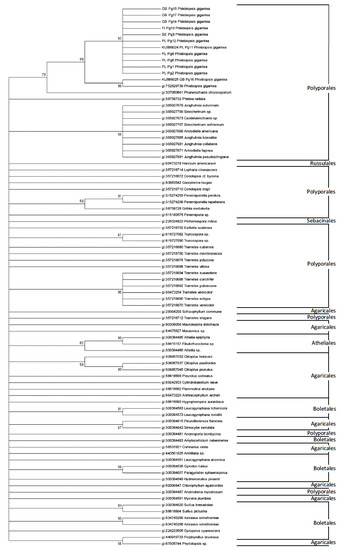
Figure 3.
Phylogenetic tree for Phlebiopsis gigantea isolates and selected Basidiomycota, based on the EF1α partial gene sequences. The tree was reconstructed using the maximum likelihood approach and tested by bootstrapping (1000 replicates).
4. Discussion
In this study, DNA partial sequence for the EF1α gene of Phlebiopsis gigantea was successfully identified and analyzed. All P. gigantea isolates yielded PCR products of similar size, indicating that the amplified partial gene region does not include large indels, and therefore is a suitable choice for phylogenetic studies. The aligned sequences showed considerable homology among P. gigantea but various species-specific nucleotide substitutions and indels were observed among all Basidiomycota species. No sequence variation was observed among all P. gigantea except of one isolate (GB Pg 16) from Great Britain and partial sequence of P. gigantea genome (gi: 752829739).
The remarkable variation was observed between different strains. This is consistent with previous studies employing ITS region and genetic fingerprinting using random amplified microsatellite (RAMS) markers in taxonomic studies of P. gigantea [21].
For the analyzed P. gigantea EF1α partial gene the occurrence of exons and introns was identified. Introns represented the so-called 3rd introns group, spliced during the maturation of RNA with the participation of spliceosome [38]. The size of introns ranged from 52 to 58 bp, at the average length of the gene from 0.75 to 1,000 bp of gene.
Among representatives of Basidiomycota six introns in gene are observed average [39]. Compared to higher Eukaryota, introns occurring in fungi are relatively short [40]. In the case of the analyzed gene, introns size corresponds to the average size of introns identified in representatives of the Fungi kingdom (50–200 bp) [41]. The length of introns of EF1α gene was slightly smaller than the average for the Puccinia graminis (0.65 kb, NCBI accession number: X73529.1) and Neurospora crassa gene (0.81 kb) [42].
Despite the randomness and variability of the structure of introns, in the case of I5 of EF1α partial gene, a larger number of segregating sites (six) was identified. In the sequences of other introns this number were from 1 to 3. Furthermore, the characteristic feature of the identified intron sequences was the presence of the sequence GT on the donor side (5’) and the sequence AG at the acceptor side (3’). They are essential in the process of identifying and splicing by spliceosome. Identified GT and AG sequences are considered as the most commonly occurring canonical dinucleotide fragments, respectively starting and ending the introns [43].
Intraspecies analysis of EF1α partial gene showed small variability within the coding and non-coding regions. Introns of analyzed EF1α partial gene were well conserved among all tested isolates of P. gigantea. The evolution of the structure of all introns and E2–E6 exon regions of EF1α partial gene was neutral. Isolates representing P. gigantea from Poland, Great Britain, Finland and Sweden had similar partial sequences of EF1α gene and were grouped together. The only British isolate (GB Pg 16 = FOC PG B 20/5) of this fungus showed coupled single mutations like single nucleotide polymorphisms (SNPs) and was very similar to P. gigantea (gi: 752829739) with identity 99% and query cover 100%.
Results of the study using DNA-RAMS markers indicated genetic similarity among isolates collected in Finland and Great Britain [10]. Additional studies using the previously mentioned markers showed that Polish isolate (PL 12) of P. gigantea was genetically analogous to FC 16 from Great Britain [11]. Vainio and Hantula [21] also showed that European and North American ITS/A alleles of P. gigantea were identical, while ITS/C alleles were different. The authors mentioned that the analysis of molecular variation and neighbor joining analysis using 28 RAMS markers revealed a considerable degree of differentiation between Europe and North America [21].
The main advantage of this study is the phylogenetic analysis of EF1α partial DNA sequence data for P. gigantea in comparing it to species belonging to the same family—Phanerochaetaceae and selected Basidiomycota species. Phylogenetic trees showed that P. gigantea is closely related to Phanerochaete chrysosporium. EF1α is also the first protein-coding gene and first single-copy gene used for phylogenetic analysis of P. gigantea. The whole genome sequence of P. gigantea (gi: 752829739, accession number: AZAG01000080) was published by Hori et al. [26]. The whole genome sequence size is approximately 30 Mbp and number of predicted genes (11,891). Sequence data from the majority of isolates belonging to the different species showed unique species-specific substitutions, allowing the isolates to be differentiated into clades representing the species.
The results of this study demonstrate that the EF1α region is useful for phylogenetic analysis and classification of Polyporales species. This is a large and taxonomically difficult order, which include several genera, for example: Phlebiopsis, Phanerochaete, Phlebia, Junghuhnia, Steccherinum, Androniella, Ganoderma, Coriolopsis, Perenniporiella and Trametes.
5. Conclusions
Our in silico studies showed a relationship between P. gigantea and selected Basidiomycota based on elongation factor 1-alpha partial DNA sequence. The partial EF1α gene sequence of P. gigantea isolates originating from Poland, Finland, Sweden and Great Britain formed a monophyletic group, except for one British isolate GB Pg 16 (FOC PG B 20/5). The results of our study proved that the partial sequence of EF1α gene is useful for phylogenetic analysis of the Phanerochaetaceae family.
Author Contributions
The manuscript was written by all authors. The general conception of the project was provided by M.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by the Department of Plant Protection, Warsaw University of Life Sciences and by the State Forest Holding in Poland through Project No. 500 426 of the Forest Research Institute in Sękocin Stary.
Acknowledgments
The authors wish to thank Marina Brandtberg (previously: Lallemand Plant Care–Verdera Oy, Finland) for two isolates of P. gigantea (VRA 1835, VRA 1984) and Katherine Tubby (Forestry Commission, UK) for four cultures of P. gigantea (FOC PG 410.3, FOC PG SP log 5, FOC BG B 20/5, FOC PG BU 3), and the anonymous reviewers for their valuable comments and suggestions.
Conflicts of Interest
The authors declare there were no personal circumstances or interest that may be perceived as inappropriately influencing the representation or interpretation of reported research results.
References
- Sierota, Z. Heterobasidion root rot in forests on former agricultural lands in Poland: Scale of threat and prevention. Sci. Res. Essays 2013, 8, 2298–2305. [Google Scholar]
- Żółciak, A.; Sikora, K.; Nowakowska, J.A.; Małecka, M.; Borys, M.; Tereba, T.; Sierota, Z. Antrodia gossypium, Phlebiopsis gigantea and Heterobasidion parviporum: In vitro growth and Norway spruce wood block decay. Biocontrol Sci. Technol. 2016, 26, 1706–1718. [Google Scholar] [CrossRef]
- Żółciak, A.; Sierota, Z.; Małecka, M. Characterization of some Phlebiopsis gigantea isolates with respect to enzymatic activity and decay of Norway spruce wood. Biocontrol Sci. Technol. 2012, 22, 777–790. [Google Scholar] [CrossRef]
- Żółciak, A.; Sikora, K.; Wrzosek, M.; Damszel, M.; Sierota, Z. Why Does Phlebiopsis gigantea not Always Inhibit Root and Butt Rot in Conifers? Forests 2020, 11, 129. [Google Scholar] [CrossRef]
- Sun, H.; Paulin, L.; Alatalo, E.; Asiegbu, F.O. Response of living tissues of Pinus sylvestris to the saprotrophic biocontrol fungus Phlebiopsis gigantea. Tree Physiol. 2011, 31, 438–451. [Google Scholar] [CrossRef]
- Sun, H.; Terhonen, E.; Koskinen, K.; Paulin, L.; Kasanen, R.; Asiegbu, F.O. The impacts of treatment with biocontrol fungus (Phlebiopsis gigantea) on bacterial diversity of Norway spruce stumps. Biological Control 2013, 64, 238–246. [Google Scholar] [CrossRef]
- Mgbeahuruike, A.C.; Karlsson, M.; Asiegbu, F.O. Differential expression of two hydrophobin genes (Pgh1 and Pgh2) from the biological control agent Phlebiopsis gigantea. Fungal Biol. 2012, 116, 620–629. [Google Scholar] [CrossRef]
- Mgbeahuruike, A.C.; Kohler, A.; Asiegbu, F.O. Expression analysis of the impact of culture filtrates from the biocontrol agent, Phlebiopsis gigantea on the conifer pathogen, Heterobasidion annosum s.s. Transcriptome. Microb. Ecol. 2013, 66, 669–681. [Google Scholar] [CrossRef]
- Garbelotto, M.; Gonthier, P. Biology, epidemiology, and control of Heterobasidion species worldwide. Annu. Rev. Phytopathol. 2013, 51, 39–59. [Google Scholar] [CrossRef]
- Sierota, Z.; Nowakowska, J.A.; Sikora, K.; Wrzosek, M.; Żółciak, A.; Małecka, M. What is important in selecting Phlebiopsis gigantea strain for commercial use? J. Agric. Sci. Technol. B 2015, 5, 55–64. [Google Scholar]
- Sierota, Z.; Nowakowska, J.A.; Sikora, K.; Wrzosek, M.; Żółciak, A.; Małecka, M. Genetic variation among Phlebiopsis gigantea strains determined by Random Amplified Microsatellite Markers. Balt. For. 2015, 21, 178–183. [Google Scholar]
- Blomquist, M.; Herrera, S.L.; Hofmann, J.; Beram, R.C.; Cleary, M.; Rönnberg, J. Size matters but is big always better? Effectiveness of urea and Phlebiopsis gigantea as treatment against Heterobasidion on Picea abies stumps of variable size. For. Ecol. Manag. 2020, 462, 117998. [Google Scholar] [CrossRef]
- Parmasto, E.; Hallenberg, N. A taxonomic study of phlebioid fungi (Basidiomycota). Nord. J. Bot. 2000, 20, 105–118. [Google Scholar] [CrossRef]
- Grillo, R.; Hantula, J.; Korhonen, K. Interfertility between North American and European strains of Phlebiopsis gigantea. For. Pathol. 2005, 35, 173–182. [Google Scholar] [CrossRef]
- Żółciak, A. Inokulacja pniaków sosnowych preparatami biologicznymi z Phlebiopsis gigantea/Scots pine stumps inoculation with Phlebiopsis gigantea biological preparations. Leśne Pr. Badaw. 2007, 2, 77–94. (In Polish) [Google Scholar]
- Kozlakidis, Z.; Hacker, C.V.; Bradley, D.; Jamal, A.; Phoon, X.; Webber, J.; Brasier, C.M.; Buck, K.W.; Coutts, R.H.A. Molecular characterization of two novel double-stranded RNA elements from Phlebiopsis gigantea. Virus Genes 2009, 39, 132–136. [Google Scholar] [CrossRef]
- Menkis, A.; Burokienė, D.; Gaitnieks, T.; Uotila, A.; Johannesson, H.; Rosling, A.; Finlay, R.D.; Stenlid, J.; Vasaitis, R. Occurrence and impact of the root-rot biocontrol agent Phlebiopsis gigantea on soil fungal communities in Picea abies forests of Northern Europe. FEMS Microbiol. Ecol. 2012, 81, 438–445. [Google Scholar] [CrossRef]
- Rehner, S. Primers for Elongation Factor 1-α (EF1-α); Insect Biocontrol Laboratory USDA, ARS, PSI: Beltsville, MD, USA, 2001; p. 4. 1p, Available online: http://ocid.NACSE.ORG/research/deephyphae/EF1primer.pdf (accessed on 20 May 2020).
- Stępień, Ł.; Koczyk, G.; Waśkiewicz, A. Genetic and phenotypic variation of Fusarium proliferatum isolates from different host species. J. Appl. Genet. 2011, 52, 487–496. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Vainio, E.J.; Hantula, J. Genetic differentiation between European and North American populations of Phlebiopsis gigantea. Mycologia 2000, 92, 436–446. [Google Scholar] [CrossRef]
- Liu, A.Z.; Samils, N.; Higgins, B.; Stenlid, J.; Slippers, B.; Nairn, C.J.; Covert, S.F. Microsatellite markers for the wood decay fungus Phlebiopsis gigantea. Conserv. Genet. 2009, 10, 1529–1532. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E.A. Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Matheny, P.B.; Wang, Z.; Binder, M.; Curtis, J.M.; Lim, Y.W.; Nilsson, R.H.; Hughes, K.W.; Hofstetter, V.; Ammirati, J.F.; Schoch, C.L.; et al. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol. Phylogenet. Evol. 2007, 43, 430–451. [Google Scholar] [CrossRef] [PubMed]
- Roger, A.J.; Sandblom, O.; Doolittle, W.F.; Philippe, H. An evaluation of elongation factor 1 alpha as a phylogenetic marker for Eukaryotes. Mol. Biol. Evol. 1999, 16, 218–233. [Google Scholar] [CrossRef]
- Hori, C.; Ishida, T.; Igarashi, K.; Samejima, M.; Suzuki, H.; Master, E.; Ferreira, P.; Ruiz-Dueñas, F.J.; Held, B.; Canessa, P.; et al. Analysis of the Phlebiopsis gigantea genome, transcriptome and secretome provides insight into its pioneer colonization strategies of wood. PLoS Genet. 2014, 10, e1004759. [Google Scholar] [CrossRef]
- Sierota, Z.; Wrzosek, M.; Małecka, M.; Zółciak, A. Decay indices for evaluating wood decomposition activity. Biocontrol Sci. Technol. 2016, 26, 163–173. [Google Scholar] [CrossRef]
- Grossbard, E. Antibiotic Production by Fungi on Organic Manures and in Soil. J. Gen. Microbiol. 1952, 6, 295–310. [Google Scholar] [CrossRef][Green Version]
- Schardl, C.L.; Craven, K.D. Interspecific hybridization in plant-associated Fungi and Oomycetes: A review. Mol. Ecol. 2003, 12, 2861–2873. [Google Scholar] [CrossRef]
- Kwaśna, H.; Chełkowski, J.; Zajkowski, P. Grzyby (Mycota), Grzyby niedoskonałe (Deuteromycetes), Strzępczakowe (Hyphomycetales), Gruzełkowate (Tuberculariaceae), Sierpik (Fusarium); Instytut Botaniki PAN: Warszawa–Kraków, Poland, 1991; Volume T.22, p. 136. (In Polish) [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Yu, J.; Hu, S.; Wang, J.; Wong, G.K.; Li, S.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7 (Suppl. 1), 10.1–1012. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000; p. 333. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Roy, S.W.; Gilbert, W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 2006, 7, 211–221. [Google Scholar]
- Loftus, B.J.; Fung, E.; Roncaglia, P.; Rowley, D.; Amedeo, P.; Bruno, D.; Vamathevan, J.; Miranda, M.; Anderson, I.J.; Fraser, J.A.; et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 2005, 307, 1321–1324. [Google Scholar] [CrossRef]
- Deutsch, M.; Long, M. Intron-exon structures of eukaryotic model organisms. Nucleic Acids Res. 1999, 27, 3219–3228. [Google Scholar]
- Wakuliński, W. Ogólna charakterystyka grzybów i ich cechy jako patogenów roślin/General characteristics of fungi and their traits as plant pathogens. In Fitopatologia. T. 1, Podstawy fitopatologii; Kryczyński, S., Weber, Z., Eds.; Wyd. zbiorowe. PWRiL: Poznań, Poland, 2010; pp. 206–244. (In Polish) [Google Scholar]
- Stajich, J.E.; Dietrich, F.S.; Roy, S.W. Comparative genomic analysis of fungal genomes reveals intron-rich ancestors. Genome Biol. 2007, 8, R223. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Silva, J.; Burstein, D.; Pupko, T.; Eyras, E.; Ast, G. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res. 2008, 18, 88–103. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).