Complete Chloroplast Genome Sequence and Phylogenetic Inference of the Canary Islands Dragon Tree (Dracaena draco L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, DNA Extraction and Sequencing
2.2. Genome Assembly, Annotation and Identification of Simple Sequence Repeats
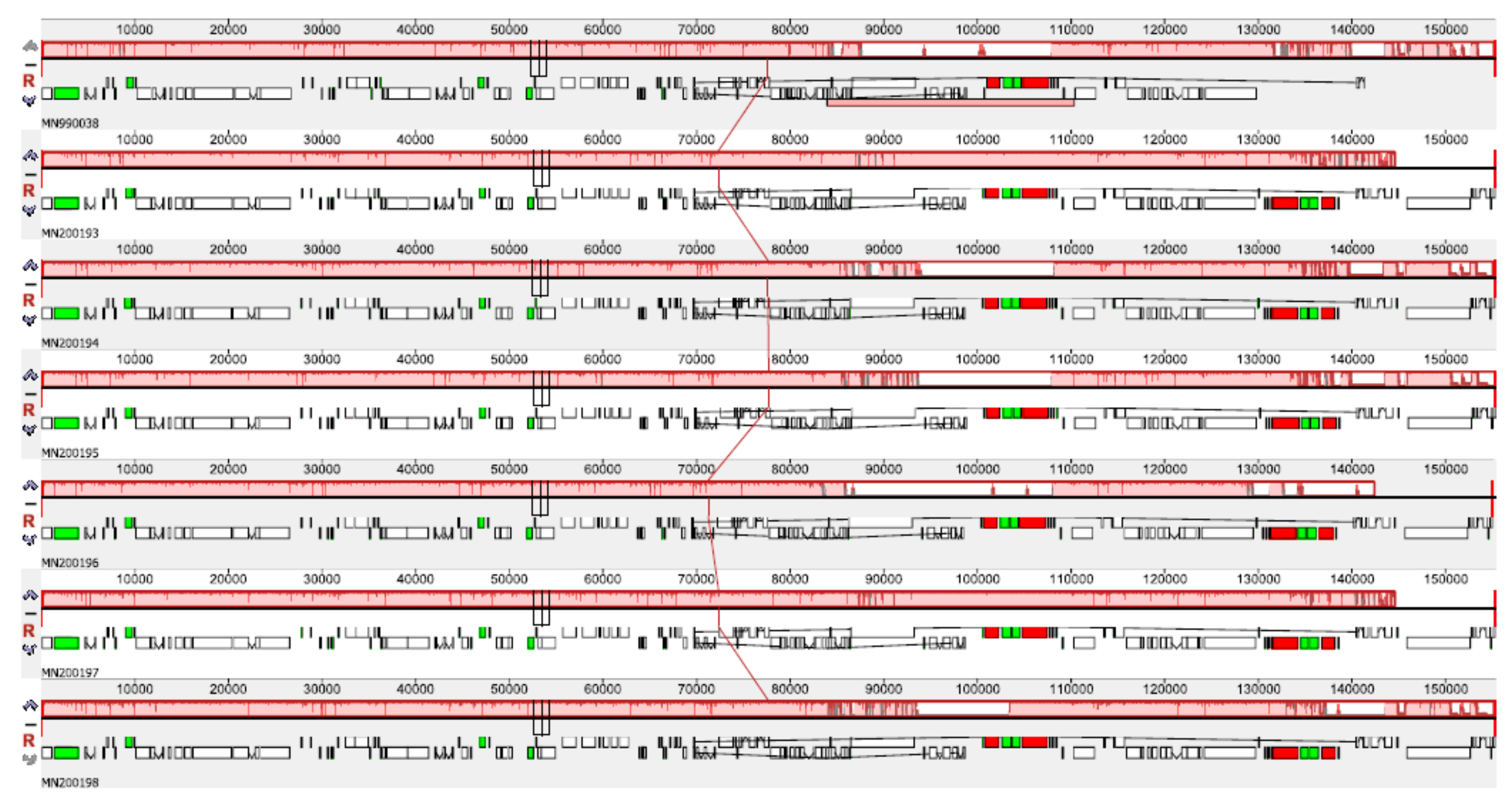
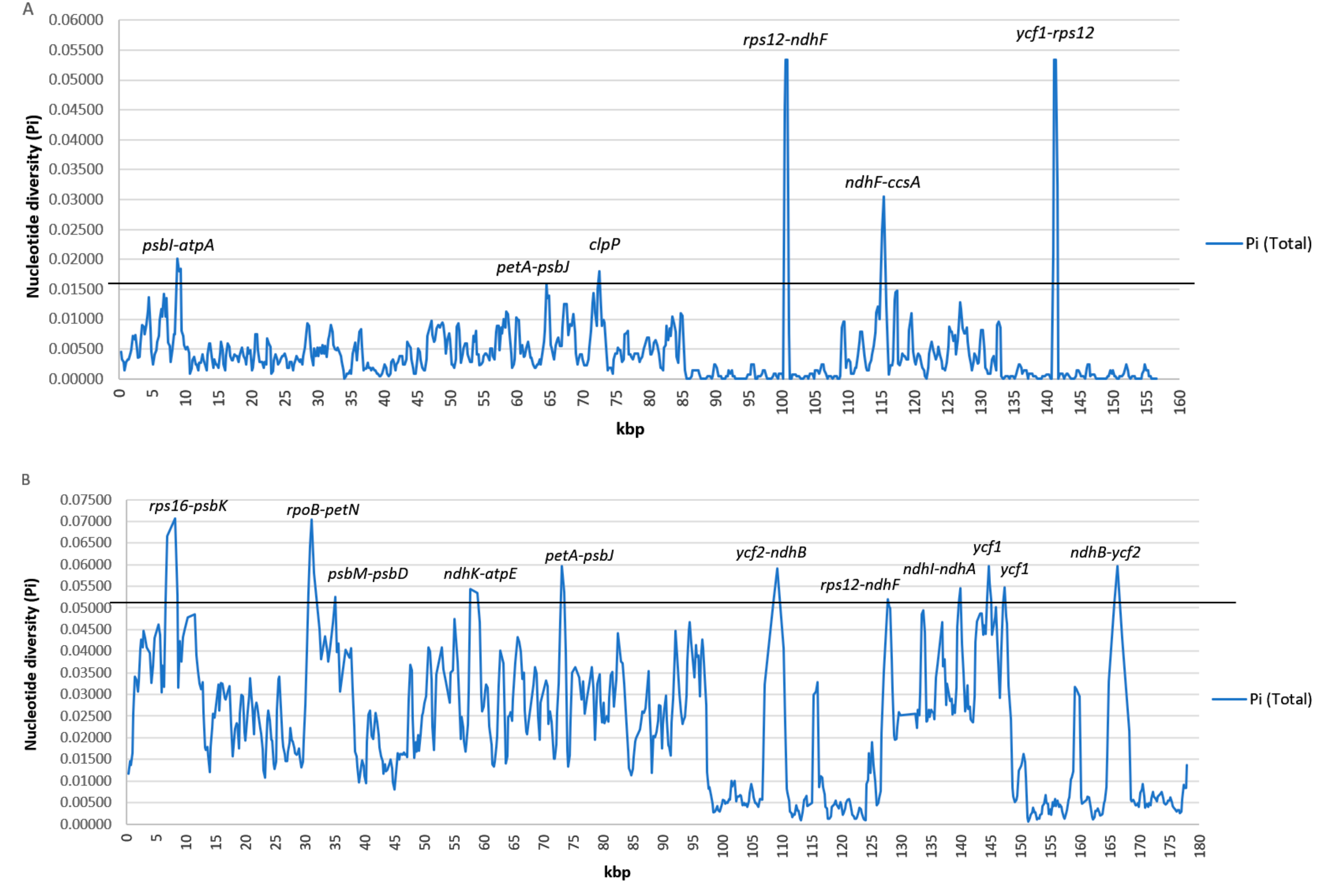
2.3. Genome Comparative Analysis and Identification of Divergent Hotspots
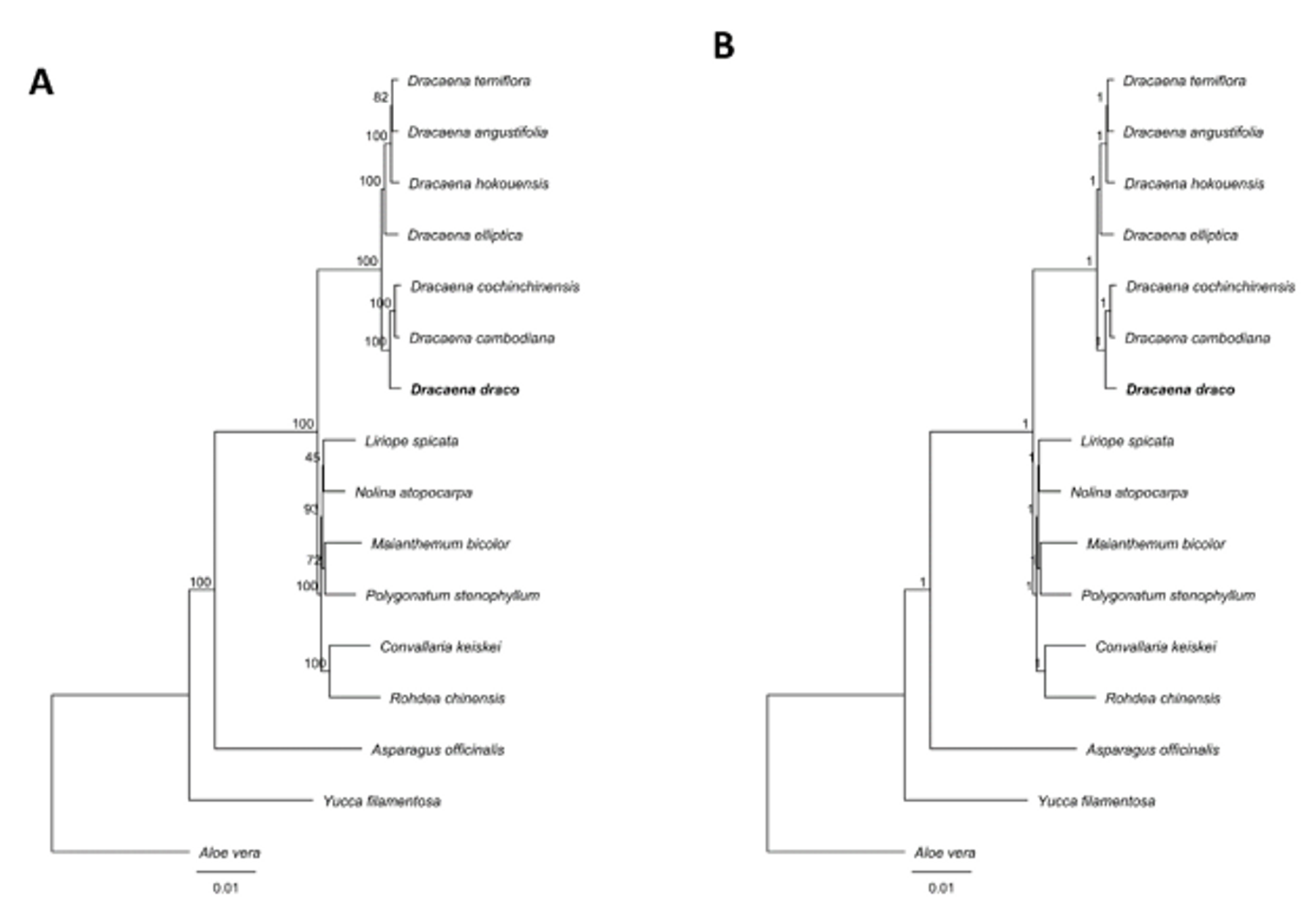
2.4. Phylogenetic Inference
3. Results and Discussion
3.1. Genome Features
3.2. Genome Comparative Analysis and Identification of Divergent Hotspots
3.3. Phylogenetic Inference
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lu, P.-L.; Morden, C.W. Phylogenetic Relationships among Dracaenoid Genera (Asparagaceae:Nolinoideae) Inferred from Chloroplast DNA Loci. Syst. Bot. 2014, 39, 90–104. [Google Scholar] [CrossRef]
- Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- Govaerts, R.; Zonneveld, B.J.M.; Zona, S.A.; World Checklist of Asparagaceae. Facilitated by the Royal Botanic Gardens, Kew. Available online: http://apps.kew.org/wcsp/ (accessed on 22 December 2019).
- Takawira-Nyenya, R.; Mucina, L.; Cardinal-Mcteague, W.; Thiele, K. Sansevieria (Asparagaceae, 1101 Nolinoideae) is a herbaceous clade within Dracaena: Inference from non-coding plastid and nuclear DNA 1102 sequence data. Phytotaxa 2018, 376, 254–276. [Google Scholar] [CrossRef]
- Marrero, A.; Almeida, S.R.; Martín-González, M. A new species of the wild Dragon Tree, Dracaena (Dracaenaceae) from Gran Canaria and its taxonomic and biogeographic Implications. Bot. J. Linn. Soc. 1998, 128, 291–314. [Google Scholar]
- Maděra, P.; Forrest, A.; Hanáček, P.; Vahalík, P.; Gebauer, R.; Plichta, R.; Jupa, R.; Rensburg, J.J.V.; Morris, M.; Nadezhdina, N.; et al. What We Know and What We Do Not Know About Dragon Trees? Forests 2020, 11, 236. [Google Scholar] [CrossRef]
- Turland, N.J. Agavaceae. In Flora of Madeira; Press, J.R., Short, M.J., Eds.; Natural History Museum (HMSO): London, UK, 1995; pp. 391–392. [Google Scholar]
- Benabid, A.; Cuzin, F. Populations de dragonnier (Dracaena draco L. subsp. aigal Benabid et Cuzin) au Maroc: Valeurs taxinomique, biogéographique et phytosociologique. C. R. Acad. Sci. Paris Sci. Vie 1997, 320, 267–277. [Google Scholar] [CrossRef]
- Marrero, A. Dracaena tamaranae, el género dracaena y otros afines: Análisis morfológico para un aproximación filogenética. Museo Canario 2000, 55, 301–334. [Google Scholar]
- Almeida Pérez, R.S. Sobre la presencia de Dracaena draco (L.) L. En gran Canaria (Islas Canarias): Aportación corológica, estado actual y significación biogeográfica. Bot. Macarónesica 2003, 24, 17–38. [Google Scholar]
- Almeida Pérez, R.S. Dracaena draco (L.). In Atlas y Libro Rojo de la Flora Vascular Amenazada de España, 2nd ed.; Bañares, A., Blanca, G., Güemes, J., Moreno, J.C., Ortiz, S., Eds.; Publicaciones de O.A.P.N.: Madrid, Spain, 2004; pp. 680–681. [Google Scholar]
- Marrero, A.; Almeida, S.R. A new subspecies, Dracaena draco (L.) L. subsp. caboverdeana Marrero Rodr. & R. Almeida (Dracaenaceae) from Cape Verde Island. Int. J. Geobot. Res. 2012, 2, 35–40. [Google Scholar]
- González-Castro, A.; Pérez-Pérez, D.; Romero, J.; Nogales, M. Unraveling the Seed Dispersal System of an Insular “Ghost” Dragon Tree (Dracaena draco) in the Wild. Front. Ecol. Evol. 2019, 7, 39. [Google Scholar] [CrossRef]
- Pütter, A. Altersbestimmung an Drachenbäumen von Tenerife. Sitzungsberichte der Heidelberger Akademie der Wissenschäften. Math.-Nat. Klasse 1925, 12, 12–18. [Google Scholar]
- Byström, K. Dracaena draco L. in the Cape Verde Islands. Acta Horti-Gotobg. 1960, 23, 179–214. [Google Scholar]
- Symon, D.E. The growth of Dracaena draco—dragon’s blood tree. J. Arnold Arbor. 1974, 55, 51–58. [Google Scholar]
- Mägdefrau, K. Das Alter der Drachenbäume auf Tenerife. Flora 1975, 164, 347–357. [Google Scholar] [CrossRef]
- Beyhl, F.E. Two different growth forms of Dracaena draco L. (Monocotyledones: Liliales: Agavaceae). Boletin Museu Municipal Funchal 1995, 4, 91–95. [Google Scholar]
- Krawczyszyn, J.; Krawczyszyn, T. Photomorphogenesis in Dracaena draco. Trees Struct. Funct. 2016, 30, 647–664. [Google Scholar] [CrossRef]
- Wiland-Szymańska, J.; Klimko, M. Differentiation of leaf anatomy of the genera Dracaena L and Sansevieria Thunb. (Dracaenaceae). In Proceedings of the XVII International Botanical Congress. 100 years after the II IBC in Vienna 1905, Vienna, Austria, 12–16 July 2005; p. 328. [Google Scholar]
- Nadezhdina, N.; Plichta, R.; Nadezhdin, V.; Gebauer, R.; Jupa, R.; Habrová, H.; Maděra, P. A comparative structural and functional study of leaf traits and sap flow in Dracaena cinnabari and Dracaena draco seedlings. Funct. Plant Biol. 2015, 42, 1092–1105. [Google Scholar] [CrossRef]
- Klimko, M.; Nowińska, R.; Wilkin, P.; Wiland-Szymańska, J. Comparative leaf micromorphology and anatomy of the dragon tree group of Dracaena (Asparagaceae) and their taxonomic implications. Plant Syst. Evol. 2018. [Google Scholar] [CrossRef]
- Jura-Morawiec, J. Formation of amphivasal vascular bundles in Dracaena draco stem in relation to rate of cambial activity. Trees Struct. Funct. 2015, 29, 1493–1499. [Google Scholar] [CrossRef]
- Jura-Morawiec, J.; Wiland-Szymańska, J. A novel insight into the structure of amphivasal secondary bundles on the example of Dracaena draco L. stem. Trees Struct. Funct. 2014, 28, 871–877. [Google Scholar] [CrossRef]
- Jura-Morawiec, J. Atypical origin, structure and arrangement of secondary tracheary elements in the stem of the monocotyledonous dragon tree, Dracaena draco. Planta 2017, 245, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Jura-Morawiec, J. Rhythmic growth and age estimation of aerial roots in Dracaena draco (Asparagaceae). Trees 2019, 33, 1513–1518. [Google Scholar] [CrossRef]
- Krawczyszyn, J.; Krawczyszyn, T. Massive aerial roots growth and form of Dracaena draco. Trees Struct. Funct. 2014, 28, 757–768. [Google Scholar] [CrossRef][Green Version]
- Brown, N.E. Notes on the genera Cordyline, Dracaena, Pleomele, Sansevieria, and Taetsia. Bull. Misc. Inf. 1914, 8, 273–279. [Google Scholar] [CrossRef]
- Klimko, M.; Nowińska, R.; Jura-Morawiec, J.; Wiland-Szymańska, J.; Wilkin, P. Pollen morphology of selected species of the genera Chrysodracon and Dracaena (Asparagaceae, subfamily Nolinoideae) and its systematic implications. Plant Syst. Evol. 2018. [Google Scholar] [CrossRef]
- Jura-Morawiec, J.; Tulik, M. Morpho-anatomical basis of dragon’s blood secret in Dracaena draco stem. Flora 2015, 213, 1–5. [Google Scholar] [CrossRef]
- Jura-Morawiec, J.; Tulik, M. Dragon’s blood secretion and its ecological significance. Chemoecology 2016, 26, 101–105. [Google Scholar] [CrossRef]
- Walter, K.S.; Gillett, H.J. (Eds.) Dracaena draco. In 1997 IUCN Red List of Threatened Plants; IUCN: Gland, Switzerland, 1998. [Google Scholar]
- Lu, P.L.; Morden, C. Phylogenetics of the plant genera Dracaena and Pleomele (Aparagaceae). Bot. Orient. J. Plant Sci. 2010, 7, 64–72. [Google Scholar] [CrossRef]
- Edwards, C.E.; Bassüner, B.; Birkinshaw, C.; Camara, C.; Lehavana, A.; Lowry, P.P.; Miller, J.S.; Wyatt, A.; Jackson, P.W. A botanical mystery solved by phylogenetic analysis of botanical garden collections: The rediscovery of the presumed-extinct Dracaena umbraculifera. Oryx 2018, 52, 427–436. [Google Scholar] [CrossRef]
- Li, D.M.; Zhao, C.Y.; Liu, X.F. Complete chloroplast genome sequences of Kaempferia galanga and Kaempferia elegans: Molecular structures and comparative analysis. Molecules 2019, 24, 474. [Google Scholar] [CrossRef]
- Vu, H.T.; Tran, N.; Nguyen, T.D.; Vu, Q.L.; Bui, M.H.; Le, M.T.; Le, L. Complete chloroplast genome of Paphiopedilum delenatii and phylogenetic relationships among Orchidaceae. Plants 2020, 9, E61. [Google Scholar] [CrossRef] [PubMed]
- Celiński, K.; Kijak, H.; Wojnicka-Półtorak, A.; Buczkowska-Chmielewska, K.; Sokołowska, J.; Chudzińska, E. Effectiveness of the DNA barcoding approach for closely related conifers discrimination: A case study of the Pinus mugo complex. Comptes Rendus Biol. 2017, 340, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Y.; Henry, R.J.; Rossetto, M.; Wang, Y.; Chen, S. Plant DNA barcoding: From gene to genome. Biol. Rev. Camb. Philos. Soc. 2015, 90, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Parks, M.; Cronn, R.; Liston, A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009, 7, 84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Y.; Song, M.; Guan, Y.; Ma, X. Species identification of Dracaena using the complete chloroplast genome as a super-barcode. Front. Pharmacol. 2019. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Mau, B.; Perna, N.T. Progressive Mauve: Multiple Genome Alignment with Gene Gain, Loss, and Rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Zhang, M.-F.; Xue, J.; Dong, R.; Du, Y.-P.; Zhang, X.-H. Chloroplast genomic resources for phylogeny and DNA barcoding: A case study on Fritillaria. Sci. Rep. 2018, 8, 1184. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Cai, L.; Bi, G.Q.; Chen, G.; Sun, W. Characterization of the complete chloroplast genomes of Buddleja colvilei and B. sessilifolia: Implications for the taxonomy of Buddleja L. Molecules 2018, 23, E1248. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sylvester, S.P.; Li, M.; Zhang, C.; Li, X.; Duan, Y.; Wang, X. The Complete Plastid Genome of Magnolia zenii and Genetic Comparison to Magnoliaceae species. Molecules 2019, 24, 261. [Google Scholar] [CrossRef] [PubMed]
- Nikbakht, H.; Xia, X.; Hickey, D.A. The evolution of genomic GC content undergoes a rapid reversal within the genus Plasmodium. Genome 2014, 57, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, E.J.; Pádua, J.G.; Zucchi, M.I.; Vencovsky, R.; Vieira, M.L.C. Origin, evolution and genome distribution of microsatellites. Genet. Mol. Biol. 2006, 29, 294–307. [Google Scholar] [CrossRef]
- Gómez, A.; González-Martínez, S.C.; Collada, C.; Climent, J.; Gil, L. Complex population genetic structure in the endemic Canary Island pine revealed using chloroplast microsatellite markers. Theor. Appl. Genet. 2003, 107, 123–131. [Google Scholar] [CrossRef]
- Urbaniak, L.; Wojnicka-Półtorak, A.; Celiński, K.; Lesiczka, P.; Pawlaczyk, E.; Aučina, A. Genetic resources of relict populations of Pinus sylvestris (L.) in Western Carpathians assessed by chloroplast microsatellites pine revealed using chloroplast microsatellite. Biologia 2019, 74, 1077–1086. [Google Scholar] [CrossRef]
- Vendramin, G.G.; Lelli, L.; Rossi, P.; Morgante, M. A set of primers for the amplification of 20 chloroplast microsatellites in Pinaceae. Mol. Ecol. 1996, 5, 595–598. [Google Scholar] [CrossRef]
- Echt, C.S.; Vendramin, G.G.; Nelson, C.D.; Marquardt, P. Microsatellite DNA as shared genetic markers among conifer species. Can. J. For. Res. 1999, 29, 365–371. [Google Scholar] [CrossRef]
- Celiński, K.; Pawlaczyk, E.M.; Wojnicka-Półtorak, A.; Chudzińska, E.; Prus-Głowacki, W. Cross-species amplification and characterization of microsatellite loci in Pinus mugo Turra. Biologia 2013, 68, 621–626. [Google Scholar] [CrossRef]
- González-Martínez, S.C.; Robledo-Arnuncio, J.J.; Collada, C.; Díaz, A.; Williams, C.G.; Alía, R.; Cervera, M.T. Cross-amplification and sequence variation of microsatellite loci in Eurasian hard pines. Theor. Appl. Genet. 2004, 109, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Li, C.; Sun, J.; Zuo, Y.; Shi, S.; Cheng, T.; Guo, J.; Zhou, S. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef] [PubMed]
- Olsson, S.; Grivet, D.; Cid-Vian, J. Species-diagnostic markers in the genus Pinus: Evaluation of the chloroplast regions matK and ycf1. For. Syst. 2018, 27, e016. [Google Scholar] [CrossRef]

| GenBank Accession | Species | Family |
|---|---|---|
| NC_035506 | Aloe vera | Asphodelaceae |
| NC_034777 | Asparagus officinalis | Asparagaceae |
| MH680946 | Convallaria keiskei | Asparagaceae |
| MN200193 | Dracaena angustifolia | Asparagaceae |
| MN200194 | Dracaena cambodiana | Asparagaceae |
| MN200195 | Dracaena cochinchinensis | Asparagaceae |
| MN990038 | Dracaena draco | Asparagaceae |
| MN200196 | Dracaena elliptica | Asparagaceae |
| MN200197 | Dracaena hokouensis | Asparagaceae |
| MN200198 | Dracaena terniflora | Asparagaceae |
| MH680945 | Liriope spicata | Asparagaceae |
| KX790362 | Maianthemum bicolor | Asparagaceae |
| KX931462 | Nolina atopocarpa | Asparagaceae |
| KX822773 | Polygonatum stenophyllum | Asparagaceae |
| MH356725 | Rohdea chinensis | Asparagaceae |
| NC_032712 | Yucca filamentosa | Asparagaceae |
| No. | Classification of Genes | Name of Genes | Number |
|---|---|---|---|
| 1 | Photosystem I | psaA, psaB, psaC, psaI, psaJ | 5 |
| 2 | Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | 15 |
| 3 | Cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN, | 6 |
| 4 | ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI, | 6 |
| 5 | NADH dehydrogenase | ndhA*, ndhB*(x2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | 12 |
| 6 | RubisCO large subunit | rbcL | 1 |
| 7 | RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 | 4 |
| 8 | Ribosomal proteins – small units (SSU) | rps2, rps3, rps4, rps7(x2), rps8, rps11, rps12*(x2), rps14, rps15, rps16*, rps18, rps19(x2), | 15 |
| 9 | Ribosomal proteins – large units (LSU) | rpl2*(x2), rpl14, rpl16*, rpl20, rpl22, rpl23(x2), rpl32, rpl33, rpl36, | 11 |
| 10 | Other genes/Miscellaneous | accD, ccsA, cemA, clpP**, infA, matK, | 6 |
| 11 | Protein of unknown function | ycf1, ycf2(x2), ycf3**, ycf4 | 5 |
| 12 | Transfer RNAs | trnA-UGC(x2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC, trnH-GUG(x2), trnI-CAU(x2), trnI-GAU*(x2), trnK-UUU*, trnL-CAA(x2), trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU(x2), trnP-UGG, trnQ-UUG, trnR-ACG(x2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(x2), trnV-UAC*, trnW-CCA, trnY-GUA | 38 |
| 13 | Ribosomal RNAs | rrn4.5(x2), rrn5(x2), rrn16(x2), rrn23(x2) | 8 |
| Total | 132 | ||
| Species | D. draco | D. cochinchinensis | D. cambodiana | D. angustifolia | D. terniflora | D. hokouensis | D. elliptica |
|---|---|---|---|---|---|---|---|
| Total length (bp) | 155,422 | 155,459 | 155,291 | 155,332 | 155,347 | 155,340 | 155,055 |
| LSC length (bp) | 83,942 | 83,907 | 83,752 | 83,807 | 83,794 | 83,796 | 83,621 |
| SSC length (bp) | 18,472 | 18,492 | 18,489 | 18,465 | 18,493 | 18,494 | 18,456 |
| IR length (bp) | 53,008 | 53,050 | 53,050 | 53,060 | 53,060 | 53,050 | 52,978 |
| Overall GC content (%) | 37.6 | 37.5 | 37.5 | 37.5 | 37.5 | 37.5 | 37.5 |
| Total gene number | 132 | 130 | 130 | 130 | 130 | 130 | 130 |
| Total SSR number | 77 | 69 | 69 | 67 | 64 | 70 | 71 |
| GenBank accession | MN990038 | MN200195 | MN200194 | MN200193 | MN200198 | MN200197 | MN200196 |
| Reference | This study | [40] | [40] | [40] | [40] | [40] | [40] |
| Repeats | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/T | - | - | - | - | - | 29 | 16 | 5 | 12 | 2 | 1 | 1 | 2 | 68 |
| C/G | - | - | - | - | - | - | - | - | 1 | - | - | - | - | 1 |
| AT/AT | - | 1 | 2 | 3 | - | - | - | - | - | - | - | - | - | 6 |
| AAT/AAT | 2 | - | - | - | - | - | - | - | - | - | - | - | - | 2 |
| D. angustifolia | D. terniflora | D. hokouensis | D. elliptica | D. cambodiana | D. cochinchinensis | D. draco | |
|---|---|---|---|---|---|---|---|
| D. angustifolia | 0.00011 | 0.00012 | 0.00016 | 0.00018 | 0.00018 | 0.00019 | |
| D. terniflora | 0.00176 | 0.00012 | 0.00016 | 0.00018 | 0.00018 | 0.00018 | |
| D. hokouensis | 0.00207 | 0.00214 | 0.00016 | 0.00018 | 0.00019 | 0.00018 | |
| D. elliptica | 0.00396 | 0.00389 | 0.00405 | 0.00018 | 0.00019 | 0.00018 | |
| D. cambodiana | 0.00507 | 0.00500 | 0.00533 | 0.00503 | 0.00010 | 0.00014 | |
| D. cochinchinensis | 0.00517 | 0.00520 | 0.00548 | 0.00534 | 0.00147 | 0.00014 | |
| D. draco | 0.00518 | 0.00524 | 0.00537 | 0.00520 | 0.00299 | 0.00317 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Celiński, K.; Kijak, H.; Wiland-Szymańska, J. Complete Chloroplast Genome Sequence and Phylogenetic Inference of the Canary Islands Dragon Tree (Dracaena draco L.). Forests 2020, 11, 309. https://doi.org/10.3390/f11030309
Celiński K, Kijak H, Wiland-Szymańska J. Complete Chloroplast Genome Sequence and Phylogenetic Inference of the Canary Islands Dragon Tree (Dracaena draco L.). Forests. 2020; 11(3):309. https://doi.org/10.3390/f11030309
Chicago/Turabian StyleCeliński, Konrad, Hanna Kijak, and Justyna Wiland-Szymańska. 2020. "Complete Chloroplast Genome Sequence and Phylogenetic Inference of the Canary Islands Dragon Tree (Dracaena draco L.)" Forests 11, no. 3: 309. https://doi.org/10.3390/f11030309
APA StyleCeliński, K., Kijak, H., & Wiland-Szymańska, J. (2020). Complete Chloroplast Genome Sequence and Phylogenetic Inference of the Canary Islands Dragon Tree (Dracaena draco L.). Forests, 11(3), 309. https://doi.org/10.3390/f11030309





