Effects of Metaxenia on Stone Cell Formation in Pear (Pyrus bretschneideri) Based on Transcriptomic Analysis and Functional Characterization of the Lignin-Related Gene PbC4H2
Abstract
1. Introduction
2. Methods and Materials
2.1. Plant Materials and Cultivation Conditions
2.2. Determination of Stone Cell and Lignin Content in Pear Fruit
2.3. RNA Extraction and Illumina Sequencing
2.4. Screening and Functional Annotation of Differentially Expressed Genes (DEGs)
2.5. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.6. Genetic Transformation of Arabidopsis thaliana
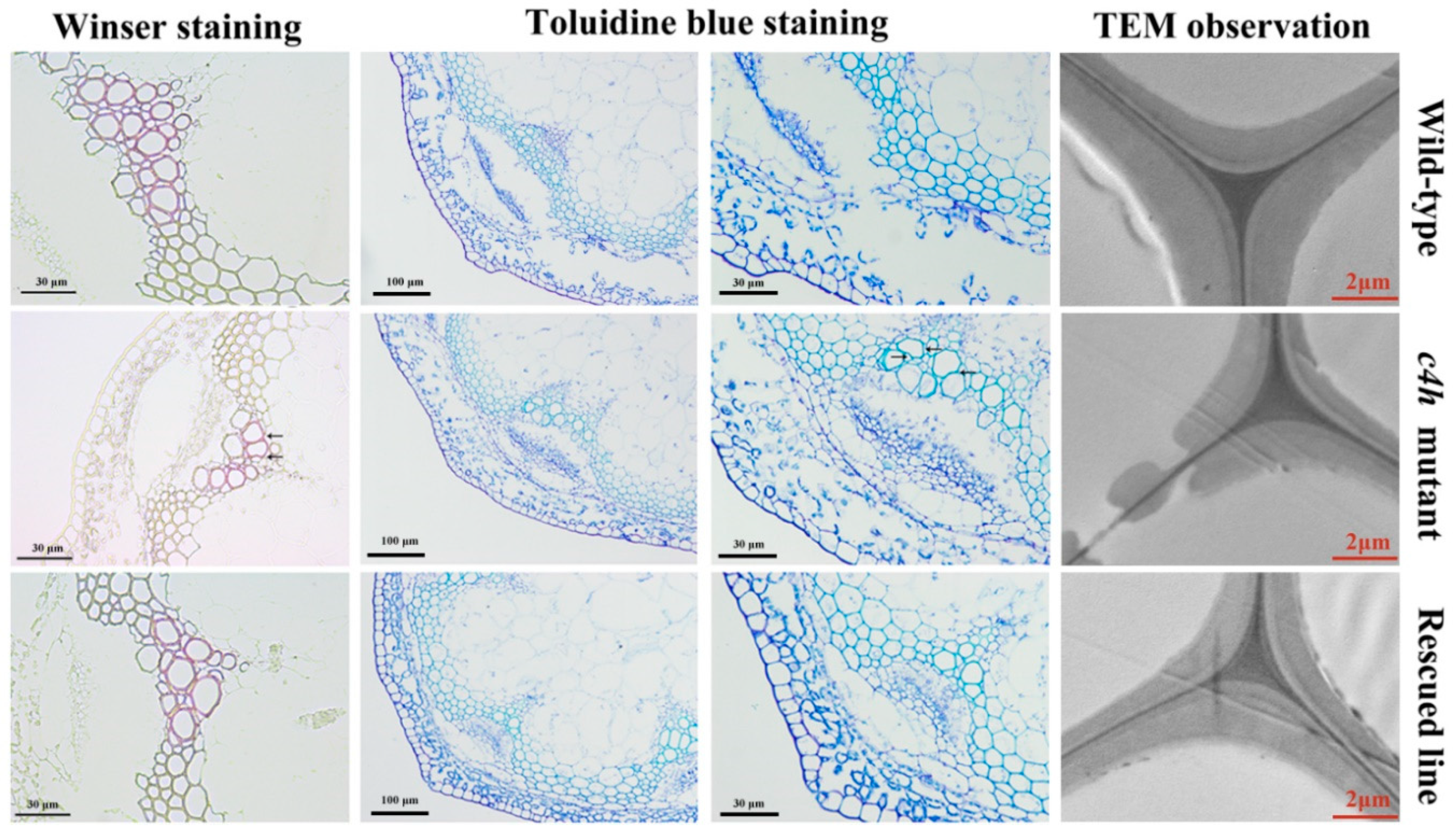
2.7. Microscopic and Ultramicroscopic Observation of the Cell Wall of Arabidopsis
2.8. Analysis of Lignin Content in the Inflorescence Stem of Arabidopsis
3. Results
3.1. Differences in Stone Cell and Lignin Content of Pear Fruit Obtained from Two Pollination Combinations
 × ‘Lianglizaosu’
× ‘Lianglizaosu’  (YL) and ‘Cuiguan’
(YL) and ‘Cuiguan’  × ‘Lianglizaosu’
× ‘Lianglizaosu’  (CL) fruits, in addition to determining the content of stone cells (Figure 1b).
(CL) fruits, in addition to determining the content of stone cells (Figure 1b).3.2. Characterization of Stone Cells in Pear Fruit after Pollination by Two Pollen Parents
3.3. Analysis of Transcriptomic Data and Mapping to Reference Genome Sequences
3.4. Identification of DEGs in Pear Fruits Pollinated by Two Pollen Parents Based on Transcriptome Sequencing
3.5. Differentially Expressed Transcription Factors (TFs) between YL and CL fruits
3.6. Differences in Monolignol Metabolism between YL and CL Fruits
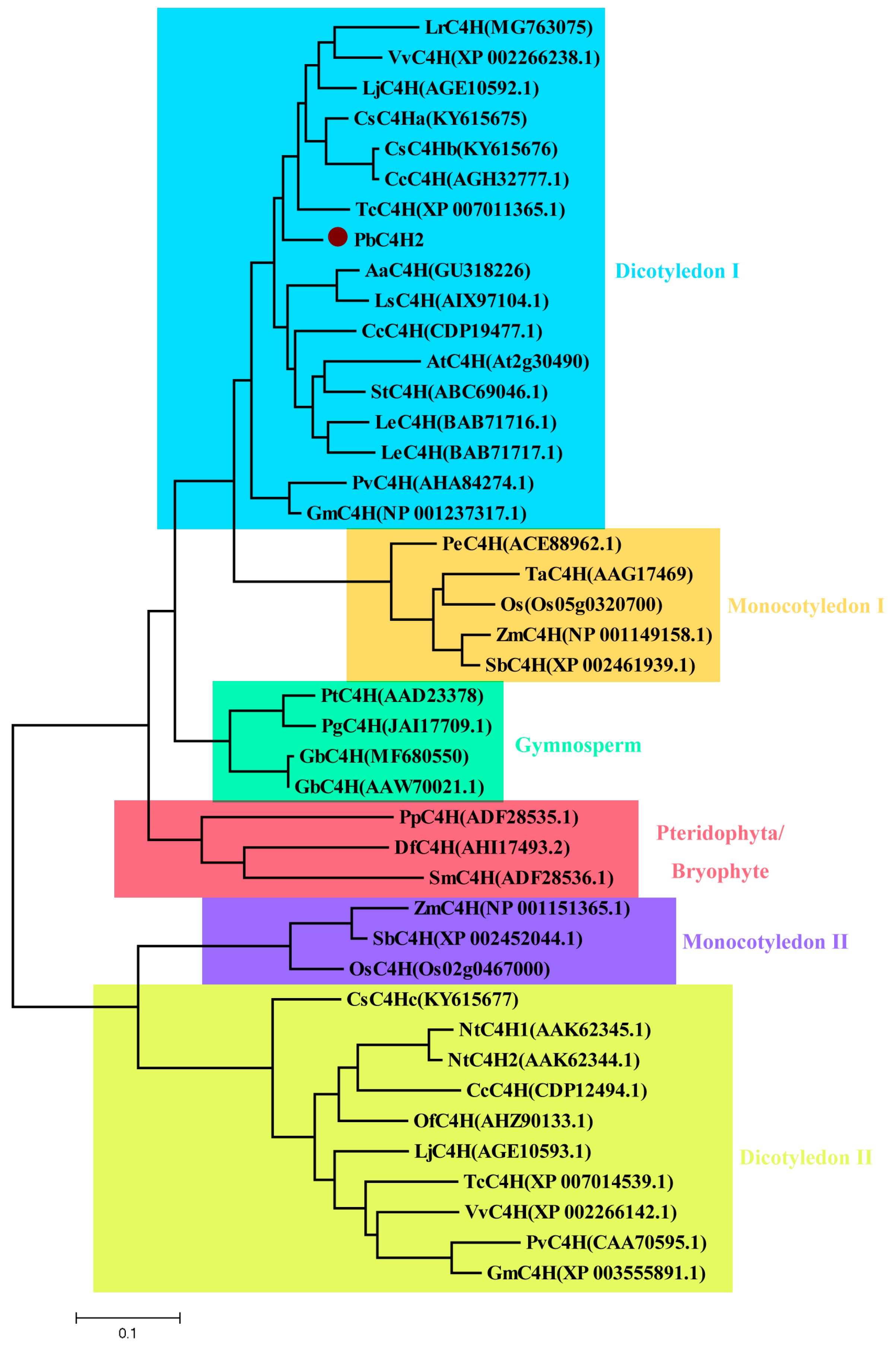
3.7. Cloning and Phylogenetic Analysis of Putative Lignin-Related PbC4H2
3.8. Complementation Analysis of the c4h Mutant with PbC4H2
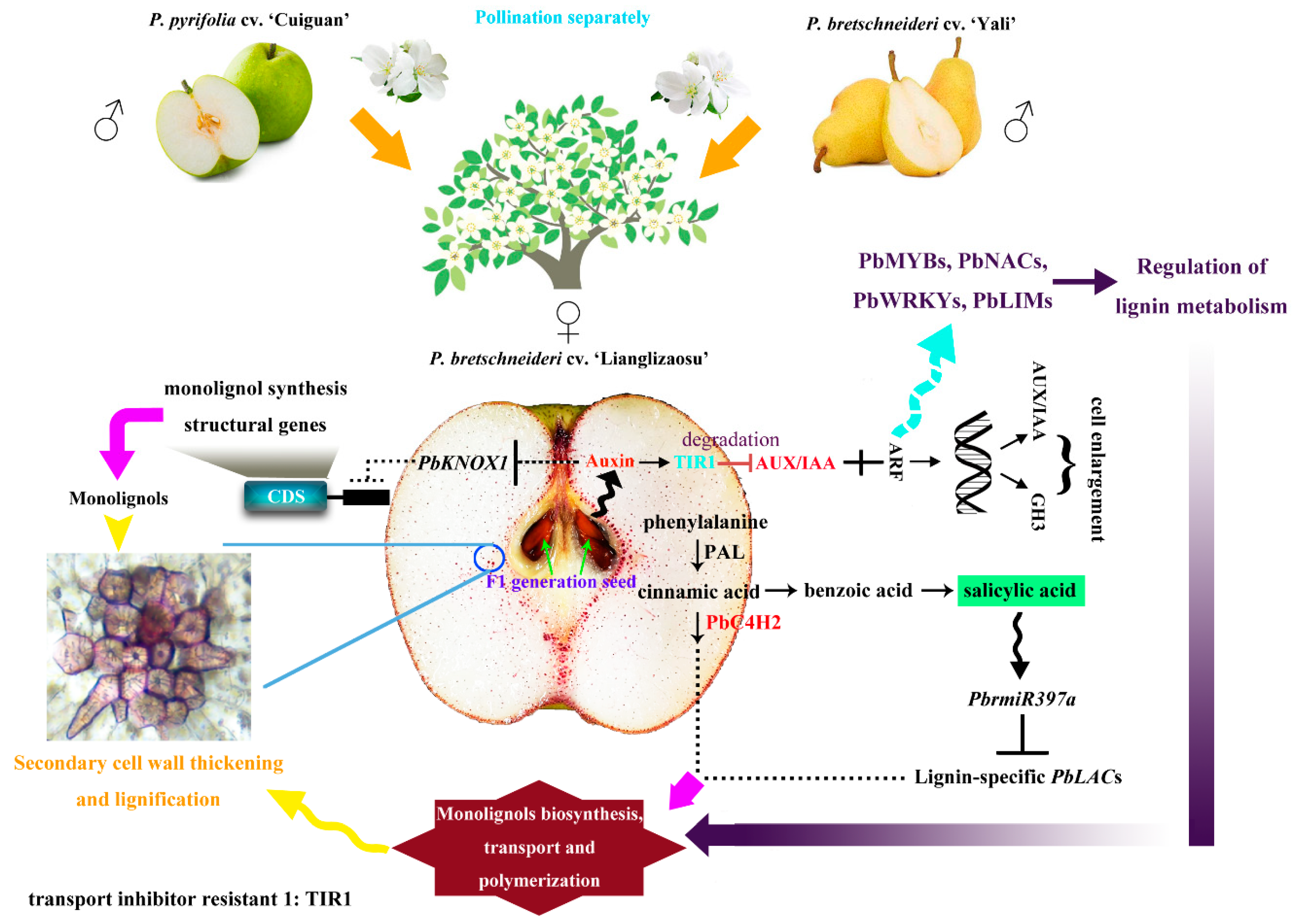
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Y.; Xu, J.; Korban, S.S.; Fei, Z.; Tao, S.; Ming, R.; Tai, S.; Khan, A.M.; Postman, J.D.; et al. Diversification and independent domestication of Asian and European pears. Genome Biol. 2018, 19. [Google Scholar] [CrossRef]
- Choi, J.; Choi, J.; Hong, K.; Kim, W. Cultivar differences of stone cells in pear flesh and their effects on fruit quality. Hortic. Environ. Biotechnol. 2007, 48, 17–31. [Google Scholar]
- Cai, Y.; Li, G.; Nie, J.; Lin, Y.; Nie, F.; Zhang, J.; Xu, Y. Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci. Hortic. 2010, 125, 374–379. [Google Scholar] [CrossRef]
- Yan, C.; Yin, M.; Zhang, N.; Jin, Q.; Fang, Z.; Lin, Y.; Cai, Y. Stone cell distribution and lignin structure in various pear varieties. Sci. Hortic. 2014, 174, 142–150. [Google Scholar] [CrossRef]
- Zhang, J.; Cheng, X.; Jin, Q.; Su, X.; Li, M.; Yan, C.; Jiao, X.; Li, D.; Lin, Y.; Cai, Y. Comparison of the transcriptomic analysis between two Chinese white pear (Pyrus bretschneideri Rehd.) genotypes of different stone cells contents. PLoS ONE 2017, 12, e0187114. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Cai, Y.; Zhang, J. Stone Cell Development in Pear. In The Pear Genome, Compendium of Plant Genomes; Korban, S., Ed.; Springer: Cham, Switzerland, 2019; pp. 201–225. ISBN 9783030110475. [Google Scholar]
- Jin, Q.; Yan, C.; Qiu, J.; Zhang, N.; Lin, Y.; Cai, Y. Structural characterization and deposition of stone cell lignin in Dangshan Su pear. Sci. Hortic. 2013, 155, 123–130. [Google Scholar] [CrossRef]
- Choi, J.; Lee, S. Distribution of stone cell in Asian, Chinese, and European pear fruit and its morphological changes. J. Appl. Bot. Food Qual. 2013, 86, 185–189. [Google Scholar]
- Cheng, X.; Li, G.; Manzoor, M.A.; Wang, H.; Abdullah, M. In Silico Genome-Wide Analysis of Respiratory Burst Oxidase Homolog (RBOH) Family Genes in Five Fruit-Producing Trees, and Potential Functional Analysis on Lignification of Stone Cells in Chinese White Pear. Cells 2019, 8, 520. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Muhammad, A.; Li, G.; Zhang, J.; Cheng, J.; Qiu, J.; Jiang, T. Family-1 UDP glycosyltransferases in pear (Pyrus bretschneideri): Molecular identification, phylogenomic characterization and expression profiling during stone cell formation. Mol. Biol. Rep. 2019, 46, 2153–2175. [Google Scholar] [CrossRef] [PubMed]
- Brahem, M.; Renard, C.M.G.C.; Gouble, B.; Bureau, S.; Le Bourvellec, C. Characterization of tissue specific differences in cell wall polysaccharides of ripe and overripe pear fruit. Carbohydr. Polym. 2017, 156, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Su, X.; Abdullah, M.; Sun, Y.; Li, G.; Cheng, X.; Lin, Y.; Cai, Y.; Jin, Q. Effects of different pollens on primary metabolism and lignin biosynthesis in pear. Int. J. Mol. Sci. 2018, 19, 2273. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Vashisth, D.; Misra, A.; Akhtar, Q.; Jalil, S.U. RNAi down-regulation of cinnamate-4-hydroxylase increases artemisinin biosynthesis in Artemisia annua. Sci. Rep. 2016, 6, 26458. [Google Scholar] [CrossRef] [PubMed]
- Barros, J.; Serrani-yarce, J.C.; Chen, F.; Baxter, D.; Venables, B.J.; Dixon, R.A. Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat. Plants 2016, 2, 16050. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. Systematic analysis of the 4-coumarate:Coenzyme A ligase (4CL) related genes and expression profiling during fruit development in the Chinese pear. Genes 2016, 7, 89. [Google Scholar] [CrossRef]
- Cheng, X.; Xiong, Y.; Li, D.H.; Cheng, J.; Cao, Y.P.; Yan, C.C.; Jin, Q.; Sun, N.; Cai, Y.P.; Lin, Y. Bioinformatic and expression analysis of the OMT gene family in Pyrus bretschneideri cv. Dangshan Su. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Cheng, X.; Li, M.; Li, D.; Zhang, J.; Jin, Q.; Sheng, L.; Cai, Y.; Lin, Y. Characterization and analysis of CCR and CAD gene families at the whole-genome level for lignin synthesis of stone cells in pear (Pyrus bretschneideri) fruit. Biol. Open 2017, 6, 1602–1613. [Google Scholar] [CrossRef]
- Xue, C.; Yao, J.-L.; Qin, M.-F.; Zhang, M.-Y.; Allan, A.C.; Wang, D.-F.; Wu, J. PbrmiR397a regulates lignification during stone cell development in pear fruit. Plant Biotechnol. J. 2019, 17, 103–117. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jin, Q.; Lin, Y. Structural, Evolutionary, and Functional Analysis of the Class III Peroxidase Gene Family in Chinese Pear (Pyrus bretschneideri). Front. Plant Sci. 2016, 7, 1874. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Wang, R.; Bai, Y.; Liu, C.; Yuan, Y.; Yang, Y.; Yang, S. Differential gene expression analysis of ‘Chili’ (Pyrus bretschneideri) fruit pericarp with two types of bagging treatments. Hortic. Res. 2017, 4, 17005. [Google Scholar] [CrossRef]
- Lee, S.H.; Choi, J.H.; Kim, W.S.; Han, T.H.; Park, Y.S.; Gemma, H. Effect of soil water stress on the development of stone cells in pear (Pyrus pyrifolia cv. ‘Niitaka’) flesh. Sci. Hortic. 2006, 110, 247–253. [Google Scholar] [CrossRef]
- Cheng, X.; Yan, C.; Zhang, J.; Ma, C.; Li, S.; Jin, Q.; Zhang, N.; Cao, Y.; Lin, Y.; Cai, Y. The effect of different pollination on the expression of Dangshan Su pear microRNA. BioMed Res. Int. 2017, 2017, 2794040. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, X.; Wang, Y.; Yang, S.; Qu, H. The changes of intracellular calcium concentration and distribution in the hard end pear (Pyrus pyrifolia cv. ‘Whangkeumbae’) fruit. Cell Calcium 2018, 71, 15–23. [Google Scholar] [CrossRef]
- Sabir, A. Xenia and metaxenia in grapes: Differences in berry and seed characteristics of maternal grape cv. “Narince” (Vitis vinifera L.) as influenced by different pollen sources. Plant Biol. 2015, 17, 567–573. [Google Scholar] [CrossRef]
- Denney, J.O. Xenia Includes Metaxenia. HortScience 1992, 27, 722–728. [Google Scholar] [CrossRef]
- Li, X.; Singh, J.; Qin, M.; Li, S.; Zhang, X.; Zhang, M.; Khan, A.; Zhang, S.; Wu, J. Development of an integrated 200K SNP genotyping array and application for genetic mapping, genome assembly improvement and genome wide association studies in pear (Pyrus). Plant Biotechnol. J. 2019. [Google Scholar] [CrossRef]
- Zhao, S.G.; Zhang, J.G.; Zhao, Y.P.; Zhang, Y.X. New discoveries of stone cell differentiation in fruitlets of ‘Yali’ pears (Pyrus bretschneideri Rehd.). J. Food Agric. Environ. 2013, 11, 937–942. [Google Scholar]
- Shi, X.; Wu, X.; Chen, Y.; Cao, P.; Bai, B.; Li, M.; Yin, H.; Zhang, S. The Biological Damage Effects of 60Co-γ Ray Radiation on Cuiguan and Yuluxiang Pear Branches. J. Nucl. Agric. Sci. 2019, 33, 5–12. [Google Scholar]
- Li, X.; Liu, L.; Ming, M.; Hu, H.; Zhang, M.; Fan, J.; Song, B.; Zhang, S.; Wu, J. Comparative Transcriptomic Analysis Provides Insight into the Domestication and Improvement of Pear. Plant Physiol. 2019, 180, 435–452. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Cheng, X.; Li, G.; Muhammad, A.; Zhang, J.; Jiang, T.; Jin, Q.; Zhao, H.; Cai, Y.; Lin, Y. Molecular identification, phylogenomic characterization and expression patterns analysis of the LIM (LIN-11, Isl1 and MEC-3 domains) gene family in pear (Pyrus bretschneideri) reveal its potential role in lignin metabolism. Gene 2019, 686, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Li, M.; Abdullah, M.; Li, G.; Zhang, J.; Manzoor, M.A.; Wang, H.; Jin, Q.; Jiang, T.; Cai, Y.; et al. In Silico Genome-Wide Analysis of the Pear (Pyrus bretschneideri) KNOX Family and the Functional Characterization of PbKNOX1, an Arabidopsis BREVIPEDICELLUS Orthologue Gene, Involved in Cell Wall and Lignin Biosynthesis. Front. Genet. 2019, 10, 632. [Google Scholar] [CrossRef]
- Pradhan Mitra, P.; Loqué, D. Histochemical Staining of Arabidopsis thaliana Secondary Cell Wall Elements. J. Vis. Exp. 2014, 87, e51381. [Google Scholar] [CrossRef]
- Anderson, N.A.; Tobimatsu, Y.; Ciesielski, P.N.; Ximenes, E.; Ralph, J.; Donohoe, B.S.; Ladisch, M.; Chapple, C. Manipulation of guaiacyl and syringyl monomer biosynthesis in an Arabidopsis cinnamyl alcohol dehydrogenase mutant results in atypical lignin biosynthesis and modified cell wall structure. Plant Cell 2015, 27, 2195–2209. [Google Scholar] [CrossRef]
- Xue, C.; Yao, J.; Xue, Y.; Su, G.; Wang, L.; Wang, L. PbrMYB169 positively regulates lignification in fruit stone cells of pear (Pyrus bretschneideri). J. Exp. Bot. 2019, 70, 1801–1814. [Google Scholar] [CrossRef]
- Li, X.; Xue, C.; Li, J.; Qiao, X.; Li, L.; Yu, L.; Huang, Y.; Wu, J. Genome-wide identification, evolution and functional divergence of MYB transcription factors in Chinese white pear (Pyrus bretschneideri). Plant Cell Physiol. 2016, 57, 824–847. [Google Scholar] [CrossRef]
- Cheng, X.; Su, X.; Muhammad, A.; Li, M.; Zhang, J.; Sun, Y.; Li, G.; Jin, Q.; Cai, Y.; Lin, Y. Molecular characterization, evolution, and expression profiling of the Dirigent (DIR) family genes in Chinese white pear (Pyrus bretschneideri). Front. Genet. 2018, 9, 136. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, H.; Li, J.; Tao, S.; Qiao, X.; Korban, S.S.; Zhang, S.; Wu, J. Genome-wide analysis and characterization of molecular evolution of the HCT gene family in pear (Pyrus bretschneideri). Plant Syst. Evol. 2017, 303, 71–90. [Google Scholar] [CrossRef]
- Liu, X.Y.; Yu, H.N.; Gao, S.; Wu, Y.F.; Cheng, A.X.; Lou, H.X. The isolation and functional characterization of three liverwort genes encoding cinnamate 4-hydroxylase. Plant Physiol. Biochem. 2017, 117, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Schilmiller, A.L.; Stout, J.; Weng, J.K.; Humphreys, J.; Ruegger, M.O.; Chapple, C. Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 2009, 60, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Hofhuis, H.; Moulton, D.; Lessinnes, T.; Routier-Kierzkowska, A.L.; Bomphrey, R.J.J.; Mosca, G.; Reinhardt, H.; Sarchet, P.; Gan, X.; Tsiantis, M.; et al. Morphomechanical Innovation Drives Explosive Seed Dispersal. Cell 2016, 166, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Jia, N.; Liu, J.; Sun, Y.; Tan, P.; Cao, H.; Xie, Y.; Wen, B.; Gu, T.; Liu, J.; Li, M.; et al. Citrus sinensis MYB transcription factors CsMYB330 and CsMYB308 regulate fruit juice sac lignification through fine-tuning expression of the Cs4CL1 gene. Plant Sci. 2018, 277, 334–343. [Google Scholar] [CrossRef]
- Piotto, F.A.; Batagin-Piotto, K.D.; De Almeida, M.; Oliveira, G.C.X. Interspecific xenia and metaxenia in seeds and fruits of tomato. Sci. Agric. 2013, 70, 102–107. [Google Scholar] [CrossRef]
- Negi, S.; Tak, H.; Ganapathi, T.R. Overexpression of MusaNAC68 reduces secondary wall thickness of xylem tissue in banana. Plant Biotechnol. Rep. 2019, 13, 151–160. [Google Scholar] [CrossRef]
- Woodward, A.W.; Bartel, B. A receptor for auxin. Plant Cell 2005, 17, 2425–2429. [Google Scholar] [CrossRef]
- Xia, J.; Liu, Y.; Yao, S.; Li, M.; Zhu, M.; Huang, K.; Gao, L.; Xia, T. Characterization and expression profiling of Camellia sinensis cinnamate 4-hydroxylase genes in phenylpropanoid pathways. Genes 2017, 8, 193. [Google Scholar] [CrossRef]
 × ‘Lianglizaosu’
× ‘Lianglizaosu’  ; CL: ‘Cuiguan’
; CL: ‘Cuiguan’  × ‘Lianglizaosu’
× ‘Lianglizaosu’  . Student’s t test was used for the statistical analyses. Four fruits were selected for analysis at each period. * indicates a significant difference (p < 0.05).
. Student’s t test was used for the statistical analyses. Four fruits were selected for analysis at each period. * indicates a significant difference (p < 0.05).
 × ‘Lianglizaosu’
× ‘Lianglizaosu’  ; CL: ‘Cuiguan’
; CL: ‘Cuiguan’  × ‘Lianglizaosu’
× ‘Lianglizaosu’  . Student’s t test was used for the statistical analyses. Four fruits were selected for analysis at each period. * indicates a significant difference (p < 0.05).
. Student’s t test was used for the statistical analyses. Four fruits were selected for analysis at each period. * indicates a significant difference (p < 0.05).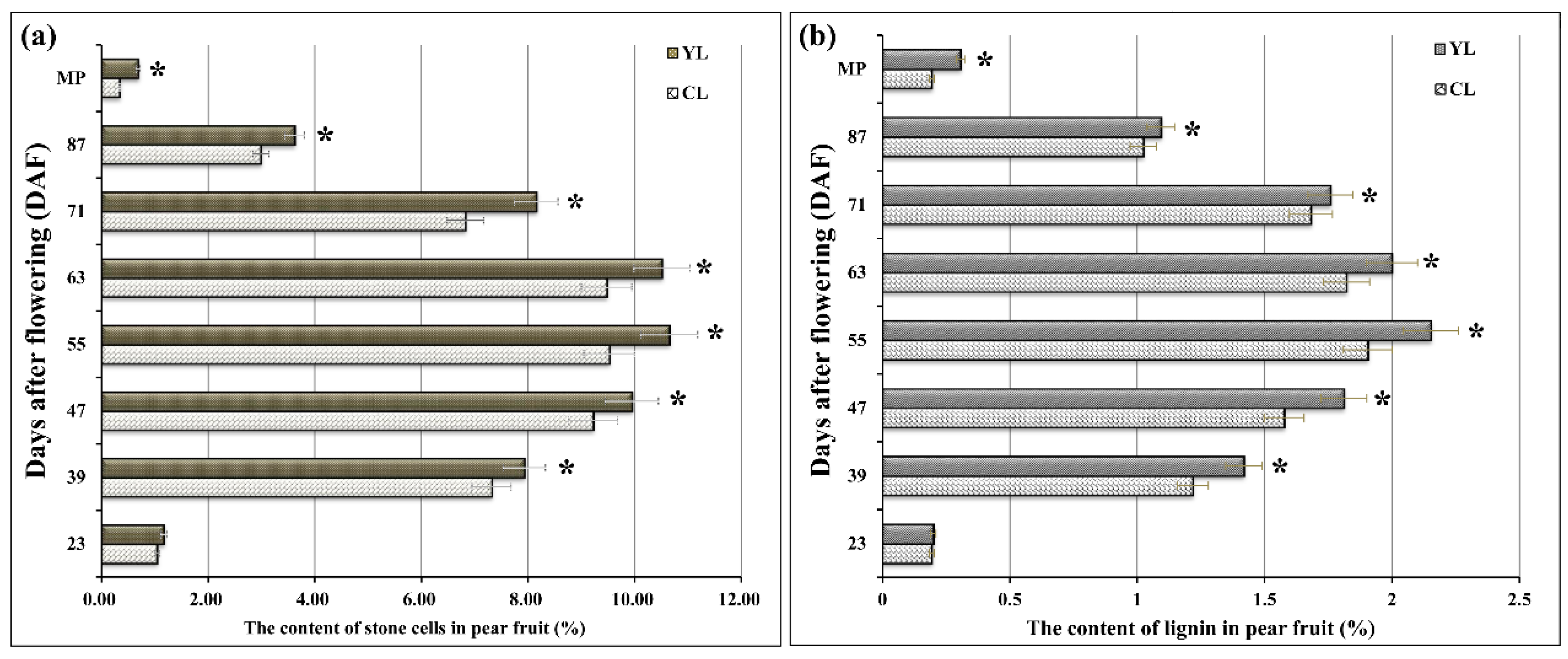
 × ‘Lianglizaosu’
× ‘Lianglizaosu’  (YL) and ‘Cuiguan’
(YL) and ‘Cuiguan’  × ‘Lianglizaosu’
× ‘Lianglizaosu’  (CL) fruit: (a) YL fruit; and, (b) CL fruit. The blue marked area is the inner flesh region, and the green marked area is the middle flesh region.
(CL) fruit: (a) YL fruit; and, (b) CL fruit. The blue marked area is the inner flesh region, and the green marked area is the middle flesh region.
 × ‘Lianglizaosu’
× ‘Lianglizaosu’  (YL) and ‘Cuiguan’
(YL) and ‘Cuiguan’  × ‘Lianglizaosu’
× ‘Lianglizaosu’  (CL) fruit: (a) YL fruit; and, (b) CL fruit. The blue marked area is the inner flesh region, and the green marked area is the middle flesh region.
(CL) fruit: (a) YL fruit; and, (b) CL fruit. The blue marked area is the inner flesh region, and the green marked area is the middle flesh region.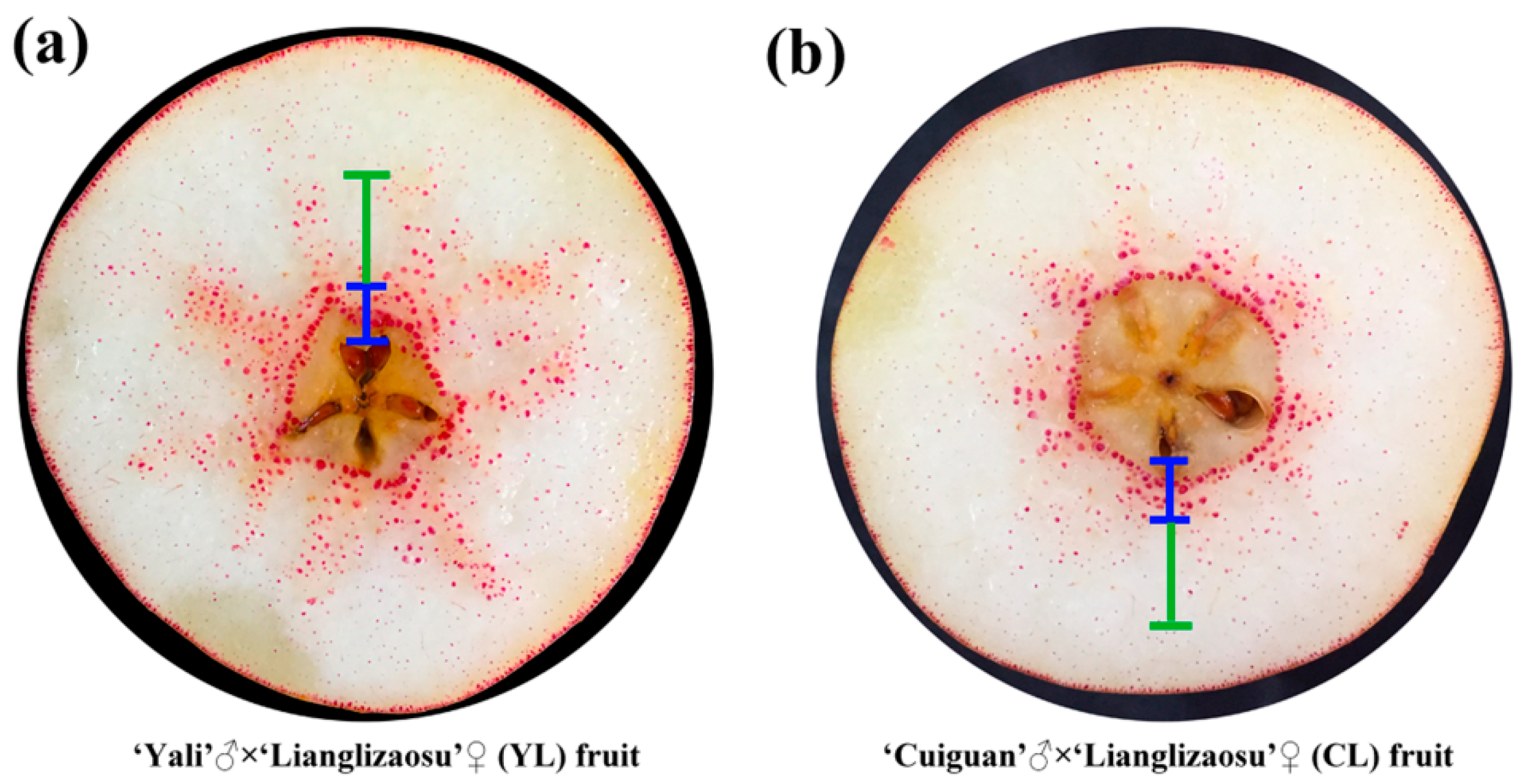

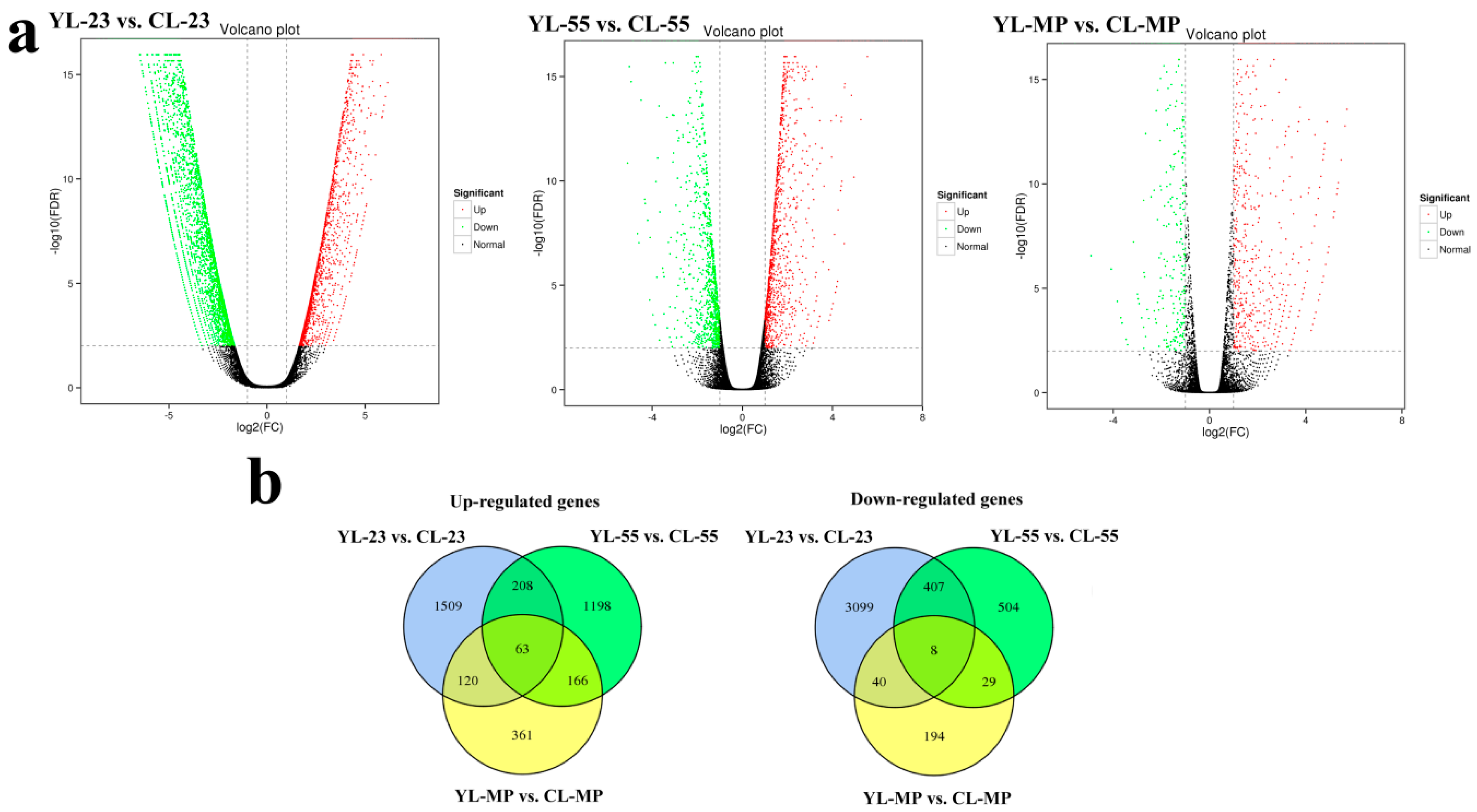
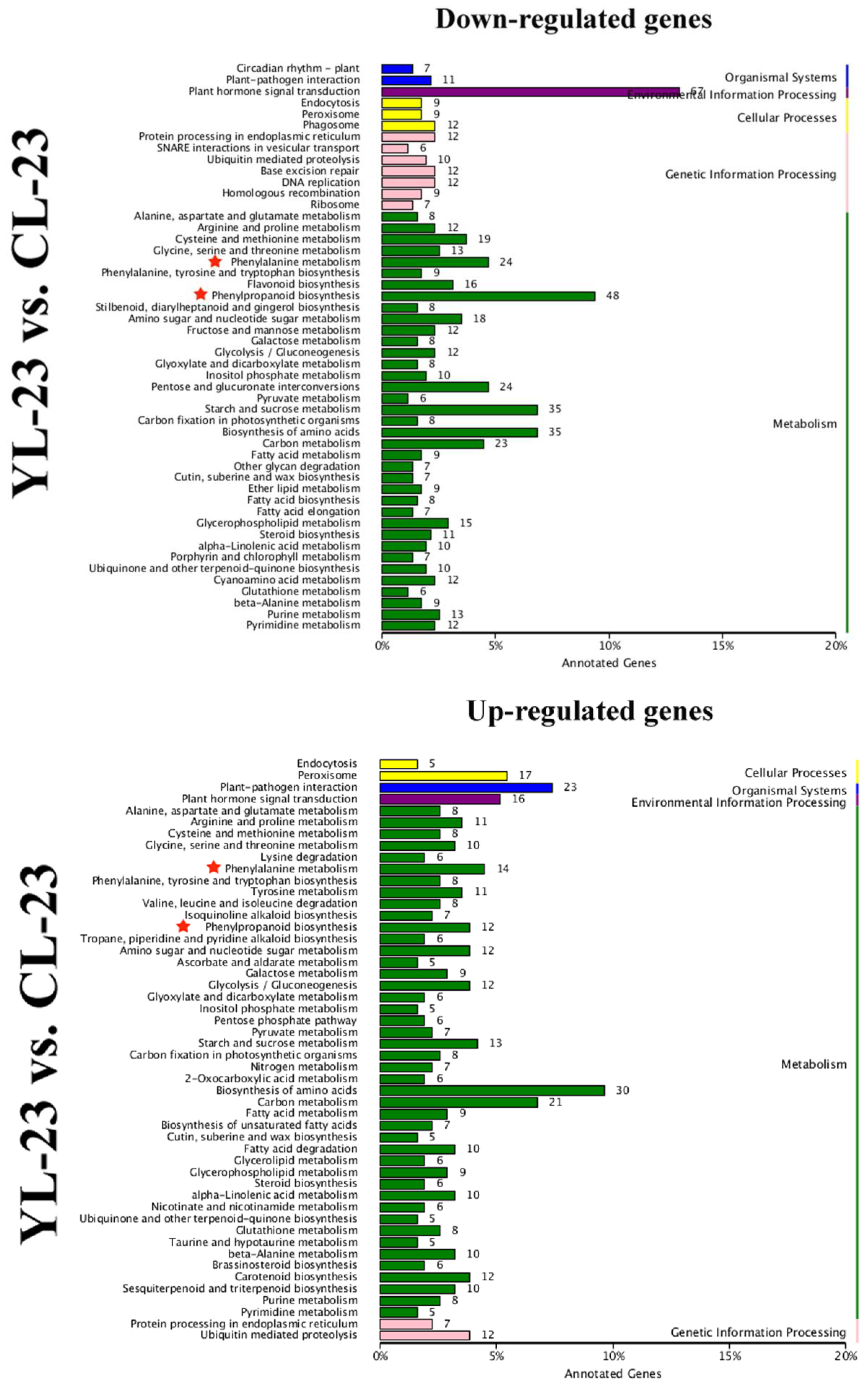
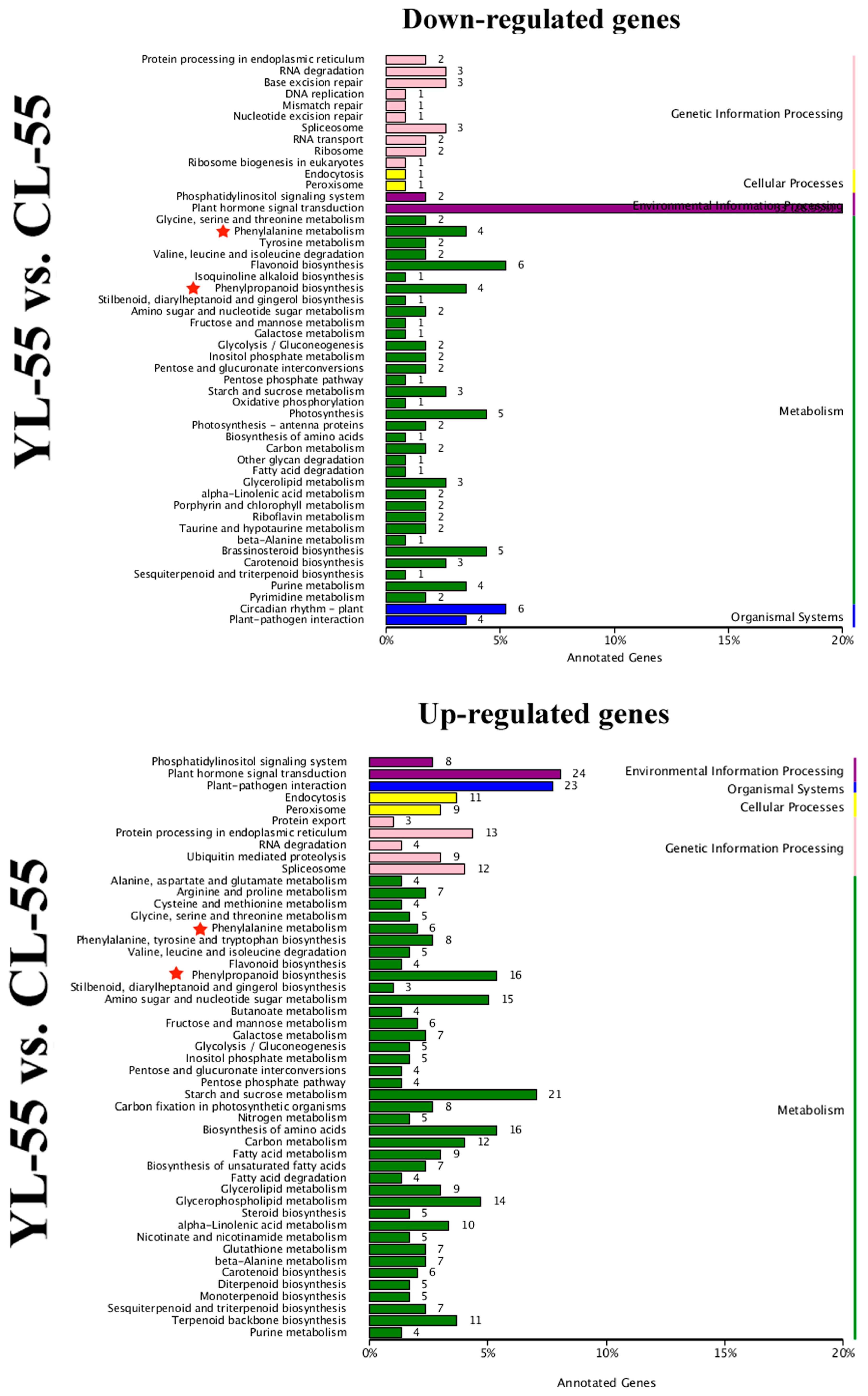
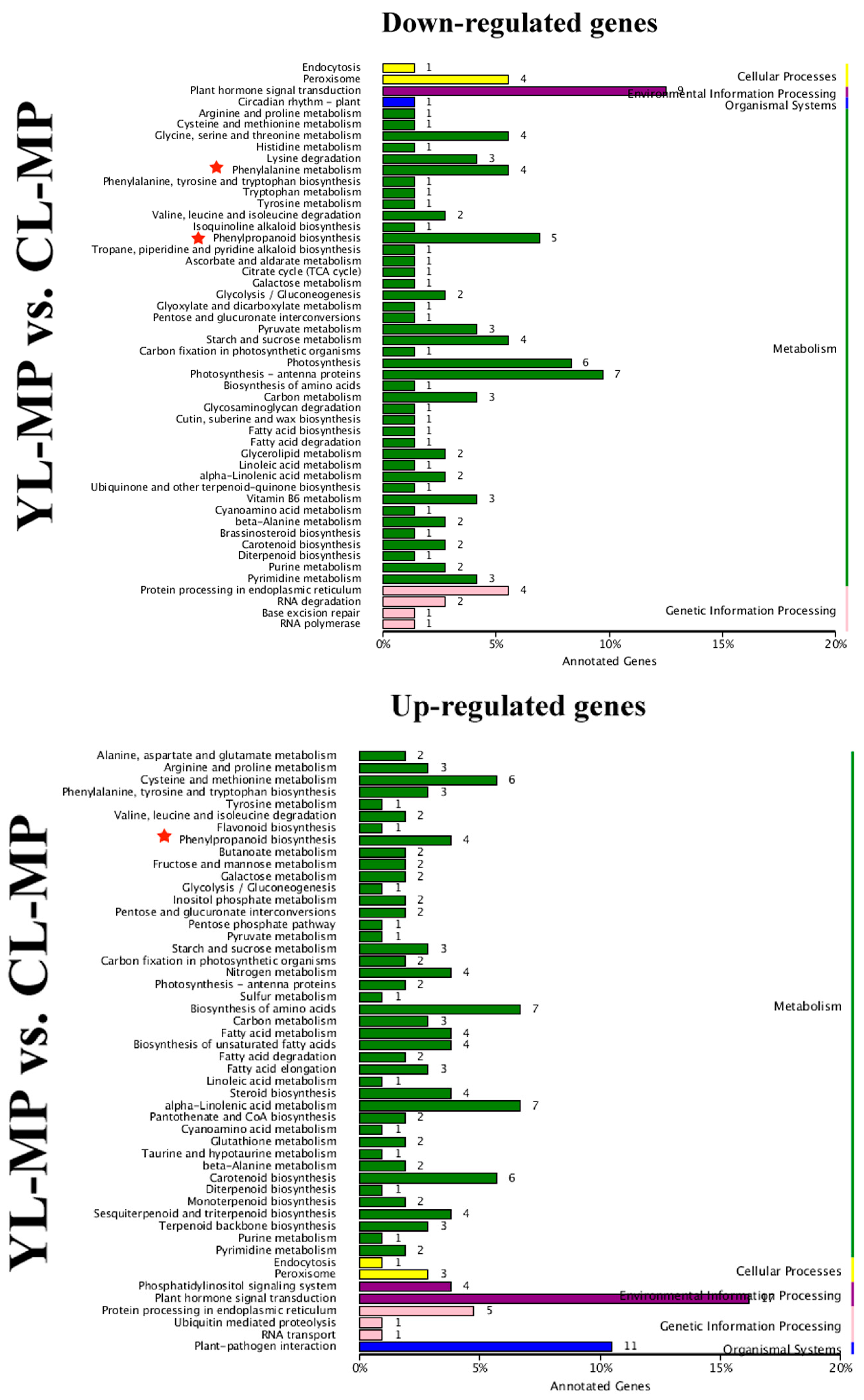
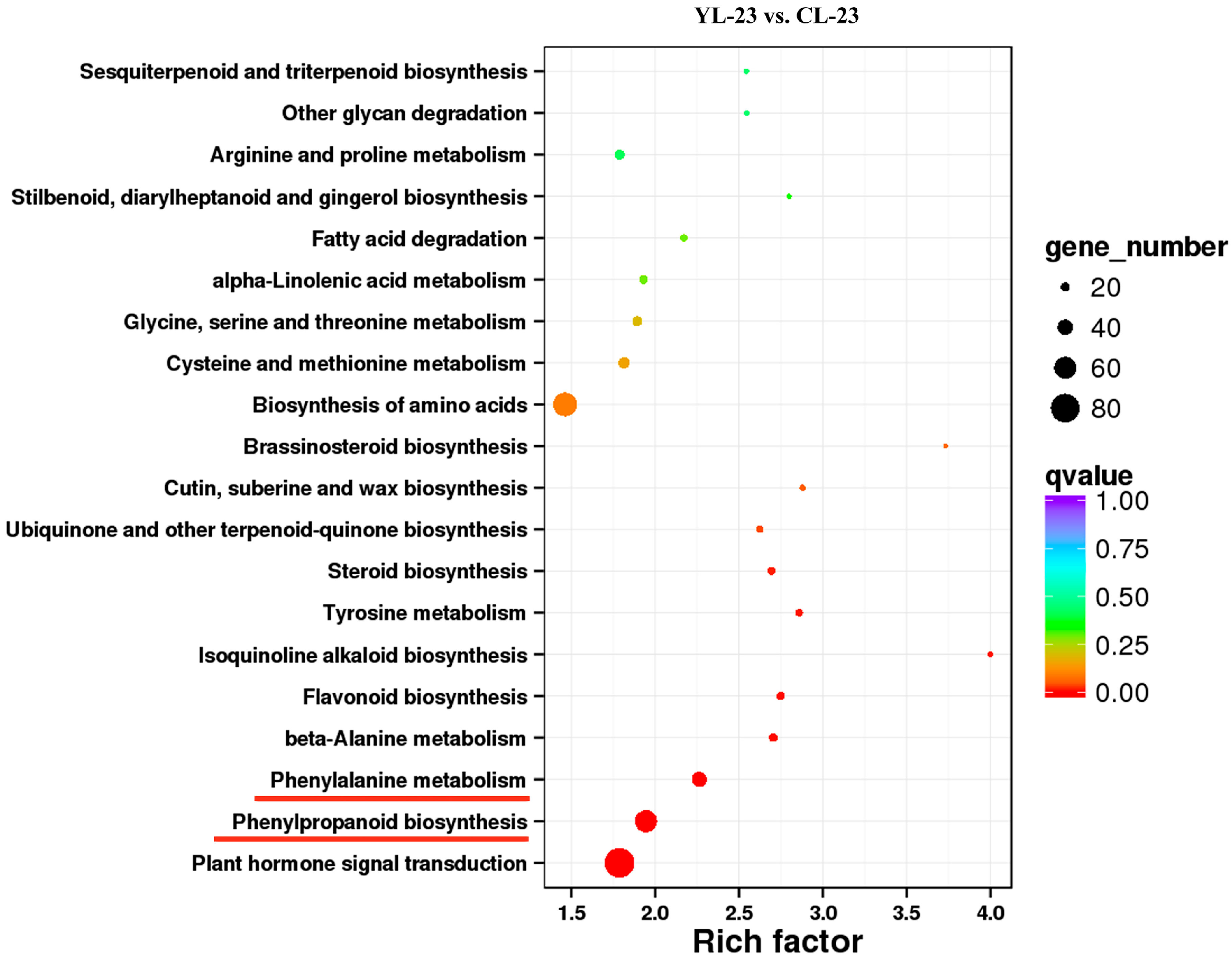
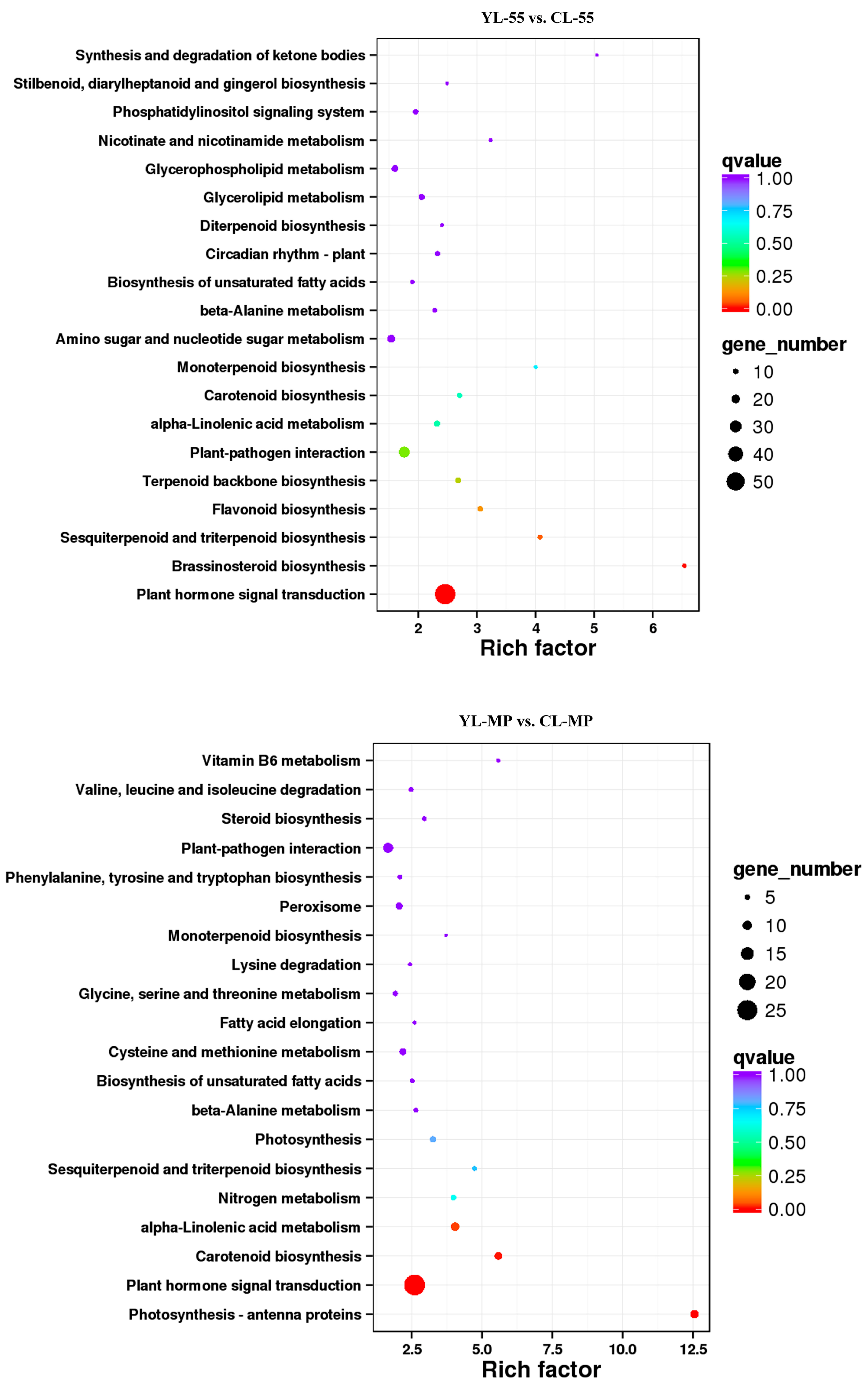
 × ‘Lianglizaosu’
× ‘Lianglizaosu’  ; CL: ‘Cuiguan’
; CL: ‘Cuiguan’  × ‘Lianglizaosu’
× ‘Lianglizaosu’  .
.
 × ‘Lianglizaosu’
× ‘Lianglizaosu’  ; CL: ‘Cuiguan’
; CL: ‘Cuiguan’  × ‘Lianglizaosu’
× ‘Lianglizaosu’  .
.
 × ‘Lianglizaosu’
× ‘Lianglizaosu’  ; CL: ‘Cuiguan’
; CL: ‘Cuiguan’  × ‘Lianglizaosu’
× ‘Lianglizaosu’  ; C23: Fruit samples from CL at 23 DAF; C55: Fruit samples from CL at 55 DAF; CMP: Fruit samples from CL at mature period; Y23: Fruit samples from YL at 23 DAF; Y55: Fruit samples from YL at 55 DAF; YMP: Fruit samples from YL at mature period. A colour scale is provided with the heat map to indicate differential pattern of expression. Red indicates a high level of expression, white signifies a medium level of expression, and blue denotes a low level of expression. FPKM values were obtained by RNA-seq analysis and presented as a heatmap. Histograms represent qRT-PCR analyses of selected candidate genes examined in this study. The ordinate of the histogram indicates the relative gene expression level. The abscissa indicates the three developmental stages (23 DAF, 55 DAF, mature period) of pear fruit.
; C23: Fruit samples from CL at 23 DAF; C55: Fruit samples from CL at 55 DAF; CMP: Fruit samples from CL at mature period; Y23: Fruit samples from YL at 23 DAF; Y55: Fruit samples from YL at 55 DAF; YMP: Fruit samples from YL at mature period. A colour scale is provided with the heat map to indicate differential pattern of expression. Red indicates a high level of expression, white signifies a medium level of expression, and blue denotes a low level of expression. FPKM values were obtained by RNA-seq analysis and presented as a heatmap. Histograms represent qRT-PCR analyses of selected candidate genes examined in this study. The ordinate of the histogram indicates the relative gene expression level. The abscissa indicates the three developmental stages (23 DAF, 55 DAF, mature period) of pear fruit.
 × ‘Lianglizaosu’
× ‘Lianglizaosu’  ; CL: ‘Cuiguan’
; CL: ‘Cuiguan’  × ‘Lianglizaosu’
× ‘Lianglizaosu’  ; C23: Fruit samples from CL at 23 DAF; C55: Fruit samples from CL at 55 DAF; CMP: Fruit samples from CL at mature period; Y23: Fruit samples from YL at 23 DAF; Y55: Fruit samples from YL at 55 DAF; YMP: Fruit samples from YL at mature period. A colour scale is provided with the heat map to indicate differential pattern of expression. Red indicates a high level of expression, white signifies a medium level of expression, and blue denotes a low level of expression. FPKM values were obtained by RNA-seq analysis and presented as a heatmap. Histograms represent qRT-PCR analyses of selected candidate genes examined in this study. The ordinate of the histogram indicates the relative gene expression level. The abscissa indicates the three developmental stages (23 DAF, 55 DAF, mature period) of pear fruit.
; C23: Fruit samples from CL at 23 DAF; C55: Fruit samples from CL at 55 DAF; CMP: Fruit samples from CL at mature period; Y23: Fruit samples from YL at 23 DAF; Y55: Fruit samples from YL at 55 DAF; YMP: Fruit samples from YL at mature period. A colour scale is provided with the heat map to indicate differential pattern of expression. Red indicates a high level of expression, white signifies a medium level of expression, and blue denotes a low level of expression. FPKM values were obtained by RNA-seq analysis and presented as a heatmap. Histograms represent qRT-PCR analyses of selected candidate genes examined in this study. The ordinate of the histogram indicates the relative gene expression level. The abscissa indicates the three developmental stages (23 DAF, 55 DAF, mature period) of pear fruit.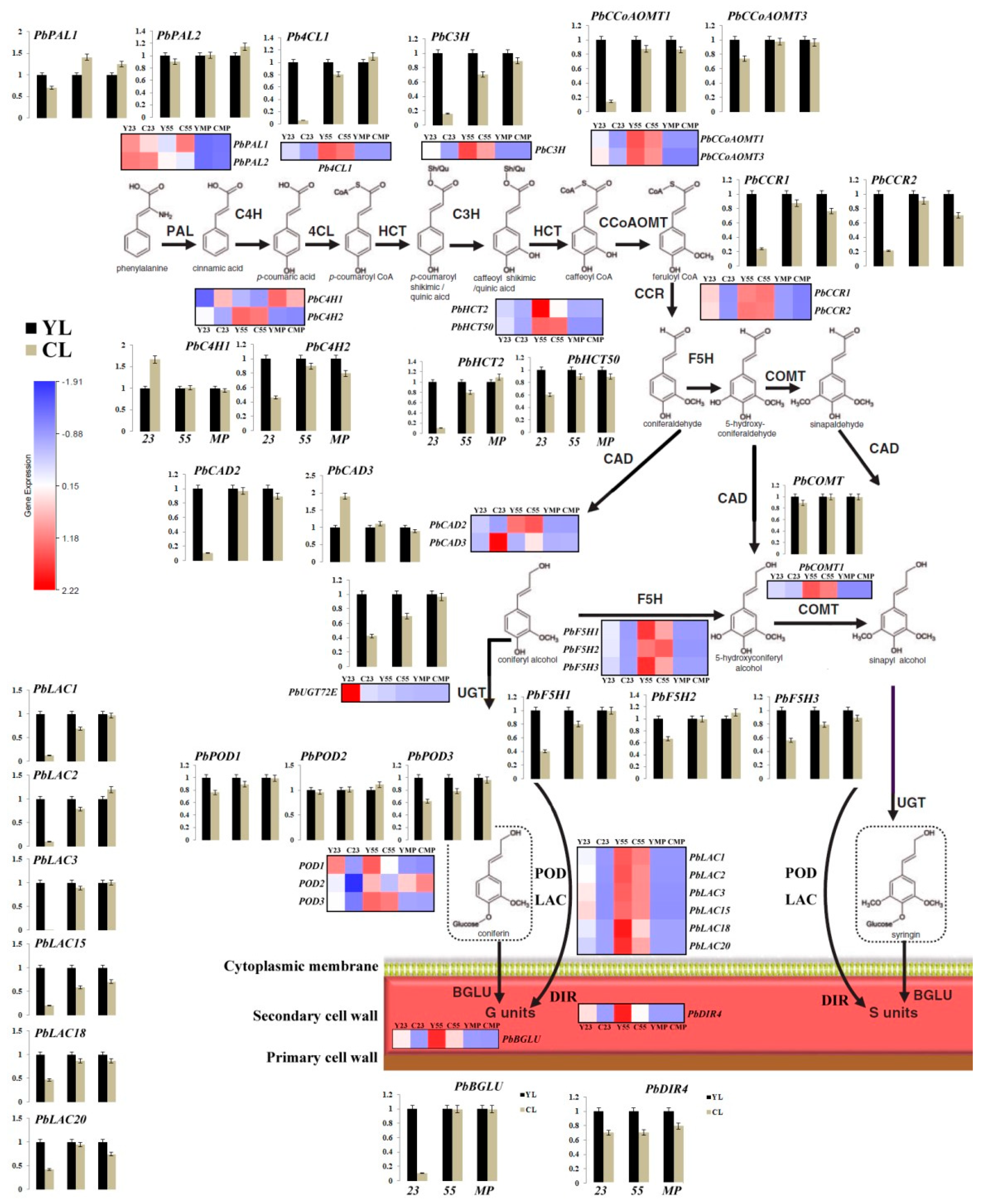

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, X.; Zhang, J.; Wang, H.; Chen, T.; Li, G.; Yan, C.; Jin, Q.; Lin, Y.; Cai, Y. Effects of Metaxenia on Stone Cell Formation in Pear (Pyrus bretschneideri) Based on Transcriptomic Analysis and Functional Characterization of the Lignin-Related Gene PbC4H2. Forests 2020, 11, 53. https://doi.org/10.3390/f11010053
Cheng X, Zhang J, Wang H, Chen T, Li G, Yan C, Jin Q, Lin Y, Cai Y. Effects of Metaxenia on Stone Cell Formation in Pear (Pyrus bretschneideri) Based on Transcriptomic Analysis and Functional Characterization of the Lignin-Related Gene PbC4H2. Forests. 2020; 11(1):53. https://doi.org/10.3390/f11010053
Chicago/Turabian StyleCheng, Xi, Jinyun Zhang, Han Wang, Tianzhe Chen, Guohui Li, Chongchong Yan, Qing Jin, Yi Lin, and Yongping Cai. 2020. "Effects of Metaxenia on Stone Cell Formation in Pear (Pyrus bretschneideri) Based on Transcriptomic Analysis and Functional Characterization of the Lignin-Related Gene PbC4H2" Forests 11, no. 1: 53. https://doi.org/10.3390/f11010053
APA StyleCheng, X., Zhang, J., Wang, H., Chen, T., Li, G., Yan, C., Jin, Q., Lin, Y., & Cai, Y. (2020). Effects of Metaxenia on Stone Cell Formation in Pear (Pyrus bretschneideri) Based on Transcriptomic Analysis and Functional Characterization of the Lignin-Related Gene PbC4H2. Forests, 11(1), 53. https://doi.org/10.3390/f11010053



