Abstract
Illegal logging and associated trade aggravate the over-exploitation of Swietenia species, of which S. macrophylla King, S. mahagoni (L.) Jacq, and S. humilis Zucc. have been listed in Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) Appendix Ⅱ. Implementation of CITES necessitates the development of efficient forensic tools to identify wood species accurately, and ideally ones readily deployable in wood anatomy laboratories across the world. Herein, a method using quantitative wood anatomy data in combination with machine learning models to discriminate between three Swietenia species is presented, in addition to a second model focusing only on the two historically more important species S. mahagoni and S. macrophylla. The intra- and inter-specific variations in nine quantitative wood anatomical characters were measured and calculated based on 278 wood specimens, and four machine learning classifiers—Decision Tree C5.0, Naïve Bayes (NB), Support Vector Machine (SVM), and Artificial Neural Network (ANN)—were used to discriminate between the species. Among these species, S. macrophylla exhibited the largest intraspecific variation, and all three species showed at least partly overlapping values for all nine characters. SVM performed the best of all the classifiers, with an overall accuracy of 91.4% and a per-species correct identification rate of 66.7%, 95.0%, and 80.0% for S. humilis, S. macrophylla, and S. mahagoni, respectively. The two-species model discriminated between S. macrophylla and S. mahagoni with accuracies of over 90.0% using SVM. These accuracies are lower than perfect forensic certainty but nonetheless demonstrate that quantitative wood anatomy data in combination with machine learning models can be applied as an efficient tool to discriminate anatomically between similar species in the wood anatomy laboratory. It is probable that a range of previously anatomically inseparable species may become identifiable by incorporating in-depth analysis of quantitative characters and appropriate statistical classifiers.
1. Introduction
In recent decades, international efforts to prohibit or limit the trade of endangered species have been made to combat illegal logging and associated trade [1,2,3,4,5], usually emphasizing the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES), which lists species in one of three appendices depending on the degree of protection required. To ensure that international trade in timber species does not threaten their survival, more than 500 tree species had been listed in the CITES appendices by 2017 [6].
Among the species protected by CITES are Swietenia macrophylla King, Swietenia mahagoni (L.) Jacq and Swietenia humilis Zucc., three commercially important species in the neotropical (South and Central America with tropical Mexico) genus Swietenia (Meliaceae) which are listed in CITES Appendix II. S. mahagoni, commonly known as American mahogany, Cuban mahogany, or small-leaf mahogany, is a species of Swietenia native to southern Florida in the United States and islands in the Caribbean including the Bahamas, Cuba, Jamaica, and the Dominican Republic and Haiti. S. mahagoni has for centuries been widely exported to European countries for use in fine furniture and musical instruments. S. macrophylla, known as big-leaf mahogany, is one of the world’s most valuable and widely traded timber species, occurring naturally from North to South America [7,8]. The wood of S. macrophylla is prized for furniture, musical instruments, decorative materials, and artisanal crafts due to its beauty, workability, and moderate decay resistance. A third Swietenia species, S. humilis, is largely restricted to Pacific Coast forests of tropical Mexico and Central America, and in more recent times has been over-exploited as a furniture wood. S. humilis is also of interest as a possibly commercial source of seed oil and pharmacologically active compounds [9]. The woods of these three species are widely considered to be unable to be discriminated between by wood anatomy [10,11]. Despite the fact that all three species are listed in CITES Appendix II, which affords them the same degree of protection and thus no direct CITES-related need for a species-level separation, there remain cultural, commercial, and scientific interests in discriminating between the woods of these species and in particular discriminating between S. macrophylla and S. mahagoni with regard to cultural property.
Implementation of CITES regulations for timber necessitates the definitive identification of wood species whether in the laboratory or in the field. Plants are typically identified at the species level by observing all their features, including leaves, fruits, seeds, and flowers. For commercial wood trade, the tree is logged and typically only boards are available, with not even the additional information that might be provided by bark available [12,13,14]. This poses additional challenges because botanists trained in field identification of plants do not by default have the necessary background in wood anatomy needed to make field identifications of wood. To overcome the general lack of human expertise in field and forensic wood identification [15], alternative methodologies have been developed for correct wood identification of CITES-listed timber species, including near-infrared spectroscopy (NIRS) [16,17,18], direct analysis in real time mass spectrometry (DART) [19,20,21,22], computer vision [23,24,25] and DNA methods [26,27,28,29,30,31,32], most of which do not depend on a human expert trained in wood anatomy. These non-anatomical techniques could provide solutions for discriminating between similar species, but not all laboratories or countries have access to these technologies and investigators with expert knowledge and skills to conduct the necessary experimental work to facilitate field deployment [14,33]. Although traditional light microscopic wood identification is fundamentally a laboratory-based endeavor typically restricted to people with access to and skill with light microscopes, reference collections, and operations involving a place of significant expertise with wood anatomy [34,35,36], many countries have at least one laboratory that maintains wood anatomical capacity, so any advances in wood anatomical methods can be widely applied at low cost.
Regarding this context, species identification technologies deployable across the whole wood product supply chain across the world are critical because many protected timbers can easily be mistaken for legally harvested species due to similar gross appearance and even similar wood anatomical structure. This is particularly the case for closely related congeneric species (e.g., species within the genus Swietenia) and can be further complicated by the potentially high degree of variability within some species. The most common wood identification method currently employed is a combination of macroscopic and microscopic identification in which many wood characters such as density, color, smell, brightness, texture, growth rings, vessels, fibers, and porosity of an unknown sample are compared with reference collection exemplars of candidate species [11,35]. An expert wood anatomist can perform an identification and provide forensically reliable results [13], but they depend on knowledge of the breadth of variability in a given species and typically require years or decades to develop that expertise. One way to investigate subtle wood anatomical divergences between species is to collect quantitative data relating to key characters and subject these data to multivariate statistical classification [36,37]. Broadly, these statistical methodologies fall into two categories, namely, unsupervised and supervised. Unsupervised methods draw inferences from datasets consisting of input data without labels, and some types of cluster analysis are used for exploratory data analysis to find hidden patterns or groupings in data [38], but this approach is virtually absent in the wood anatomical literature, perhaps in part because unsupervised methods perform most reliably with large amounts of data which are difficult to generate in wood anatomy. Supervised classification is the data mining task of inferring a function from labeled training data, and includes Naïve Bayes (NB), Support Vector Machine (SVM), Artificial Neural Network (ANN) and Decision Tree C5.0 (DT) methods, among others, each of which is widely used for classification tasks [39]. Such statistical analysis in wood discrimination has been recently applied to data from non-traditional wood identification methods [22,29,32,40,41,42], but not for quantitative wood anatomy.
In his 1933 paper, Panshin wrote that “the woods of the Swietenia species cannot be separated anatomically with any degree of certainty” and no publications since that time have overturned this judgment [10]. The aim of this study was to test Panshin’s claim using quantitative wood anatomy data in combination with machine learning models to establish an identification method that could discriminate between three Swietenia species, namely, S. marcophylla, S. mahagoni, and S. humilis, based on a large number of specimens from wood collections. This was approached using three steps: (1) investigating the intra- and inter-specific variations of the three species, (2) testing multivariate supervised classifiers (DT, NB, SVM, and ANN) on all characters for their discrimination ability regarding the three species, and (3) investigating a two-species model to separate the historically more important S. macrophylla and S. mahagoni given the relative paucity of Swietenia humilis specimens available for study.
2. Materials and Methods
2.1. Reference Specimens
Mature wood of 278 Swietenia specimens (198 Swietenia macrophylla, 65 Swietenia mahagoni, and 15 Swietenia humilis) from the Samuel J. Record Collection (SJRw), the USDA Forest Products Laboratory Wood Collection (MADw) of Madison, Wisconsin, and the wood collection (RBw) of the Botanic Garden of Rio de Janeiro in Brazil was selected. Wood microscopy and microtomy were performed as in [43] and images of three planes of sections of S. macrophylla, S. mahagoni, and S. humilis are shown in Figure 1.
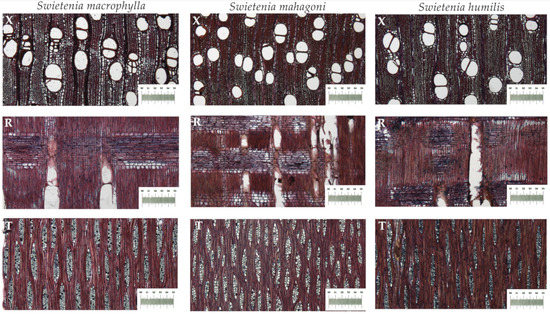
Figure 1.
The three planes of sections of S. macrophylla, S. mahagoni, and S. humilis, showing the similarity of the three species with regard to wood anatomical features. Legend: X, cross section; R, radial section; T, tangential section.
2.2. Quantitative Characters
Measured and calculated quantitative character definitions are listed in Table 1 and generally correspond to the IAWA list [44]. Vessel element length (VEL) and fiber length (FL) are reported as the average of 25 and 50 measurements, respectively. The ratio of fiber length to vessel length (F/V) was calculated. Mean tangential vessel diameter (TVD) was calculated from measurements taken at the widest point of 50 vessels using ImageJ version 1.47 (National Institute of Health, Bethesda, MD, USA) [45]. Frequency of vessels (FOV) was measured by dividing the total number of vessels in one picture by its area of 11.2 mm2 (4612 microns in width and 2432 microns in height), allowing for more than the IAWA-recommended use of 10 fields of view with 1 mm2 each. Rays per linear mm (RPMM) was measured by counting the number of rays transected by a 2 mm reference line in five different fields of view. Ray height (RHEIGHT) and ray width (RWIDTH) were also measured in microns using ImageJ. Rays of every width category (determined as the number of cells at maximum width) were measured in random fields of view until at least five rays per width category (whenever possible) were obtained, and were indexed according to the area of the field of view to provide a rays per mm2 value. Ray area index (RAI) was calculated as described in [43]. As rays are fusiform in shape to varying degrees, the RAI is, by virtue of being a rectangular metric, an overestimate of total ray area.

Table 1.
Definitions of and number of measured and calculated characters.
2.3. Statistical Analysis
All statistical work was performed in R version 3.4.4. Intra- and inter-specific quantitative variations were examined using descriptive statistical data analysis with Bonferroni corrections in all t-tests to account for multiple comparisons.
To implement supervised machine learning algorithms, we used a 10-fold cross-validation at the specimen level for both the three-species and two-species models. We tested the performance of four classifiers, namely, DT, NB, SVM, and ANN, which were implemented with R packages C50, e1071, kernlab, and nnet, respectively [46,47,48,49]. Decision Tree is a branching model similar to a traditional identification key that separates classes by splitting data at decision nodes and ending at the separated classes [50]. Naïve Bayes employs Bayes’ theorem, assuming independence between features among the classes, to calculate a class membership probability [51]. An SVM model maps the input data into a higher dimensional space and then determines optimal class-separating hyperplanes [52]. ANN is an information processing system with interconnected “neurons” that receive, process, and transmit input signals using feature weights, the reticulated structure of which is inspired by the biological processes of the human brain [53].
3. Results
3.1. Quantitative Wood Anatomy Data
Table 2 presents mean values and standard deviations for each of the characters measured in the three Swietenia species, as well as the results of t-tests of the mean values. The results of t-tests indicated that there were significant differences among three Swietenia species in the mean values of FL, FOV, and RHEIGHT. Nevertheless, the calculated RAI failed to show any divergence across the three species.

Table 2.
Mean values and standard deviations with t-tests for the nine measured and calculated indices in three Swietenia species.
Smoothed data density histograms of the nine characters are presented in Figure 2. Not surprisingly, none of the three Swietenia species could be separated by any single quantitative character because the data distribution within and between species showed overlap for all characters.
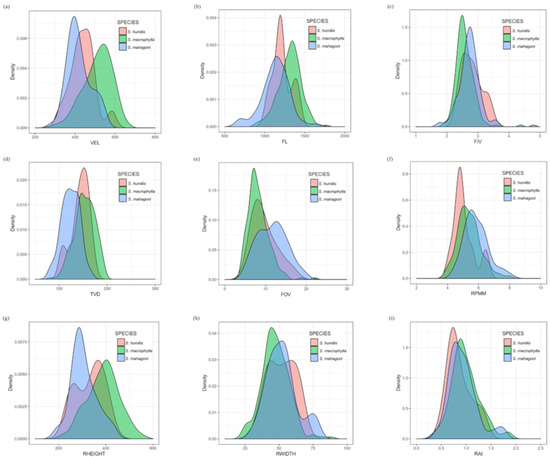
Figure 2.
Smoothed data density histograms of the nine characters: (a) VEL, (b) FL, (c) F/V, (d) TVD, (e) FOV, (f) RPMM, (g) RHEIGHT, (h) RWIDTH, (i) RAI.
3.2. Machine Learning Classifiers for Discrimination between the Three Swietenia Species
Confusion matrices reporting performance metrics for the four supervised classifiers, namely, DT, NB, SVM, and ANN are presented in Figure 3. The true species are reported on the left and the predicted species are given in the columns, such that on-diagonal predictions are correct and all other predictions are incorrect. The value in each cell is the proportion of the total predictions, and each row always sums to 1.0. Of the four supervised classifiers, SVM showed the best overall performance (91.4%), followed by ANN (83.8%), NB (79.5%), and DT (78.4%). Using the SVM classifier, the three Swietenia species could be separated with a relatively higher accuracy, i.e., 66.7% for S. humilis, 95.0% for S. macrophylla, and 80.0% for S. mahagoni. However, the DT and ANN classifiers failed to predict S. humilis correctly.
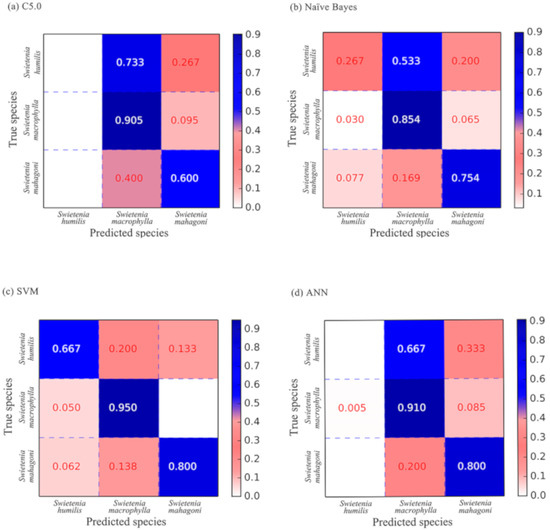
Figure 3.
Confusion matrices generated by the four supervised classifiers: (a) Decision Tree C5.0, (b) Naïve Bayes, (c) Support Vector Machine, (d) Artificial Neural Network.
3.3. Machine Learning Classifiers for Discrimination of S. mahagoni and S. macrophylla
Figure 4 presents the confusion matrices for the two-species model discriminating between Swietenia mahagoni and Swietenia macrophylla. The four supervised classifiers exhibited overall accuracy values of over 80%, being i.e., 93.9% (SVM), 85.2% (ANN), 81.7% (NB), and 81.4% (DT). SVM was able to discriminate between S. macrophylla and S. mahagoni with thresholds of 94.9% and 90.8%, respectively. The results indicated that the two-species model performed slightly better than the three-species model.
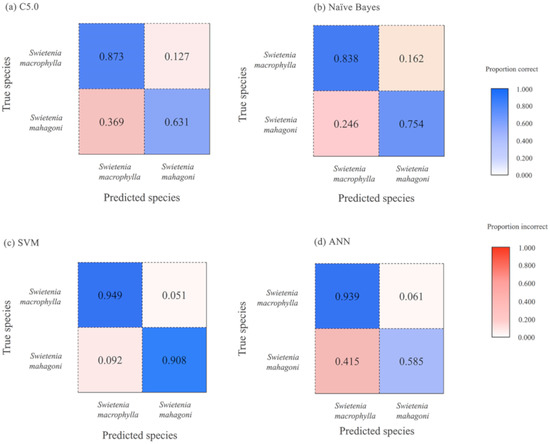
Figure 4.
Confusion matrices for discriminating between Swietenia mahagoni and Swietenia macrophylla: (a) Decision Tree C5.0, (b) Naïve Bayes, (c) Support Vector Machine, (d) Artificial Neural Network.
4. Discussion
4.1. Intra- and Inter-Species Variations of the Three Swietenia Species
In traditional wood identification, species-level resolution is often impossible because intra-species variation is as large as inter-species variation within a genus [54,55,56]. All characters showed overlap in intra- and inter-species variation for the three Swietenia species, and, therefore, no single character was sufficient to separate them. Swietenia macrophylla showed relatively higher within-species variation across all characters, which could be related to the wider geographic range it spans and thus the greater likelihood of genetic diversity, or this could be due to the larger number of specimens of this taxon included in this study [14]. Comparing the variance in each of the nine characters across the three species, VEL, TVD, and RHEIGHT showed the largest variance for S. macrophylla (Table 2), which also had the most specimens in this study. Four characters, namely, F/V, RPMM, RWIDTH, and RAI, were nearly invariant between species. As variance is independent of sample size, we conclude that the wider geographic range of S. macrophylla and not the larger number of specimens likely accounts for the larger variations within this species [57,58].
It is not surprising that no single character was sufficient to separate the three species—traditional wood identification employs combinations of many characters sequentially in written, diagnostic keys [15,36]. However, for quantitative wood anatomy data, the complexity of the data distributions within and between species when combining characters can confound human ability to visualize important patterns that might be sufficient to discriminate between species [59]. To overcome this, we fed all the data into supervised machine learning classifiers to separate these species.
4.2. Discrimination between the Three Swietenia Species
According to the confusion matrices generated by the four supervised classifiers, S. macrophylla was always correctly classified with the highest accuracy (85.4%–95.0%), followed by S. mahagoni (60.0%–80.0%), and S. humilis (0–66.7%, Figure 3). Misclassified specimens of S. humilis and S. mahagoni were typically predicted as S. macrophylla, although never in proportion with the abundance of S. macrophylla in the dataset, indicating that class imbalance in the dataset was not the proximate cause of any inaccuracy. The high wood anatomical variability within S. macrophylla encompasses much of the variability in the other two species, and even machine learning methods did not suffice to provide full forensic certainty [22,60] but did greatly exceed prior reported accuracy [10]. When separating similar species using quantitative wood anatomy data, it is recommended that a sufficiently large number of specimens be studied to ensure that the full range of variation across the species is incorporated [56].
Although all three Swietenia are listed in CITES Appendix II, there remain historical and cultural demands to separate S. mahagoni and S. macrophylla, without reference to S. humilis, which by virtue of its more inland and Pacific distribution is presumed not to appear in older Western cultural property. In this study, SVM exhibited the highest accuracy of all the supervised classifiers with a correct threshold of over 90.0% when separating the two species (Figure 4). Machine learning models have shown considerably better performance than expert wood anatomists when separating S. macrophylla and S. mahagoni [10,11,61]. The results demonstrated in this study provide a pathway to discriminate between similar woods, including CITES species, using quantitative wood anatomy data in combination with machine learning models [36,55].
4.3. Evaluation of Machine Learning Methods for Wood Identification
Historically, traditional wood identification relies on wood anatomists who spend decades to gain their expertise and experience, and such identifications typically only reach the genus level. The main challenge for traditional wood identification around the world is the paucity of trained wood anatomists [15], especially for large-scale challenges like combating illegal logging and associated trade. This study has demonstrated the feasibility of using quantitative wood anatomy data with machine learning models to discriminate between CITES-listed species. To use these models, the only requirement is to collect quantitative wood anatomy data and input them to the machine learning models using free software. The machine learning models then output the classification results and show better discrimination ability than wood anatomists.
In previous studies, ANN and NB have been widely reported to identify species using quantitative wood anatomy data coupled with principle components analysis [62,63]. In this study, SVM outperformed ANN and NB in discrimination ability for both the three-species and two-species models. This is consistent with results from other works in which a supervised SVM classifier was used for wood species classification with images [64,65] and DNA data [32,42,66,67].
5. Conclusions
Traditional wood identification relying on qualitative wood anatomy is typically accurate only to the genus level. Non-anatomical technologies capable of species-level identification are quite expensive and also require comprehensive reference data libraries, and, in some cases, specialized technical expertise. By contrast, preparation and measurement of microscope slides is a familiar task in a wood anatomy laboratory. By analyzing wood anatomy data collected using standard tools, we have presented a highly effective machine-learning-based classifier able to separate the three Swietenia species, as well as discriminate between S. macrophylla and S. mahagoni. This study shows that quantitative traditional wood anatomy data can discriminate between anatomically similar species, shown here with CITES-listed species, and thus provide an easily-implemented technique for wood anatomy laboratories to aid in efforts to eliminate illegal logging.
Author Contributions
Conceptualization, A.C.W., J.M., and T.H.; data curation, T.H. and J.M.; formal analysis, T.H. and J.M.; funding acquisition, A.C.W.; investigation, A.C.W. and J.M.; methodology, A.C.W., T.H., and J.M.; project administration, A.C.W.; resources, R.S.; software, T.H.; supervision, A.C.W. and Y.Y.; validation, A.C.W.; visualization, T.H.; writing—original draft, T.H., writing—review and editing, A.C.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported in part by a grant from the US Department of State via Interagency Agreement number 19318814Y0010.
Acknowledgments
We express our gratitude to Prabu Ravindran, Adriana Costa, Rafael Arévalo, and Xiaomei Jiang for their assistance with this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dormontt, E.E.; Boner, M.; Braun, B.; Breulmann, G.; Degen, B.; Espinoza, E.; Gardner, S.; Guillery, P.; Hermanson, J.C.; Koch, G.; et al. Forensic timber identification: it’s time to integrate disciplines to combat illegal logging. Biol. Conserv. 2015, 191, 790–798. [Google Scholar] [CrossRef]
- Lewis, S.L.; Edwards, D.P.; Galbraith, D. Increasing human dominance of tropical forests. Science 2015, 349, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.K.S.; Lee, S.L.; Tnah, L.H.; Nurul-Farhanah, Z.; Ng, H.C.; Lee, T.C.; Tani, N.; Diway, B.; Sing, L.P.; Khoo, E. Forensic timber identification: A case study of a CITES listed species, Gonystylus bancanus (Thymelaeaceae). Forensic Sci. Int. Genet. 2016, 23, 197–209. [Google Scholar] [CrossRef] [PubMed]
- United Nations Office on Drug and Crime. Best Practice Guide for Forensic Timber Identification; Laboratory and Scientific Section, Global Programme for Combating Wildlife and Forest Crime: Vienna, Austria; New York, NY, USA, 2016; pp. 1–226. Available online: https://www.unodc.org/documents/Wildlife/Guide_Timber.pdf (accessed on 24 August 2016).
- Brancalion, P.H.S.; Almeida, D.R.A.; Vidal, E.; Molin, P.G.; Sontag, V.E.; Souza, S.E.X.F.; Schulze, M. Fake legal logging in the Brazilian Amazon. Sci. Adv. 2018, 4, 1192. [Google Scholar] [CrossRef]
- Baas, P. CITES and timbers- a guide to CITES listed tree species. IAWA J. 2017, 38, 135–136. [Google Scholar] [CrossRef]
- Dünisch, O.; Montóia, V.R.; Bauch, J. Dendroecological investigations on Swietenia macrophylla King and Cedrela odorata L. (Meliaceae) in the central Amazon. Trees 2003, 17, 244–250. [Google Scholar] [CrossRef]
- Grogan, J.; Barreto, P. Big-leaf mahogany on CITES appendix II: Big challenge, big opportunity. Conserv. Biol. 2005, 19, 973–976. [Google Scholar]
- Ovalle-Magallanes, B.; Madariaga-Mazon, A.; Navarrete, A.; Mata, R. Mechanisms of action of antihyperglycemic mexicanolides isolated from Swietenia humillis: In vivo and in silico approaches. Planta Med. 2016, 82, 136. [Google Scholar] [CrossRef]
- Panshin, A.J. Comparative anatomy of the woods of the Meliaceae, sub-family Swietenioideae. Am. J. Bot. 1933, 20, 638–668. [Google Scholar] [CrossRef]
- Gasson, P. How precise can wood identification be? Wood anatomy’s role in support of the legal timber trade, especially CITES. IAWA J. 2011, 32, 137–154. [Google Scholar] [CrossRef]
- Gasson, P.; Baas, P.; Wheeler, E. Wood anatomy of CITES-listed tree species. IAWA J. 2011, 32, 155–197. [Google Scholar] [CrossRef]
- Ruffinatto, F.; Crivellaro, A.; Wiedenhoeft, A.C. Review of macroscopic features for hardwood and softwood identification and a proposal for a new character list. IAWA J. 2015, 36, 208–241. [Google Scholar] [CrossRef]
- Bergo, M.C.J.; Pastore, T.C.M.; Coradin, V.T.R.; Wiedenhoeft, A.C.; Braga, J.W.B. NIRS identification of Swietenia macrophylla is robust across specimens from 27 countries. IAWA J. 2016, 37, 420–430. [Google Scholar] [CrossRef]
- Wiedenhoeft, A.C.; Simeone, J.; Smith, A.; Parker-Forney, M.; Soares, R.; Fishman, A. Fraud and misrepresentation in retail forest products exceeds U.S. forensic wood science capacity. PLoS ONE 2019, 14, e0219917. [Google Scholar] [CrossRef] [PubMed]
- Pastore, T.C.M.; Braga, J.W.B.; Coradin, V.T.R.; Magalhães, W.L.E.; Okino, E.Y.A.; Camargos, J.A.A.; de Muñiz, G.I.B.; Bressan, O.A.; Davrieux, F. Near infrared spectroscopy (NIRS) as a potential tool for monitoring trade of similar woods: Discrimination of true mahogany, cedar, andiroba, and curupixá. Holzforschung 2010, 65, 73–80. [Google Scholar] [CrossRef]
- Braga, J.W.B.; Pastore, T.C.M.; Coradin, V.T.R.; Camargos, J.A.A.; Silva, A.R. The use of near infrared spectroscopy to identify solid wood specimens of Swietenia macrophylla (CITES Appendix II). IAWA J. 2011, 32, 285–296. [Google Scholar] [CrossRef]
- Snel, F.A.; Braga, J.W.B.; Silva, D.; Wiedenhoeft, A.C.; Costa, A.; Soares, R.; Coradin, V.T.R.; Pastore, T.C.M. Potential field-deployable NIRS identification of seven Dalbergia species listed by CITES. Wood Sci. Technol. 2018, 52, 1411–1427. [Google Scholar] [CrossRef]
- Cody, R.B.; Dane, A.J.; Andoh, B.J.; Adedipe, E.O.; Nkansahb, K. Rapid classification of white oak (Quercus alba) and Northern red oak (Quercus rubra) by using pyrolysis direct analysis in real time (DART™) and time-of-flight mass spectrometry. J. Anal. Appl. Pyrolysis 2012, 95, 134–137. [Google Scholar] [CrossRef]
- Lancaster, C.; Espinoza, E.O. Analysis of select Dalbergia and trade timber using direct analysis in real time and time-of-flight mass spectrometry for CITES enforcement. Rapid Commun. Mass Spectrom. 2012, 26, 1147–1156. [Google Scholar] [CrossRef]
- Espinoza, E.O.; Wiemann, M.C.; Morales, J.B.; Chavarria, G.D.; McClure, P.J. Forensic analysis of CITES-protected Dalbergia timber from the Americas. IAWA J. 2015, 36, 311–325. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, G.; Liu, B.; He, T.; Guo, J.; Jiang, X.; Yin, Y. Wood discrimination analyses of Pterocarpus tinctorius and endangered Pterocarpus santalinus using DART-FTICR-MS coupled with multivariate statistics. IAWA J. 2019, 40, 58–74. [Google Scholar] [CrossRef]
- Hwang, S.K.; Kobayashi, K.; Zhai, S.; Sugiyama, J. Automated identification of Lauraceae by scale-invariant feature transform. J. Wood Sci. 2017, 64, 69–77. [Google Scholar] [CrossRef]
- Ibrahim, I.; Khairuddin, A.S.M.; Arof, H.; Yusof, R.; Hanafi, E. Statistical feature extraction methods for wood recognition system. Euro. J. Wood Prod. 2018, 76, 345–356. [Google Scholar] [CrossRef]
- Ravindran, P.; Costa, A.; Soares, R.; Wiedenhoeft, A.C. Classification of CITES-listed and other neotropical Meliaceae wood images using convolutional neural networks. Plant Meth. 2018, 14, 25. [Google Scholar] [CrossRef] [PubMed]
- White, G.; Powell, W. Isolation and characterization of microsatellite loci in Swietenia humilis (Meliaceae): An endangered tropical hardwood species. Mol. Ecol. 2008, 6, 851–860. [Google Scholar] [CrossRef]
- Muellner, A.N.; Schaefer, H.; Lahaye, R. Evaluation of candidate DNA barcoding loci for economically important timber species of the mahogany family (Meliaceae). Mol. Ecol. Resour. 2011, 11, 450–460. [Google Scholar] [CrossRef]
- Höltken, A.M.; Schröder, H.; Wischnewski, H.; Degen, B.; Magel, E.; Fladung, M. Development of DNA based methods to identify CITES-protected timber species: A case study in the Meliaceae family. Holzforschung 2012, 66, 97–104. [Google Scholar]
- Hartvig, I.; Czako, M.; Kjær, E.D.; Nielsen, L.R.; Theilade, I. The use of DNA barcoding in identification and conservation of rosewood (Dalbergia spp.). PLoS ONE 2015, 10, e0138231. [Google Scholar] [CrossRef]
- Yu, M.; Jiao, L.; Guo, J.; Wiedenhoeft, A.C.; He, T.; Jiang, X.; Yin, Y. DNA barcoding of vouchered xylarium wood specimens of nine endangered Dalbergia species. Planta 2017, 246, 1165–1176. [Google Scholar] [CrossRef]
- Jiao, L.; Yu, M.; Wiedenhoeft, A.C.; He, T.; Li, J.; Liu, B.; Jiang, X.; Yin, Y. DNA barcode authentication and library development for the wood of six commercial Pterocarpus species: The critical role of Xylarium specimens. Sci. Rep. 2018, 8, 1945. [Google Scholar] [CrossRef]
- He, T.; Jiao, L.; Wiedenhoeft, A.C.; Yin, Y. Machine learning approaches outperform distance- and tree-based methods for DNA barcoding of Pterocarpus wood. Planta 2019, 249, 1617–1625. [Google Scholar] [CrossRef] [PubMed]
- Ugochukwu, A.; Hobbs, J.; Phillips, P.; Kerr, W. Technological solutions to authenticity issues in international trade: The case of CITES listed endangered species. Ecol. Econ. 2018, 146, 730–739. [Google Scholar] [CrossRef]
- Wheeler, E.A.; Bass, P.; Rodgers, S. Variations in dicot wood anatomy: A global analysis based on the insidewood database. IAWA J. 2007, 28, 229–258. [Google Scholar] [CrossRef]
- Koch, G.; Haag, V.; Heinz, I.; Richter, H.; Schmitt, U. Control of international traded timber-the role of macroscopic and microscopic wood identification against illegal logging. J. Forensic Res. 2016, 6, 317. [Google Scholar]
- Gasson, P.; Miller, R.; Stekel, D.J.; Whinder, F.; Ziemińska, K. Wood identification of Dalbergia nigra (CITES Appendix I) using quantitative wood anatomy, principal components analysis and Naïve Bayes classification. Ann. Bot. 2010, 105, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Lowe, A.J.; Dormontt, E.E.; Bowie, M.J.; Degen, B.; Gardner, S.; Thomas, D.; Clarke, C.; Rimbawanto, A.; Wiedenhoeft, A.; Yin, Y.; et al. Opportunities for improved transparency in the timber trade through scientific verification. Bioscience 2016, 66, 990–998. [Google Scholar] [CrossRef]
- Von-Arx, G.; Crivellaro, A.; Prendin, A.L.; Čufar, K.; Carrer, M. Quantitative wood anatomy-practical guidelines. Front. Plant Sci. 2016, 7, 781. [Google Scholar] [CrossRef]
- Hofmann, T. Unsupervised learning by probabilistic latent semantic analysis. Mach. Learn. 2001, 42, 177–196. [Google Scholar] [CrossRef]
- Møller, M.F. A scaled conjugate gradient algorithm for fast supervised learning. Neural Net. 1993, 6, 525–533. [Google Scholar] [CrossRef]
- Filho, P.L.P.; Oliveira, L.S.; Nisgoski, S.; Britto, J.A.S. Forest species recognition using macroscopic images. Mach. Vis. Appl. 2014, 25, 1019–1031. [Google Scholar] [CrossRef]
- He, T.; Jiao, L.; Yu, M.; Guo, J.; Jiang, X.; Yin, Y. DNA barcoding authentication for the wood of eight endangered Dalbergia timber species using machine learning approaches. Holzforschung 2018, 73, 277–285. [Google Scholar] [CrossRef]
- Wiedenhoeft, A.C.; Arévalo, R.; Ledbetter, C.; Jakes, J.E. Structure-property characterization of the crinkle-leaf peach wood phenotype: A future model system for wood properties research? JOM 2016, 68, 2405–2412. [Google Scholar] [CrossRef]
- IAWA Committee. IAWA list of microscopic features for hardwood identification. IAWA Bull. 1989, 10, 219–332. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, P.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Meth. 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Kuhn, M.; Quinlan, R. C50:C5.0 Decision Trees and Rule-Based Models. Available online: https://CRAN.R-project.org/package=C50 (accessed on 15 December 2018).
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. e1071: Misc Functions of the Department of Statistics, Probability. Available online: https://CRAN.R-project.org/package=e1071 (accessed on 15 December 2018).
- Karatzoglou, A.; Smola, A.; Hornik, K.; Zeileis, A. kernlab-An S4 package for kernel methods in R. J. Stat. Softw. 2004, 11, 1–20. [Google Scholar] [CrossRef]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with R; Springer: New York, NY, USA, 2002. [Google Scholar]
- Gupta, B.; Rawat, A.; Jain, A. Analysis of various decision tree algorithms for classification in data mining. Int. Comput. Appl. 1997, 27, 5–44. [Google Scholar] [CrossRef]
- Lewis, D.D. Naive (Bayes) at forty: The independence assumption in information retrieval. Mach. Learn. 1998, 98, 4–15. [Google Scholar]
- Suykens, J.A.K.; Vandewalle, J. Least squares support vector machine classifiers. Neural Proc. Lett. 1999, 9, 293–300. [Google Scholar] [CrossRef]
- Jiang, J.; Trundle, P.; Ren, J. Medical image analysis with artificial neural networks. Comput. Med. Imaging Graph. 2010, 34, 617–631. [Google Scholar] [CrossRef]
- Cai, J.; Tyree, M.T. The impact of vessel size on vulnerability curves: Data and models for with-species variability in saplings of aspen, Populus tremuloides Michx. Plant Cell Environ. 2010, 33, 1059–1069. [Google Scholar] [CrossRef]
- Wiedenhoeft, A.C.; Baas, P. Wood science for promoting legal timber harvest. IAWA J. 2011, 32, 121–297. [Google Scholar]
- Li, S.; Link, R.; Li, R.; Deng, L.; Schuldt, B.; Jiang, X.; Zhao, R.; Zheng, J.; Li, S.; Yin, Y. Influence of cambial age and axial height on the spatial patterns of xylem traits in Catalpa bungei, a ring-porous tree species native to China. Forests 2019, 10, 662. [Google Scholar] [CrossRef]
- Lemes, M.R.; Gribel, R.; Proctor, J.; Grattapaglia, D. Population genetic structure of mahogany (Swietenia macrophylla King, Meliaceae) across the Brazilian Amazon, based on variation at microsatellite loci: Implications for conservation. Mol. Ecol. 2003, 12, 2875–2883. [Google Scholar] [CrossRef]
- Degen, B.; Ward, S.E.; Lemes, M.R.; Navarro, C.; Cavers, S.; Sebbenn, A.M. Verifying the geographic origin of mahogany (Swietenia macrophylla King) with DNA-fingerprints. Forensic Sci. Int. Genet. 2013, 7, 55–62. [Google Scholar] [CrossRef]
- Arévalo, R.; Ee, B.W.; Riina, R.; Berry, P.E.; Wiedenhoeft, A.C. Force of habit: Shrubs, trees and contingent evolution of wood anatomical diversity using Croton (Euphorbiaceae) as a model system. Ann. Bot. 2017, 119, 563–579. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, H.H.; Wang, X. Machine learning for Big Data analysis. Trends Plant Sci. 2015, 19, 798–808. [Google Scholar] [CrossRef]
- Deklerck, V.; Mortier, T.; Goeders, N.; Cody, R.B.; Waegeman, W.; Espinoza, E.; Acker, J.V.; Bulcke, J.V.; Beeckman, H. A protocol for automated timber species identification using metabolome profiling. Wood Sci. Technol. 2019, 53, 953–965. [Google Scholar] [CrossRef]
- Esteban, L.G.; Fernández, F.G.; Palacios, P.; Romero, R.M.; Cano, N.N. Artificial neural networks in wood identification: The case of two Juniperus species from the Canary Islands. IAWA J. 2009, 30, 87–94. [Google Scholar] [CrossRef]
- Esteban, L.G.; Palacios, P.; Conde, M.; Fernández, F.G.; García-Iruela, A.; González-Alonso, M. Application of artificial neural networks as a predictive method to differentiate the wood of Pinus sylvestris L. and Pinus nigra Arn subsp. salzmannii (Dunal) Franco. Wood Sci. Technol. 2017, 51, 1249–1258. [Google Scholar] [CrossRef]
- Mallik, A.; Saavedra, J.T.; Fernández, M.F.; Naya, S. Classification of wood micrographs by image segmentation. Chem. Intell. Lab. Syst. 2011, 107, 351–362. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, G.; Qi, H. Wood recognition using image texture features. PLoS ONE 2013, 8, e76101. [Google Scholar] [CrossRef]
- Weitschek, E.; Fiscon, G.; Felici, G. Supervised DNA barcodes species classification: Analysis, comparisons and results. BioData Min. 2014, 7, 4. [Google Scholar] [CrossRef]
- More, R.P.; Mane, R.C.; Purohit, H.C. MatK-QR classifier: A patterns based approach for plant species identification. BioData Min. 2016, 9, 39. [Google Scholar] [CrossRef][Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).