Differential Expression of Genes Related to the Formation of Giant Leaves in Triploid Poplar
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Total RNA Isolation, cDNA Preparation, and Transcriptome Sequencing
2.3. RT-PCR Validation of Differentially Expressed Genes
3. Results
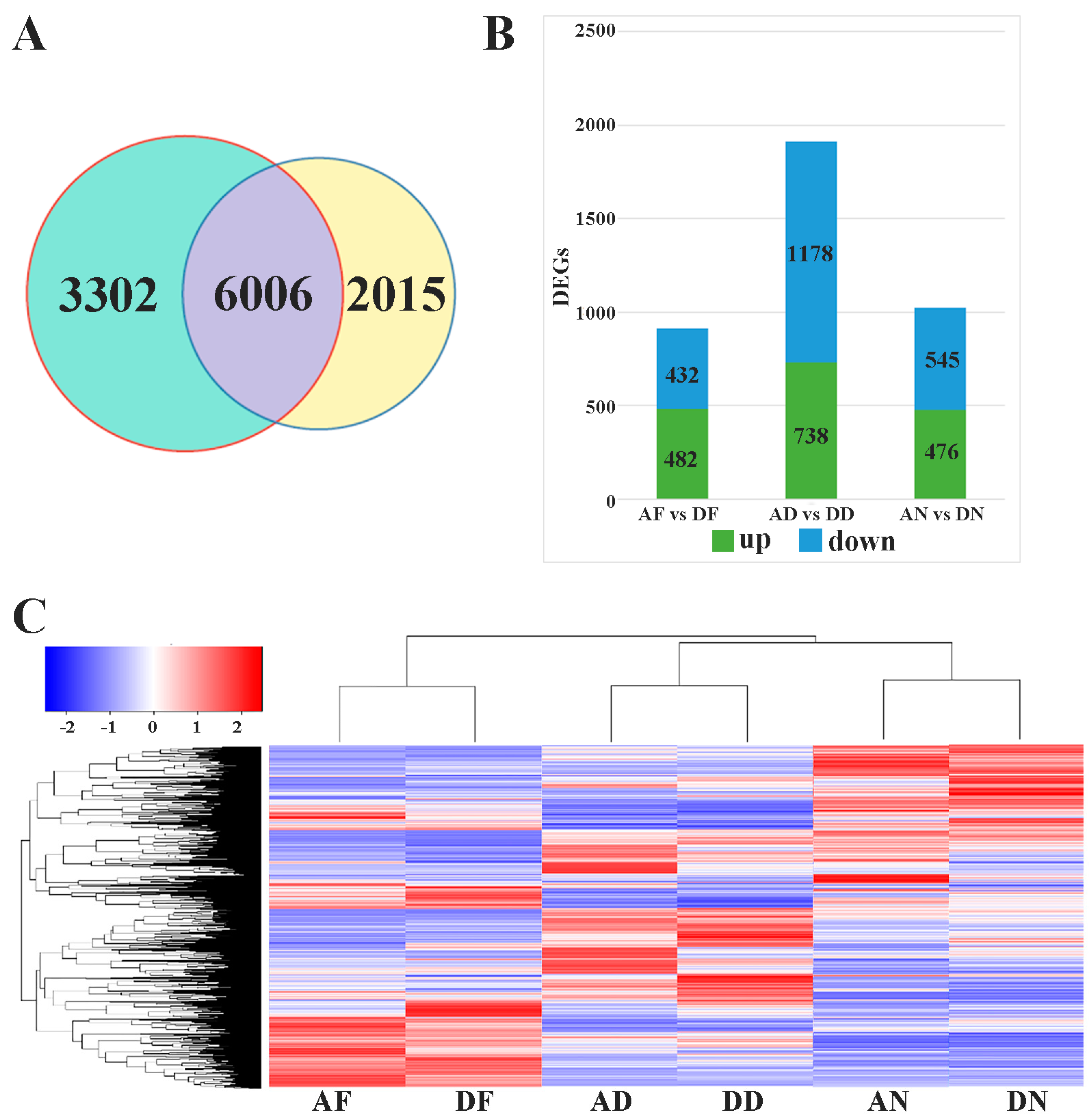
3.1. Transcriptome Sequencing and Data Analysis
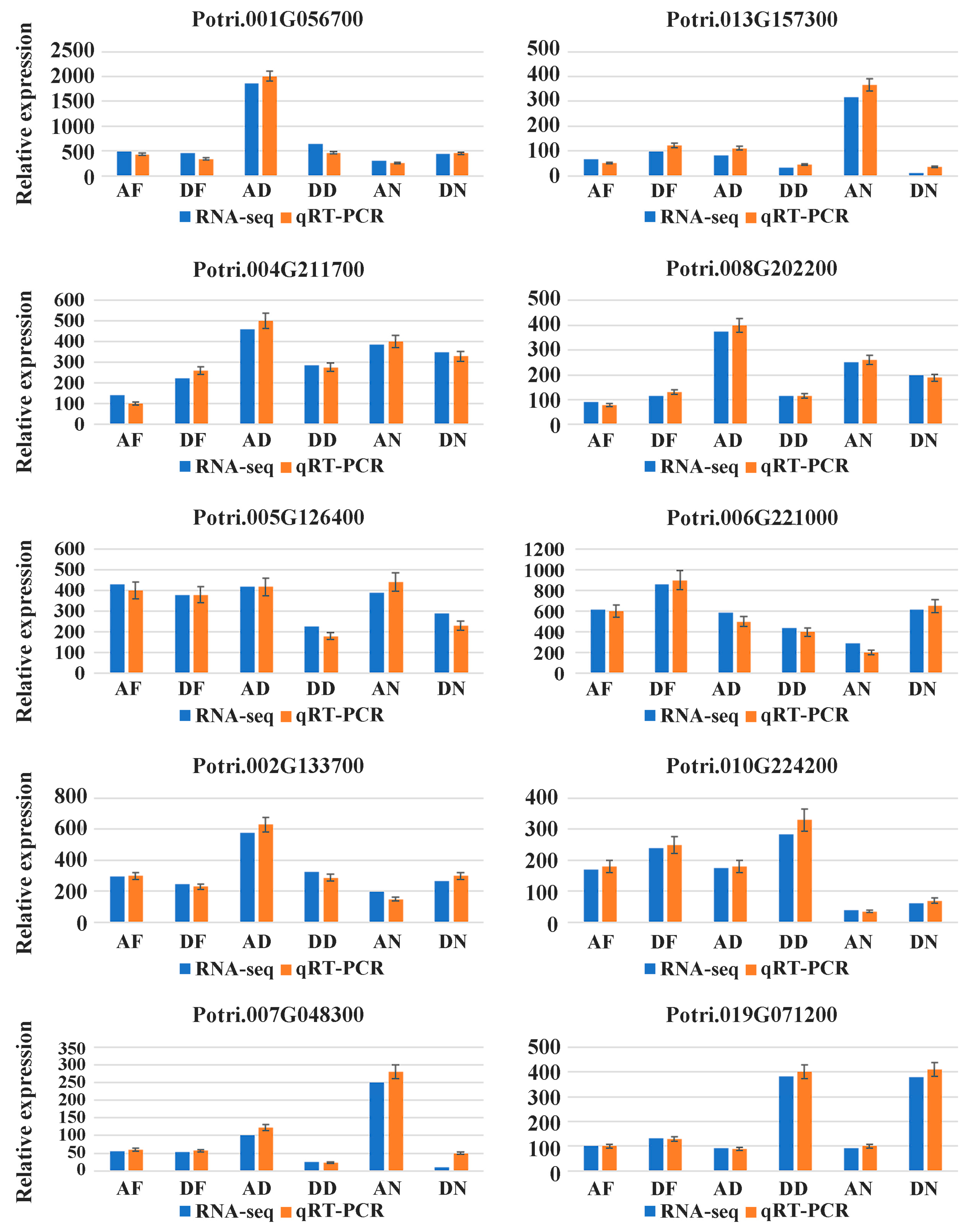
3.2. Differentially Expressed Genes and RT-PCR Validation
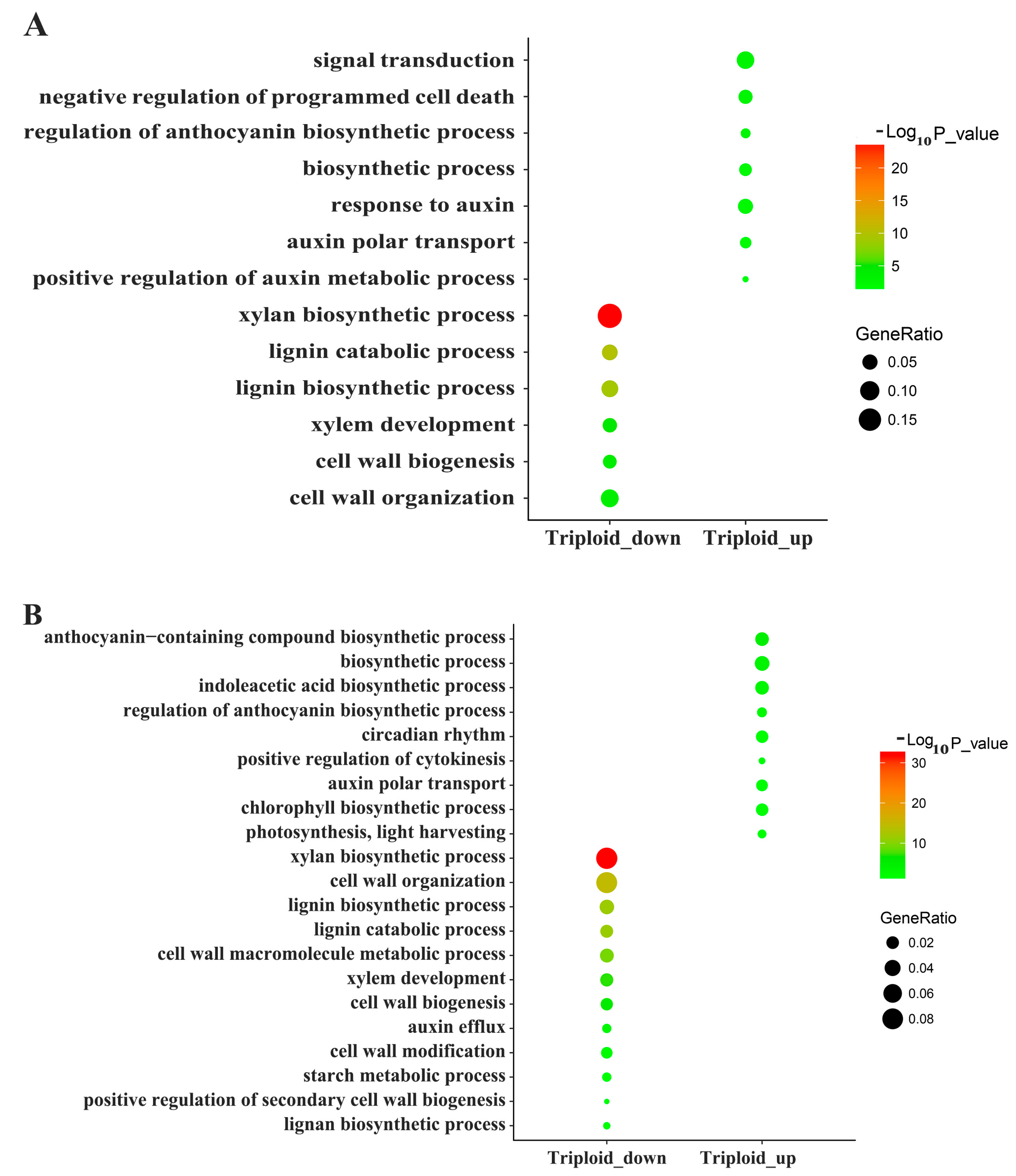
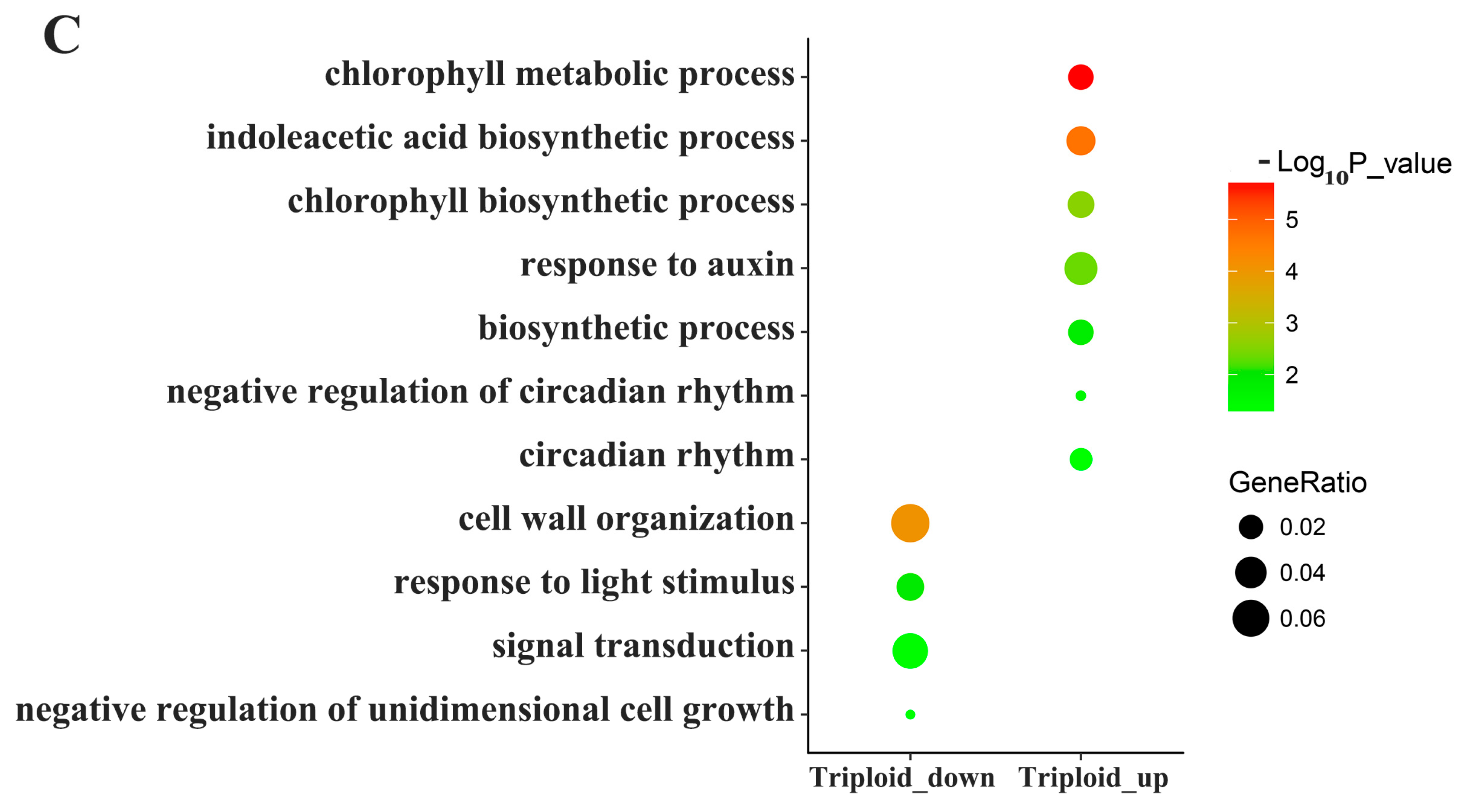
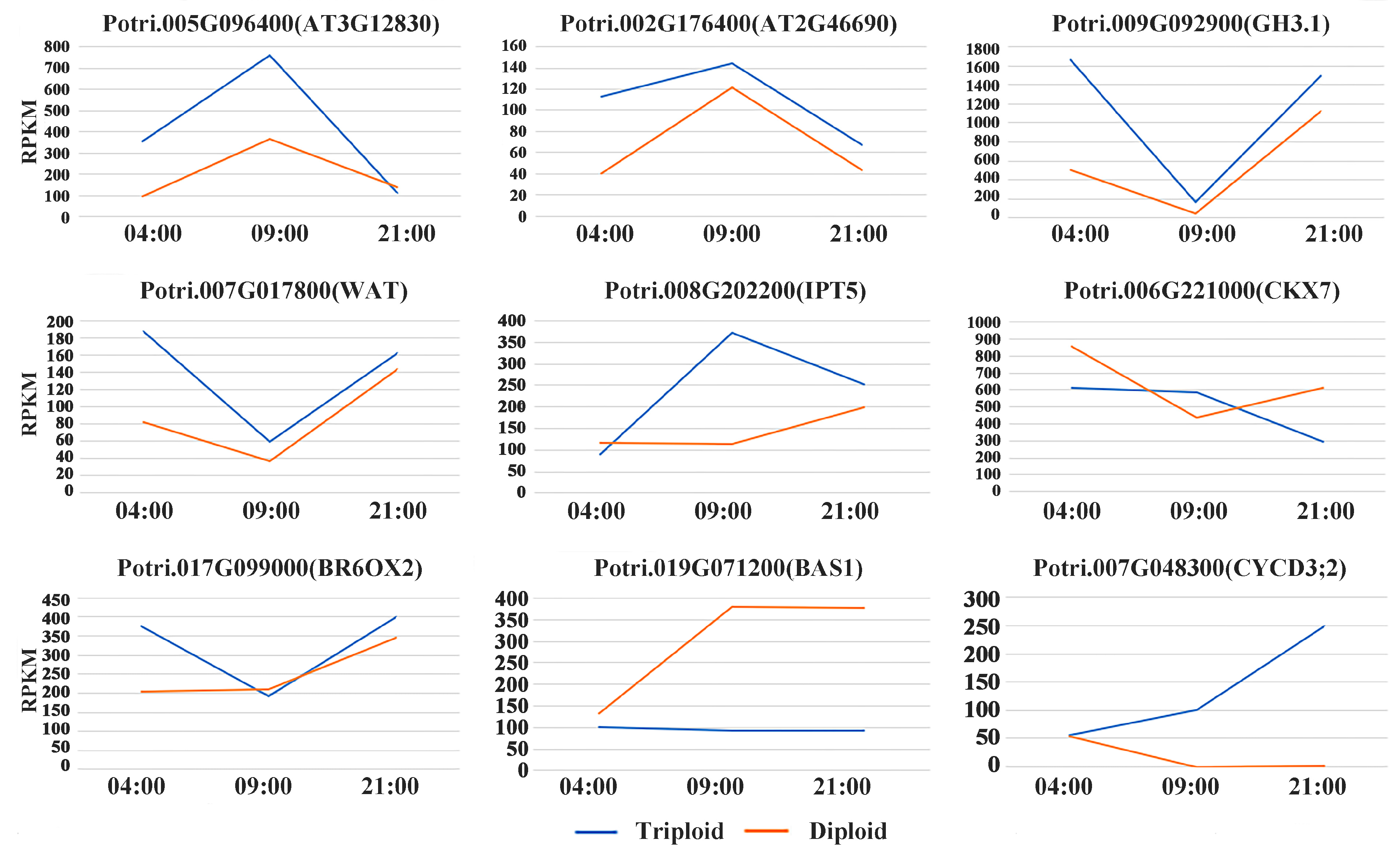
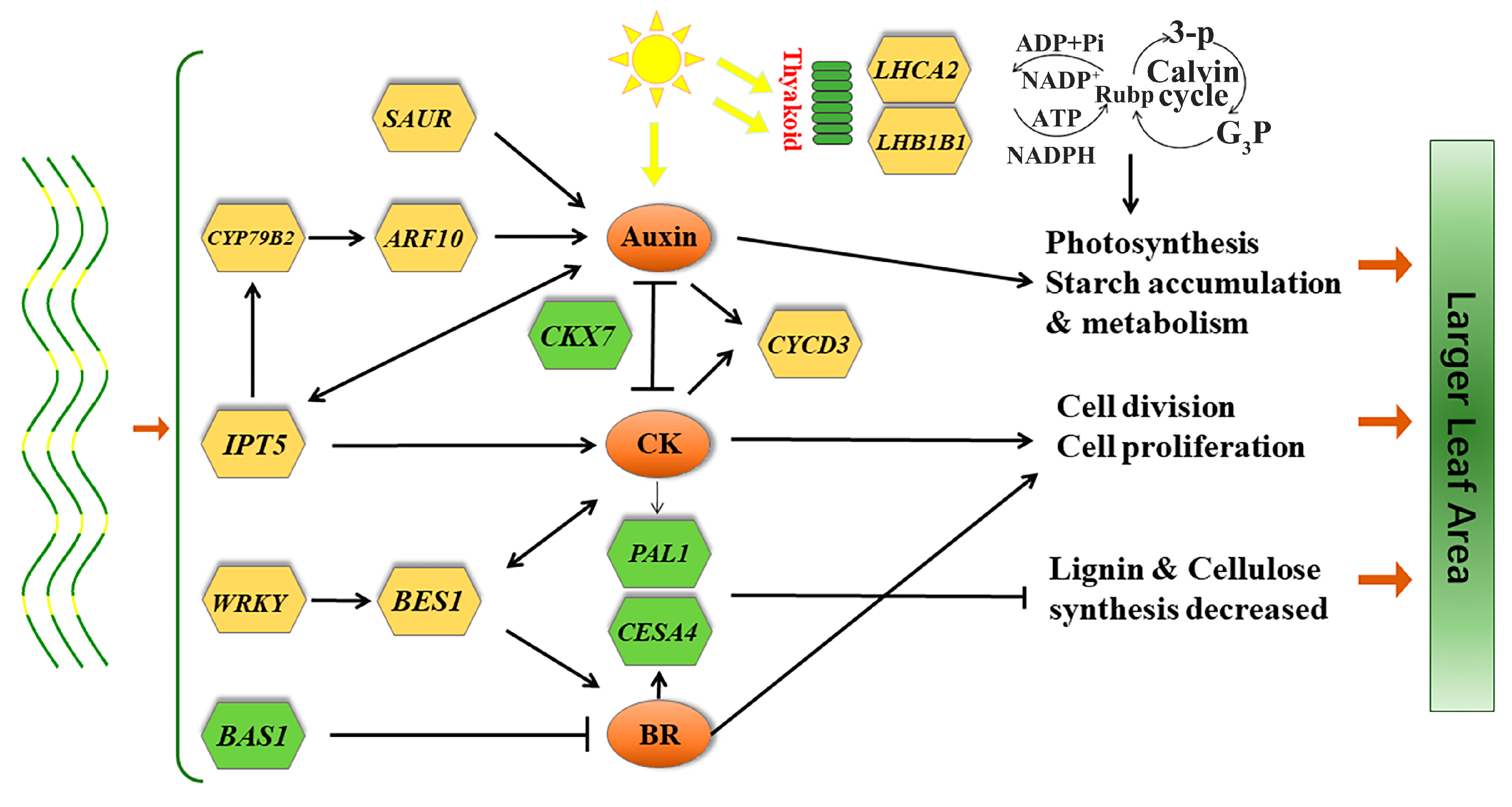
3.3. Differentially Expressed Genes Related to Plant Hormones
3.4. Differentially Expressed Genes Related to Photosynthesis and Energy Metabolism
3.5. Differentially Expressed Genes Related to Lignin and Cellulose
4. Discussion
4.1. Endogenous Hormone-Related DEGs
4.2. Lignin and Cellulose-Related DEGs
4.3. Photosynthesis and Energy Metabolism Related DEGs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Adams, K.L.; Wendel, J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005, 8, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Ann. Rev. Plant Biol. 2007, 58, 377–406. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Kim, E.D.; Ha, M.; Lackey, E.; Liu, J.; Zhang, Y.; Sun, Q.; Chen, Z.J. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 2009, 457, 327–331. [Google Scholar] [CrossRef]
- Miller, M.; Zhang, C.; Chen, Z.J. Ploidy and Hybridity Effects on Growth Vigor and Gene Expression in Arabidopsis thaliana Hybrids and Their Parents. G3 Genes Genomes Genet. 2012, 2, 505–513. [Google Scholar] [CrossRef]
- Wang, Z.; Fan, G.; Dong, Y.; Zhai, X.; Deng, M.; Zhao, Z.; Liu, W.; Cao, Y. Implications of polyploidy events on the phenotype, microstructure, and proteome of Paulownia australis. PLoS ONE 2017, 12, e0172633. [Google Scholar] [CrossRef]
- Sugiyama, S. Polyploidy and cellular mechanisms changing leaf size: Comparison of diploid and autotetraploid populations in two species of Lolium. Ann. Bot. 2005, 96, 931–938. [Google Scholar] [CrossRef]
- Einspahr, D.W. Production and utilization of triploid hybrid aspen [Populus tremuloides, Populus tremula]. Iowa State J. Res. 1984, 58, 401–409. [Google Scholar]
- Zhu, Z.; Kang, X.; Zhang, Z. Studies on selection of natural triploids of Populus tomentosa. Sci. Silvae Sin. 1998, 34, 22–31. [Google Scholar]
- Mu, H.Z.; Liu, Z.J.; Lin, L.; Li, H.Y.; Jiang, J.; Liu, G.F. Transcriptomic Analysis of Phenotypic Changes in Birch (Betula platyphylla) Autotetraploids. Int. J. Mol. Sci. 2012, 13, 13012–13029. [Google Scholar] [CrossRef]
- Zhang, X.; Deng, M.; Fan, G. Differential transcriptome analysis between Paulownia fortunei and its synthesized autopolyploid. Int. J. Mol. Sci. 2014, 15, 5079–5093. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Birchler, J.A. Polyploid and Hybrid Genomics; Wiley-Blackwell: Hoboken, NJ, USA, 2013; pp. 323–333. [Google Scholar]
- Thirulogachandar, V.; Alqudah, A.M.; Koppolu, R.; Rutten, T.; Graner, A.; Hensel, G.; Kumlehn, J.; Brautigam, A.; Sreenivasulu, N.; Schnurbusch, T.; et al. Leaf primordium size specifies leaf width and vein number among row-type classes in barley. Plant J. 2017, 91, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.; Dewitte, W.; Murray, J.A. D-type cyclins control cell division and developmental rate during Arabidopsis seed development. J. Exp. Bot. 2012, 63, 3571–3586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, C.; Ling, C.; Sheng, W.; Yongpeng, L.; Xianzong, S.; Han, L.; Lixia, L.; Zhengli, Z.; Fowke, L.C.; Hong, W.J. Downregulation of multiple CDK inhibitor ICK/KRP genes upregulates the E2F pathway and increases cell proliferation, and organ and seed sizes in Arabidopsis. Plant J. 2013, 75, 642–655. [Google Scholar]
- Zhou, Y.M.; Wang, H.S.; Whitwill, S.; Fowke, L.C. Effects of co-expressing the plant CDK inhibitor ICK1 and D-type cyclin genes on plant growth, cell size and ploidy in Arabidopsis thaliana. Planta 2003, 216, 604–613. [Google Scholar]
- Hu, Y.; Xie, Q.; Chua, N.H. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 2003, 15, 1951–1961. [Google Scholar] [CrossRef]
- Kim, G.T.; Tsukaya, H.; Uchimiya, H.J. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 1998, 12, 2381–2391. [Google Scholar] [CrossRef]
- Li, X.; Wang, L.; Wang, S.; Yang, Q.; Zhou, Q.; Huang, X. A preliminary analysis of the effects of bisphenol A on the plant root growth via changes in endogenous plant hormones. Ecotoxicol. Environ. Saf. 2017, 150, 152. [Google Scholar] [CrossRef]
- Wang, S.; Chen, W.; Yang, C.; Yao, J.; Xiao, W.; Xin, Y.; Qiu, J.; Hu, W.; Yao, H.; Ying, W. Comparative proteomic analysis reveals alterations in development and photosynthesis-related proteins in diploid and triploid rice. BMC Plant Biol. 2016, 16, 199. [Google Scholar] [CrossRef]
- Liao, T.; Cheng, S.; Zhu, X.; Min, Y.; Kang, X. Effects of triploid status on growth, photosynthesis, and leaf area in Populus. Trees 2016, 30, 1137–1147. [Google Scholar] [CrossRef]
- Wang, W.; Meng, M.; Zhang, Y.; Wei, C.; Xie, Y.; Jiang, L.; Wang, C.; Yang, F.; Tang, W.; Jin, X. Global transcriptome-wide analysis of CIK cells identify distinct roles of IL-2 and IL-15 in acquisition of cytotoxic capacity against tumor. BMC Med. Genom. 2014, 7, 49. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Sjödin, A.; Street, N.R.; Sandberg, G.; Gustafsson, P.; Jansson, S. The Populus Genome Integrative Explorer (PopGenIE): a new resource for exploring the Populus genome. New Phytol. 2009, 182, 1013–1025. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J.; Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef] [Green Version]
- Draghici, S.; Khatri, P.; Tarca, A.L.; Amin, K.; Done, A.; Voichita, C.; Georgescu, C.; Romero, R. A systems biology approach for pathway level analysis. Genome Res. 2007, 17, 1537–1545. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Santner, A.; Calderonvillalobos, L.I.; Estelle, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 2009, 5, 301–307. [Google Scholar] [CrossRef]
- Rademacher, W. Plant Growth Regulators: Backgrounds and Uses in Plant Production. J. Plant Growth Regul. 2015, 34, 845–872. [Google Scholar] [CrossRef]
- Mroue, S.; Simeunovic, A.; Robert, H.S. Auxin production as an integrator of environmental cues for developmental growth regulation. J. Exp. Bot. 2018, 69, 201. [Google Scholar] [CrossRef]
- Saini, S.; Sharma, I.; Kaur, N.; Pati, P.K. Auxin: A master regulator in plant root development. Plant Cell Rep. 2013, 32, 741–757. [Google Scholar] [CrossRef]
- Hull, A.K.; Vij, R.; Celenza, J.L. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 2379–2384. [Google Scholar] [CrossRef]
- Zheng, H.; Pan, X.; Deng, Y.; Wu, H.; Liu, P.; Li, X.J. AtOPR3 specifically inhibits primary root growth in Arabidopsis under phosphate deficiency. Sci. Rep. 2016, 6, 24778. [Google Scholar] [CrossRef]
- Ranocha, P.; Dima, O.; Nagy, R.; Felten, J.; Corratge-Faillie, C.; Novak, O.; Morreel, K.; Lacombe, B.; Martinez, Y.; Pfrunder, S.; et al. Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 2013, 4, 2625. [Google Scholar] [CrossRef]
- Qiao, L.; Zhang, W.; Li, X.; Zhang, L.; Zhang, X.; Li, X.; Guo, H.; Ren, Y.; Zheng, J.; Chang, Z. Characterization and Expression Patterns of Auxin Response Factors in Wheat. Front. Plant Sci. 2018, 9, 1395. [Google Scholar] [CrossRef]
- Van, H.M.; Van, A.D.; Stortenbeker, N.; Angenent, G.C.; Bemer, M. Divergent regulation of ArabidopsisSAURgenes: A focus on theSAUR10-clade. BMC Plant Biol. 2017, 17, 245. [Google Scholar]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef] [Green Version]
- Akiyoshi, D.E.; Klee, H.; Amasino, R.M.; Nester, E.W.; Gordon, M.P. T-DNA of Agrobacterium tumefaciens encodes an enzyme of cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 1984, 81, 5994–5998. [Google Scholar] [CrossRef]
- Gan, S.; Amasino, R.M. Inhibition of Leaf Senescence by Autoregulated Production of Cytokinin. Science 1995, 270, 1986–1988. [Google Scholar] [CrossRef]
- Zeng, C.J.; Lee, Y.R.; Liu, B. The WD40 repeat protein NEDD1 functions in microtubule organization during cell division in Arabidopsis thaliana. Plant Cell 2009, 21, 1129–1140. [Google Scholar] [CrossRef]
- Kieffer, M.; Master, V.; Waites, R.; Davies, B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 2011, 68, 147–158. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Gan, S. AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant. Physiol. 2011, 156, 1612–1619. [Google Scholar] [CrossRef]
- Köllmer, I.; Novák, O.; Strnad, M.; Schmülling, T.; Werner, T. Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. 2014, 78, 359. [Google Scholar] [CrossRef]
- Khripach, V.; Zhabinskii, V.; Groot, A.D. Twenty Years of Brassinosteroids: Steroidal Plant Hormones Warrant Better Crops for the XXI Century. Ann. Bot. 2000, 86, 441–447. [Google Scholar] [CrossRef] [Green Version]
- Sairam, R.K. Effects of homobrassinolide application on plant metabolism and grain yield under irrigated and moisture-stress conditions of two wheat varieties. Plant Growth Regul. 1994, 14, 173–181. [Google Scholar] [CrossRef]
- Chen, J.; Nolan, T.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54 and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Response. Plant Cell 2017, 29, 1425. [Google Scholar] [CrossRef]
- Neff, M.M.; Nguyen, S.M.; Malancharuvil, E.J.; Fujioka, S.; Noguchi, T.; Seto, H.; Tsubuki, M.; Honda, T.; Takatsuto, S.; Yoshida, S. BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA 1999, 96, 15316–15323. [Google Scholar] [CrossRef]
- Katsumata, T.; Hasegawa, A.; Fujiwara, T.; Komatsu, T.; Notomi, M.; Abe, H.; Natsume, M.; Kawaide, H. Arabidopsis CYP85A2 catalyzes lactonization reactions in the biosynthesis of 2-deoxy-7-oxalactone brassinosteroids. J. Agric. Chem. Soc. Jpn. 2008, 72, 2110–2117. [Google Scholar] [CrossRef]
- Chang, S.C.; Takatsuto, S. Arabidopsis CYP85A2, a Cytochrome P450, Mediates the Baeyer-Villiger Oxidation of Castasterone to Brassinolide in Brassinosteroid Biosynthesis. Plant Cell 2005, 17, 2397–2412. [Google Scholar]
- Tanaka, K.; Okamoto, S. Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Phys. 2005, 138, 1117. [Google Scholar] [CrossRef]
- Zhiponova, M.K.; Vanhoutte, I.; Boudolf, V.; Betti, C.; Dhondt, S.; Coppens, F.; Mylle, E.; Maes, S.; Gonzalez-Garcia, M.P.; Cano-Delgado, A.I.; et al. Brassinosteroid production and signaling differentially control cell division and expansion in the leaf. New Phytol. 2013, 197, 490–502. [Google Scholar] [CrossRef]
- Dewitte, W.; Scofield, S.; Alcasabas, A.A.; Maughan, S.C.; Menges, M.; Braun, N.; Collins, C.; Nieuwland, J.; Prinsen, E.; Sundaresan, V.; et al. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 2007, 104, 14537–14542. [Google Scholar] [CrossRef]
- Machakova, I.; Zazimalova, E.; George, E.F.; George, E.F.; Hall, M.A.; Klerk, G.J.D. Plant growth regulators I: Introduction; auxins, their analogues and inhibitors. In Plant Propagation by Tissue Culture; Springer: Medford, MA, USA, 2008; pp. 175–204. [Google Scholar]
- Hu, Y.; Bao, F.; Li, J. Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J. 2000, 24, 693–701. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, B.; Qi, S.; Dong, M.; Wang, Z.; Li, Y.; Chen, S.; Li, B.; Zhang, J. Ploidy and hybridity effects on leaf size, cell size and related genes expression in triploids, diploids and their parents in Populus. Planta 2018, 249, 635–646. [Google Scholar] [CrossRef]
- Hancock, J.E.; Loya, W.M.; Giardina, C.P.; Li, L.; Chiang, V.L.; Pregitzer, K.S. Plant growth, biomass partitioning and soil carbon formation in response to altered lignin biosynthesis in Populus tremuloides. New Phytol. 2007, 173, 732–742. [Google Scholar] [CrossRef]
- Humphreys, J.M.; Chapple, C. Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 2002, 5, 224–229. [Google Scholar] [CrossRef]
- Yang, C.; Xu, Z.; Song, J.; Conner, K.; Barrena, G.V.; Wilson, Z.A. Arabidopsis MYB26/MALE STERILE35 Regulates Secondary Thickening in the Endothecium and Is Essential for Anther Dehiscence. Plant Cell 2007, 19, 534–548. [Google Scholar] [CrossRef]
- Ko, J.H.; Yang, S.H.; Park, A.H.; Lerouxel, O.; Han, K.H. ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J. 2010, 50, 1035–1048. [Google Scholar] [CrossRef]
- Hahlbrock, K.; Scheel, D. Physiology and Molecular Biology of Phenylpropanoid Metabolism. Ann. Rev. Plant Physiol. Plant Mol. Biol 1989, 40, 347–369. [Google Scholar] [CrossRef]
- Hu, W.J.; Kawaoka, A.; Tsai, C.J.; Lung, J.; Osakabe, K.; Ebinuma, H.; Chiang, V.L. Compartmentalized expression of two structurally and functionally distinct 4-coumarate:CoA ligase genes in aspen (Populus tremuloides). Proc. Natl. Acad. Sci. USA 1998, 95, 5407–5412. [Google Scholar] [CrossRef]
- Hu, W.J.; Harding, S.A.; Jrhau, L.; Popko, J.L.; Ralph, J.; Stokke, D.D.; Chungjui, T.; Chiang, V.L. Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat. Biotechnol. 1999, 17, 808–812. [Google Scholar] [CrossRef] [Green Version]
- Rohde, A.; Morreel, K.; Ralph, J.; Goeminne, G.; Hostyn, V.; Rycke, R.D.; Kushnir, S.; Doorsselaere, J.V.; Joseleau, J.P.; Vuylsteke, M. Molecular Phenotyping of the pal1 and pal2 Mutants of Arabidopsis thaliana Reveals Far-Reaching Consequences on Phenylpropanoid, Amino Acid, and Carbohydrate Metabolism. Plant Cell 2004, 16, 2749–2771. [Google Scholar] [CrossRef]
- Olsen, K.M.; Lea, U.S.; Slimestad, R.; Verheul, M.; Lillo, C. Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J. Plant Physiol. 2008, 165, 1491–1499. [Google Scholar] [CrossRef]
- Meyermans, H.; Morreel, K.; Lapierre, C.; Pollet, B.; Bruyn, A.D.; Busson, R.; Herdewijn, P.; Devreese, B.; Beeumen, J.V.; Marita, J.M. Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J. Biol. Chem. 2000, 275, 36899. [Google Scholar] [CrossRef]
- Zhong, R.; Iii, W.H.; Negrel, J.; Ye, Z.H. Dual methylation pathways in lignin biosynthesis. Plant Cell 1998, 10, 2033–2045. [Google Scholar] [CrossRef]
- Feng, W.; Zhang, P.; Pei, J.; Kang, X. Genotypic parameters of wood density and fiber traits in triploid hybrid clones of Populus tomentosa at five clonal trials. Ann. For. Sci. 2013, 70, 751–759. [Google Scholar]
- Kim, C.; Apel, K. Arabidopsis light-dependent NADPH: Protochlorophyllide oxidoreductase A (PORA) is essential for normal plant growth and development: An addendum. Plant Mol. Biol. 2012, 80, 237–240. [Google Scholar] [CrossRef]
- Wientjes, E.; Croce, R. The light-harvesting complexes of higher-plant Photosystem I: Lhca1/4 and Lhca2/3 form two red-emitting heterodimers. Biochem. J. 2011, 433, 477–485. [Google Scholar] [CrossRef] [Green Version]
- Ytterberg, A.J.; Peltier, J.B.; van Wijk, K.J. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 2006, 140, 984–997. [Google Scholar] [CrossRef]
| Gene Name | AF | DF | Transcription Factors | Description |
|---|---|---|---|---|
| up | ||||
| Potri.005G098200 | 27.234 | 3.46619 | NAC1 | NAC(transcription factor NAC) domain containing protein 1 |
| Potri.004G181900 | 545.664 | 218.295 | NAC036 | NAC domain containing protein 36 |
| Potri.009G141600 | 736.5 | 296.549 | NAC036 | NAC domain containing protein 36 |
| Potri.016G137900 | 3589.25 | 1505.05 | WRKY70 | WRKY(transcription factor WRKY) DNA-binding protein 70 |
| Potri.006G109100 | 3168.18 | 1614.5 | WRKY70 | WRKY DNA-binding protein 70 |
| Potri.002G168700 | 107.436 | 58.9119 | WRKY46 | WRKY DNA-binding protein 46 |
| down | ||||
| Potri.T044100 | 101.378 | 207.353 | TCP20 | TEOSINTE BRANCHED 1, TCP (transcription factor TCP)-domain family protein 20 |
| Potri.011G153300 | 100.003 | 219.598 | NAC012 | NAC domain containing protein 12 |
| Gene Name | AD | DD | Transcription Factors | Description | |
|---|---|---|---|---|---|
| up | |||||
| Potri.004G211700 | 462.101 | 284.943 | ARF10 | auxin response factor 10 | |
| Potri.005G126400 | 418.002 | 226.958 | BZR1 | Brassinosteroid signaling positive regulator (BZR1) family protein | |
| Potri.002G133700 | 576.321 | 326.778 | BZR1 | Brassinosteroid signaling positive regulator (BZR1) family protein | |
| Potri.002G142800 | 75.7639 | 42.1725 | GATA2 | GATA(transcription factor GATA) transcription factor 2 | |
| Potri.009G123400 | 588.683 | 357.448 | GATA5 | GATA transcription factor 5 | |
| Potri.005G116800 | 30.0181 | 4.70138 | VND1 | Vascular related NAC-domain protein 1 | |
| Potri.002G168700 | 379.122 | 94.068 | WRKY46 | WRKY DNA-binding protein 46 | |
| Potri.006G109100 | 3542.13 | 1256.65 | WRKY70 | WRKY DNA-binding protein 70 | |
| down | |||||
| Potri.003G050100 | 48.6193 | 101.898 | ATHB-15 | Homeobox-leucine zipper family protein | |
| Potri.005G182900 | 76.5924 | 143.312 | AT1G21580 | Zinc finger C-x8-C-x5-C-x3-H type family protein | |
| Potri.004G095100 | 2.89458 | 20.2711 | AT1G66810 | Zinc finger C-x8-C-x5-C-x3-H type family protein | |
| Potri.010G118700 | 0.3343 | 22.2819 | AT1G68200 | Zinc finger C-x8-C-x5-C-x3-H type family protein | |
| Potri.001G079800 | 41.3335 | 86.3043 | AT5G51190 | Integrase-type DNA-binding superfamily protein | |
| Gene Name | AN | DN | Transcription Factors | Description | |
|---|---|---|---|---|---|
| up | |||||
| Potri.005G236700 | 121.671 | 50.9552 | MP | Transcriptional factor B3 family protein/auxin-responsive factor AUX/IAA-related | |
| Potri.018G029500 | 182.423 | 103.379 | HY5 | Basic-leucine zipper (bZIP) transcription factor family proteisn | |
| Potri.014G117000 | 25.0831 | 1.97366 | MYB2 | Myb domain protein 2 | |
| Potri.002G180800 | 397.419 | 111.409 | LHY | Homeodomain-like superfamily protein | |
| Potri.005G116800 | 27.7685 | 4.70297 | VND1 | Vascular related NAC-domain protein 1 | |
| Potri.014G108100 | 334.082 | 153.051 | TIP | TCV-interacting protein | |
| Potri.002G168700 | 294.105 | 125.42 | WRKY46 | WRKY DNA-binding protein 46 | |
| down | |||||
| Potri.019G112000 | 207.012 | 368.607 | AT1G05710 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein | |
| Potri.008G161800 | 234.024 | 436.951 | BHLH92 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein | |
| Potri.003G139300 | 180.925 | 325.849 | AT1G64380 | Integrase-type DNA-binding superfamily protein | |
| Potri.001G092400 | 86.1233 | 159.747 | AT1G64380 | Integrase-type DNA-binding superfamily protein | |
| Potri.007G076800 | 81.6476 | 160.502 | ABR1 | Integrase-type DNA-binding superfamily protein | |
| Potri.002G142800 | 134.963 | 291.083 | GATA2 | GATA transcription factor 2 | |
| Potri.004G078800 | 472.198 | 830.782 | SCL8 | SCARECROW-like 8 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, K.; Han, Q.; Zhang, Y.; Kang, X. Differential Expression of Genes Related to the Formation of Giant Leaves in Triploid Poplar. Forests 2019, 10, 920. https://doi.org/10.3390/f10100920
Du K, Han Q, Zhang Y, Kang X. Differential Expression of Genes Related to the Formation of Giant Leaves in Triploid Poplar. Forests. 2019; 10(10):920. https://doi.org/10.3390/f10100920
Chicago/Turabian StyleDu, Kang, Qiang Han, Ying Zhang, and Xiangyang Kang. 2019. "Differential Expression of Genes Related to the Formation of Giant Leaves in Triploid Poplar" Forests 10, no. 10: 920. https://doi.org/10.3390/f10100920
APA StyleDu, K., Han, Q., Zhang, Y., & Kang, X. (2019). Differential Expression of Genes Related to the Formation of Giant Leaves in Triploid Poplar. Forests, 10(10), 920. https://doi.org/10.3390/f10100920





