Abstract
Cinnamomum camphora is a valuable broad-leaf tree indigenous to South China and East Asia and has been widely cultivated and utilized by humans since ancient times. However, owing to its overutilization for essential oil extraction, the Transplanting Big Trees into Cities Program, and over deforestation to make furniture, its wild populations have been detrimentally affected and are declining rapidly. In the present study, the genetic diversity and population structure of 180 trees sampled from 41 populations in South China were investigated with 22 expressed sequence tag-simple sequence repeat (EST-SSR) markers. In total, 61 alleles were harbored across 180 individuals, and medium genetic diversity level was inferred from the observed heterozygosity (Ho), expected heterozygosity (He), and Nei’ gene diversity (GD), which were 0.45, 0.44, and 0.44, respectively. Among the 41 wild populations, C. camphora had an average of 44 alleles, 2.02 effective alleles, and He ranging from 0.30 (SC) to 0.61 (HK). Analysis of molecular variance (AMOVA) showed that 17% of the variation among populations and the average pairwise genetic differentiation coefficient (FST) between populations was 0.162, indicating relatively low genetic population differentiations. Structure analysis suggested two groups for the 180 individuals, which was consistent with the principal coordinate analysis (PCoA) and unweighted pair-group method with arithmetic means (UPGMA). Populations grouped to cluster I were nearly all distributed in Jiangxi Province (except population XS in Zhejiang Province), and cluster II mainly comprised populations from other regions, indicating a significant geographical distribution. Moreover, the Mantel test showed that this geographical distance was significantly correlated with genetic distance. The findings of this research will assist in future C. camphora conservation management and breeding programs.
1. Introduction
The Cinnamomum L. (Lauraceae) genus of tree species is ecologically and economically important and includes approximately 250 species that are widely distributed in the tropical and subtropical regions of Asia and South America [1,2]. Most species have long been utilized as aromatic medicinal plants throughout human history because of their essential oils. Among these species, Cinnamomum camphora is a broad-leaf tree species indigenous to South China, including Fujian, Zhejiang, Guangdong, Hubei, Guizhou, and especially Jiangxi Provinces [3,4], and East Asia [5]. It has also been introduced into a number of countries—including Australia, southeastern France, Madagascar, the Canary Islands, and the United States [6,7,8,9]. It is one of the most valuable arbor species in Asia and has been widely cultivated and utilized by humans since ancient times [10]. C. camphora can be used to make solid wood flooring boards, high-grade furniture, handicrafts, etc. because of its brownish-yellow wood, yellowish sapwood, and fragrant smell [11]. It is one of the top four most valuable trees for timber production in China, and camphor timber was too expensive for people to use for building whole houses in the past. In addition, it has long been prescribed in traditional medicine in India [12], Korea [13], and China [14]. The fruits, wood, roots, bark, and leaves have been used in traditional herbal medicine as dietary supplements, perfume, health care products, and incense since the Han Dynasty [11]. Moreover, this species has important ornamental values because of its good structure, large canopy, evergreen color, fragrance, and elegance. It has been widely used in landscape architecture in recent decades in China [15]. Furthermore, camphor trees have significant cultural and religious values to Chinese people because of their longevity. Many heritage trees were discovered in the so-called ‘geomantic forest’ in addition to temples or villages, and some ancient trees were planted 2000 years ago. For example, three camphor trees were cultivated during the Han Dynasty (BC 202–AD 220) in Anfu county, Jiangxi Province [15]. Moreover, 2 provinces and 36 cities selected C. camphora as the city tree, which include 172.81 million people in South China [11].
However, owing to over deforestation and overutilization for essential oil extraction in the past half century [4,16], the Transplanting Big Trees (defined as over 100 cm in diameter at breast height, DBH) into Cities Program in South China during recent decades, and illegal logging to make furniture, wild populations of C. camphora have been detrimentally affected and are declining rapidly; currently, the camphor tree is listed in “China Species Red List” [17]. Hence, there is an urgent need to carry out research on its conservation systematically and strengthen the conservation of its natural habitat.
In previous studies, Yao et al. studied phenotypic variation in the seedling bud germination of C. camphora in South China [18]. Li et al. [19] developed 21 simple sequence repeat (SSR) primers from the transcriptome and investigated the genetic diversity of three Chinese Cinnamomum populations and the cross-species transferability of six other related species in Cinnamomum. Kameyama et al. [20] developed 22 pairs of SSR primers and investigated the genetic diversity of three C. camphora populations in Japan. Furthermore, 11 primers were used to investigate the genetic differentiation among the 6 natural populations from Japan and China and how geographical isolation and human activity mediated the gene flow between these populations [16]. However, no studies have reported the genetic diversity and population structure of this species in South China.
Molecular markers are often used to investigate the genetic diversity and population genetic structure in tree species, especially for those species with less genetic information [21,22,23,24,25]. SSR markers are the ideal choices for wild tree populations because of their high variability and codominance [26,27]. In the present study, we aimed to evaluate the genetic diversity of wild C. camphora populations, elucidate the phylogenic relationships of this species, and determine the spatiotemporal impact of natural factors on its genetic composition. These findings will be useful for revealing the history of C. camphora and assisting in future C. camphora conservation and breeding management.
2. Materials and Methods
2.1. Plant Material and DNA Isolation
In total, 180 wild individuals representing 41 C. camphora populations covering its whole native distribution in South China were sampled (Figure 1, Table 1). Only trees with a DBH larger than 30 cm were included and sampled randomly, ensuring the distance between them was above 30 m. Detailed information on sampling location, site name, and DBH is summarized in Table S1. Five leaves were collected from each individual and dried with silica gel. Total genomic DNA was isolated using the Hi-DNAsecure Plant Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. The quality and concentration of DNA were determined using a NanoDrop 2000 (Thermo Fisher Scientific Inc., Waltham, MA, USA).
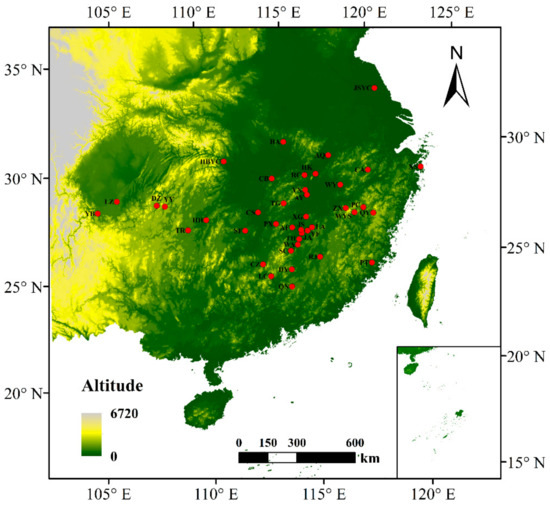
Figure 1.
Geographical distribution of the plant materials. Solid dots indicate the populations that were collected in South China.

Table 1.
Geographical location, diameter at breast height (DBH), and genetic diversity for 41 Cinnamomum camphora populations in South China.
2.2. SSR Development, Identification, and Analysis
To develop SSRs, young camphor leaves were used for RNA sequencing [28], and then expressed sequence tag (EST)-SSR loci were identified using the MISA search module. Of the identified SSRs, 100 SSR loci were selected for primer pair design [19]. Subsequently, DNA from four samples (randomly selected from 180 individuals) was used to validate the selected 100 EST-SSR primers. The PCR conditions were as follows: an initial denaturation at 94 °C for 5 min; followed by 30 cycles of 30 s at 94 °C, 30 s at 59 °C, and 30 s at 72 °C; and a final extension 3 min at 72 °C. A typical 10 μL reaction included the following reagents: 1 × PCR buffer, 0.75 mM MgCl2, 0.1 mM dNTPs, 0.25 U of Taq DNA polymerase, 0.4 μM each primer, and 10–25 ng genomic DNA. The PCR products were resolved on 8% denaturing polyacrylamide gels, which were silver-stained to detect the SSR bands. To estimate allele sizes, the lengths of the bands were compared with a 50 bp DNA ladder.
2.3. Statistical Analysis
POPGENE 1.31 (University of Alberta, Edmonton, Canada) [29] was used to evaluate the following genetic diversity parameters: allele frequency; number of alleles (Na) and effective number of alleles (Ne) per locus; observed (Ho) and expected (He) heterozygosity per locus; Nei’s gene diversity index (GD); the percentage of polymorphic loci (PPB); Wright’s (1978) fixation index (F); and gene flow (Nm); and then, the deviation from Hardy–Weinberg equilibrium (HWE) was investigated using the chi-squared test for each population. The polymorphic information content (PIC) was calculated for each loci using the online program PICcalc [30]. F-statistics, including the genetic differentiation coefficient among populations (FST), inbreeding coefficient among individuals (FIS), and pairwise FST and hierarchical analyses of molecular variance (AMOVA), were assessed by GenAlEx version 6.5 [31]. The genetic structure of the C. camphora populations was investigated using STRUCTURE 2.0 [32]. To evaluate the discontinuous group (K) numbers, the K number was set between 1 and 41 with 10 replicates, and the length of the burn-in period was set to 100,000 times. Then, the best K numbers were selected according to the principle of the highest value of DK by STRUCTURE Harvester online (http:taylor0.biology.ucla.edu/struct_harvest/) [33]. Repeated sampling analysis and genetic structural plots were analyzed by CLUMPP 1.1.2 (Stanford University, Stanford, CA, USA) [34]. Population clustering based on GD by the unweighted pair-group method with arithmetic means (UPGMA) method was performed with NTSYS 2.1 [35]. To summarize the patterns of variation in the multilocus dataset, principal coordinate analysis (PCoA) was performed by GenAlEx version 6.5 based on the pairwise FST matrix. The isolation-by-distance pattern (IBD) was detected by Mantel tests [36] with 1000 permutations based on matrices of pairwise genetic distance (FST/1 − FST) and geographic distance among populations performed in GenAlEx 6.5.
3. Results
3.1. Development of Polymorphic EST-SSR Markers
We selected 100 SSR loci to design primers and to test the specificity of the amplification from four samples and the native information of these SSR markers. Of these, 65 primer pairs (65%) amplified clear bands with the expected size of 100–300 bp, while 35 pairs of primers did not produce either any products or expected products. Furthermore, we randomly selected 30 primers for polymorphism detection, and 22 (73.33%) showed polymorphism (Table 2). Among these polymorphic SSR loci, nine (40.91%) were located in 5’-untranslated regions (5’-UTR), three (13.64%) were located in 3’-untranslated regions (3’-UTR), and only one (4.55%) was located in coding sequences.

Table 2.
Characteristics of 22 EST-SSR markers developed for C. camphora.
3.2. Polymorphism of 22 SSR Loci
In total, 61 alleles were amplified by 22 SSR loci among 180 C. camphora individuals (Table S1), and the Na detected per locus ranged from two to four, with an allele frequency range of 0.011–0.991 (Table 3, Table S2). The loci CcSSR3 and CcSSR13 both harbored the largest Na (four), while the locus CcSSR3 harbored the largest Ne (3.60), He (0.72), GD (0.72), and PIC (0.72). However, the locus CcSSR13 showed lower than average Ne, He, GD, and PIC values, which were 1.64, 0.39, 0.39, and 0.48, respectively. In terms of PIC values, seven loci (CcSSR3, CcSSR11, CcSSR18, CcSSR1, CcSSR12, CcSSR14, and CcSSR5) had highly informative alleles with values higher than 0.50, while only CcSSR2 had less informative alleles with a value less than 0.25; the remaining 14 loci had moderately informative alleles with PIC values between 0.25–0.50. The average FST, Nm, and F were 0.22, 1.20, and −0.41, respectively. Furthermore, the HWE test across the 180 C. camphora individuals showed significant deviations for many EST-SSR loci; only four loci (CcSSR2, CcSSR9, CcSSR20, and CcSSR21) revealed no significant deviations (Table 3), indicating that almost all populations were affected by factors of interference, such as selection by migration, mutation, and introgression.

Table 3.
Statistics diversity of the 22 EST-SSR loci across 180 C. camphora individuals.
3.3. Genetic Diversity in C. camphora
Among the 41 wild C. camphora populations investigated, the PPB ranged from 59.09% to 95.45% (Table 1). Presenting the largest PPB, the RJ population also had the largest Na (50), whereas the YX population had the fewest alleles (39), with the smallest PPB. The Ho and He ranged from 0.30 (SC) to 0.61 (HK) and 0.31 (JSYC) to 0.48 (HK), with an average of 0.45 and 0.40, respectively. Additionally, the GD ranged from 0.27 (JSYC) to 0.42 (HK), and the FIS ranged from −0.56 (YX) to 0.10 (SC). Population HK, which exhibited that the largest GD, also possessed the highest Ho and He. At the species level, C. camphora exhibited relatively high genetic diversity, and the GD, Ho and He were 0.44, 0.45, and 0.44, respectively (Table 1). Both AMOVA and pairwise FST analysis were conducted to investigate the genetic variations among these populations and groups. Hierarchical AMOVA revealed that 17% of the total genetic variation occurred among three populations, and 83% of the total variation was distributed within the populations (Table 4). This was also confirmed by the small overall FST and large overall Nm, which were 0.22 and 1.20, respectively (Table 3). Moreover, the pairwise FST varied from 0.028 to 0.439 (Table S3). The highest level appeared between populations JA and YY, whereas the lowest level appeared between populations YX and DY.

Table 4.
Analysis of molecular variance (AMOVA) for 180 individuals in 41 populations of C. camphora.
3.4. Population Genetic Structure of C. camphora
AMOVA revealed a strong population genetic structure at the species level (FIS = −0.207, p < 0.001). We also used STRUCTURE software to analyze the genetic structure of C. camphora populations. The result showed that ΔK was the highest when K = 2 (Figure 2A), indicating that the 41 C. camphora populations could be divided into 2 groups (Figure 2B). The proportion of cluster members of each individual from 41 populations is shown in Figure 2B. Individuals with a proportion higher than 0.75 were considered pure, and those with a score lower than 0.75 were considered admixed. In the first group, the red cluster included 86 individuals with 82 pure and 4 admixed individuals, while the green cluster included 94 individuals, with 88 pure and 6 admixed. Nearly all populations from Jiangxi and XS—except the RJ, JS, SC, and ZX populations—entirely consisted of individuals from the red cluster, whereas other populations consisted of individuals from the green cluster (Figure 2B). Based on the Q values, we also graphed the proportion of cluster members for each population at K = 2 (Figure 3). The graphs showed that the geographical distribution was relatively clear. The first cluster (red), including 19 populations from Jiangxi Province and the XS population from Zhejiang Province, had a larger proportion than the second cluster. In the second cluster, including 20 populations from other provinces and the ZX population from Jiangxi Province, the green cluster had a larger proportion than the red cluster, with a very small proportion from the red cluster (Figure 3). This result is consistent with that of UPGMA clustering analysis (Figure 4). Similarly, the two clusters were clearly distinguished by PCoA analysis: the populations from Jiangxi Province (except ZX) and XS from Zhejiang were grouped as cluster I, and the other 21 populations were grouped as cluster II (Figure 5). We further analyzed the correlation between pairwise FST and geographical distance (Figure 6) using the Mantel test. The results showed that there was a significant correlation between genetic distance and geographic distance (r = 0.159, p = 0.026), indicating a clear geographic origin-based structuring or predominate isolation by distance among the investigated populations.
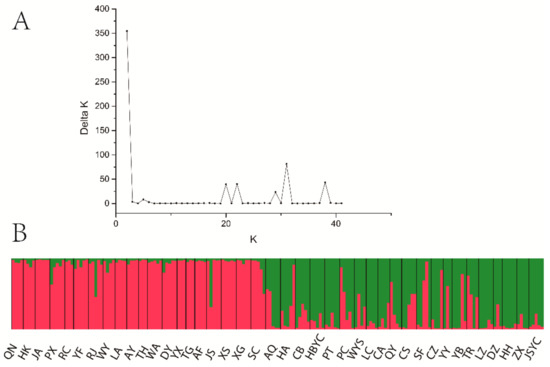
Figure 2.
Population genetic structure. (A) Relations between the rational groups number K and estimated value ΔK. (B) Genetic structural plot of 41 C. camphora populations based on structure analysis. Each individual is represented by a single vertical bar, which is partitioned into two different colors.
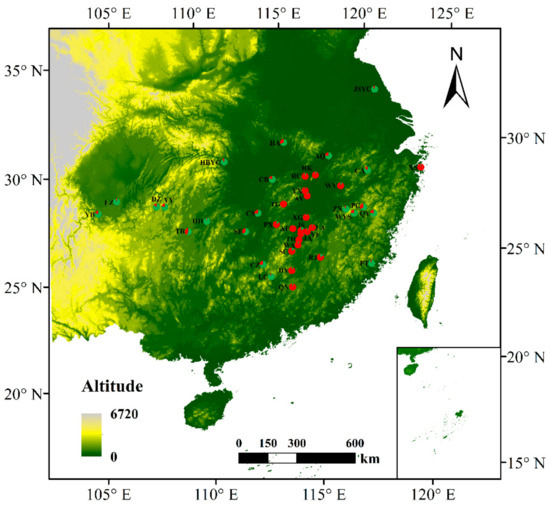
Figure 3.
Mean proportions of cluster memberships of analyzed individuals in each of the 41 C. camphora populations based on structure at K = 2.
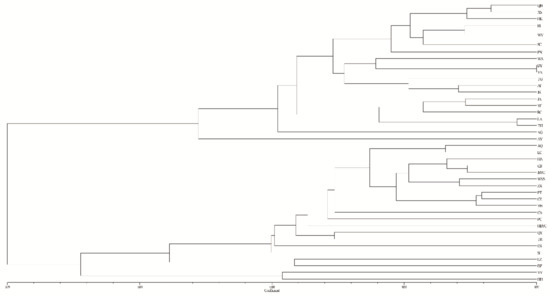
Figure 4.
Genetic divergence among 41 populations of C. camphora based on UPGMA clustering analysis.
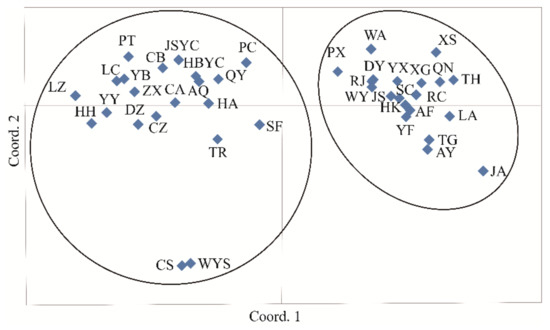
Figure 5.
Principal coordinate analysis (PCoA) of 41 C. camphora populations.
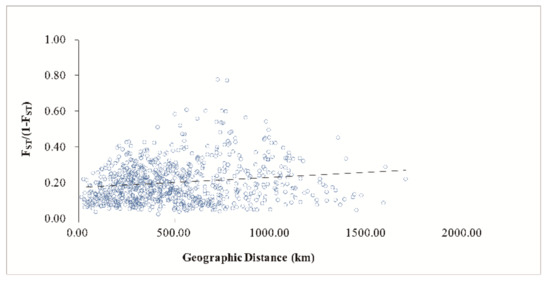
Figure 6.
Mantel between genetic distance and geographical distance of 41 C. camphora populations.
4. Discussion
4.1. Development of EST-SSR Markers for C. camphora
EST-SSR markers have been extensively used in the study of genetic diversity and population structure because of several advantages over studies of noncoding nuclear genomes, such as increased transferability to related species [37], increased accuracy when accessing differences in adaptation among populations [38], increased amplification success, conserved primer sites and a simplified analysis [39]. However, even though whole-genome sequencing has been completed [1], only a few EST-SSR markers have been developed in earlier studies [9,16,20], and a number of polymorphic and informative EST-SSR markers in particular are still lacking. In the present study, we developed 100 EST-SSR markers based on sequencing data [28], and 65% (65) yielded a clear and strong single band, indicating that no introns existed within these regions. Then, 22 of 30 randomly selected markers from validated primers showed polymorphism among 180 wild C. camphora individuals. The average Na, Ne, Ho, He, and GD were 2.77, 1.93, 0.45, 0.44, and 0.44, respectively, which were slightly higher than the values detected in the study by Li et al. [19] on C. camphora based on EST-SSR data (Na = 2.68, Ho = 0.34, and He = 0.41), much lower than those values from the report by Kameyama et al. [20] based on genic-SSR data (Na = 6.01, Ho = 0.56 and He = 0.63) and comparable to or slightly lower than those values based on SSR or inter-simple sequence repeat (ISSR) markers in other Cinnamomum species, such as C. kanehirae (Na = 4.00, Ho = 0.39, and He = 0.59) [40], C. camphora var. nominale (Na = 3.73, Ho = 0.54 and He = 0.60) [40] and C. chago (Na = 1.83 and GD = 0.35) [41]. The markers with both high and low values are useful to study population genetic diversity [42,43]. Therefore, the EST-SSR markers we developed in this study appear to be useful for genetic studies of C. camphora populations.
4.2. Genetic Diversity of C. camphora
Genetic diversity plays an important role in genetic breeding programs, especially for long-term species survival. The genetic diversity of wild plant species is often positively correlated with geographic range and population size [44,45,46]. Previously, a medium genetic diversity level (Nanjing: Ho = 0.35 and He = 0.42; Anqing: Ho = 0.32 and He = 0.43; Ruihong: Ho = 0.37 and He = 0.43) in three Chinese C. camphora populations, including 15 individuals in each population, was reported by Li et al. [19]. However, Kameyama et al. indicated relatively high levels in Fujian (Ho= 0.73 and He = 0.86), Shanghai (Ho= 0.79 and He = 0.84) and Taiwan (Ho = 0.69 and He = 0.87) populations from China and slightly lower population genetic diversity in east Japan (Ho = 0.62 and He = 0.71), west Japan (Ho = 0.57 and He = 0.69) and Kyushu (Ho = 0.69 and He = 0.72), which sampled 187, 41, 76, 179, 219, and 115 individuals, respectively [16]. Our results indicated only a medium genetic diversity level in 41 C. camphora populations from China (Ho range from 0.30 to 0.61 and He range from 0.31 to 0.48). This is consistent with the different populations [46] and population size used in those studies [21].
Generally, the genetic diversity of species with a widespread distribution is higher than that of species with a narrow or endemic distribution [47,48]. Considering that the native habitant of C. camphora is widespread in the whole region of South China, it is not surprising that the genetic diversity in this study (Ho = 0.45 and He = 0.40) is much higher than that of other narrow or endemic species, such as Dalbergia odorifera (Ho = 0.28 and He = 0.37) [21], Gastrodia elata (Ho = 0.05 and He = 0.46) [24], and Ottelia acuminata (Ho = 0.28 and He = 0.37) [49]. However, the genetic diversity of C. camphora is much lower than that of other widespread tree species, such as Xanthoceras sorbifolia (Ho = 0.72 and He = 0.53) [50], Albanian olive (Ho = 0.75 and He = 0.60) [51], and Eugenia dysenterica DC. (Ho = 0.545 and He = 0.62) [52], and even lower than some fragment habitant species, such as Erythrophleum fordii Oliv. (Ho = 0.52 and He = 0.56) [22], Liriodendron chinense (Ho = 0.70 and He = 0.68) [53], and Alnus cremastogyne (Ho = 0.63 and He = 0.74) [54]. Although C. camphora is a tree species with great longevity and has been cultivated since ancient times, the impact of artificial clonal propagation and human-mediated transfer of seedlings or seeds on the genetic diversity of the natural population may be substantial [16,55]. In addition, due to the over deforestation and overutilization over recent decades, those factors could be associated with the slightly low genetic diversity level in this study.
4.3. Genetic Differentiation and Population Genetic Structure of C. camphora
According to Wright, a coefficient of genetic differentiation less than 0.25 indicates low genetic variation among populations [56]. In the present study, the genetic differentiation among C. camphora populations was low (FST = 0.22, Table 3). Similarly, AMOVA analysis showed that only 17% of the genetic variation existed among populations, while 83% existed within populations. This is consistent with previous reports on species from South China, such as Dalbergia cochinchinensis (FST = 0.23) [57] and Erythrophleum fordii Oliv. (FST = 0.18) [22]. These findings also indicated that the Nm (1.20) among C. camphora populations was frequent, suggesting that Nm replaced the genetic drift effects to a certain extent [58], consistent with its widespread distribution in South China. However, we cannot ignore the fact that entomophilous plants with a predominately outcrossing mating system will have reduced pollen flow among populations by long-term habitat destruction, thus increasing the genetic differentiation and erosion and even moderating Nm among populations [59].
For population classification, 41 wild C. camphora populations were divided into two clusters. The populations from Jiangxi Province (except ZX) and population XS from Zhejiang Province were grouped as cluster I, and the other 21 populations were grouped as cluster II. This suggests that certain geographical distribution characteristics exist in those populations. Similar results were also shown from both the PCoA analysis and UPGMA clustering. Interestingly, genetic differentiation among the investigated populations was only weakly correlated but marginally significant with the geographical locations. This is possibly because C. camphora prefers river bank habitats, and its seeds are easily affected by regional water systems, such as the Ganjiang River, Fu River, and Rao River in Jiangxi Province. This is also possibly because a large number of ancient camphor trees exists in nearly all villages of Jiangxi Province, and human disturbance is seldom an issue as C. camphora is treated as a sacred tree in these areas compared to other regions in South China [11].
4.4. Conservation Strategies for C. camphora
Knowledge of genetic diversity and population structure is essential to establish a suitable strategy for the conservation of C. camphora and the elucidation of mechanisms of endangerment [60]. There are many ancient camphor trees in rural and urban areas of South China. However, the present study showed a medium level of genetic diversity in the wild populations of C. camphora, although it is widespread in South China. This may be mainly attributed to the forests being severely disturbed by human activity, such as the occurrence of over deforestation and overutilization in recent decades, and the large DBH of adult trees, most being ancient trees, that were sampled in this study. Furthermore, considering the longevity of this species, domestication processes and the transference of seeds or saplings have severely affected its genetic diversity and ecological fates. Moreover, as an entomophilous plant, genetic erosion, and genetic drift might influence genetic diversity in subsequent generations. To date, C. camphora has been promoted to a second-grade state-protected species by the Chinese government and protected carefully by customary laws. However, this is not sufficient because of its wild populations are declining rapidly. It is urgent to carry out in situ conservation for natural populations and ancient trees, especially for populations with high genetic diversity and unique alleles, such as HK population from Jiangxi Province. Simultaneously, ex situ conservation should also be conducted for as many families and individuals as possible via seeds or branches, as genetic diversity mainly existed within populations. Additionally, human-mediated sapling transfer, such as the Transplanting Big Trees into Cities Program, should be prohibited to conserve the genetic composition within populations.
5. Conclusions
This study provides an initial assessment of the genetic diversity and population structure of C. camphora in South China using 22 EST-SSR markers. The results showed medium genetic diversity and low genetic differentiation among populations. These 41 C. camphora populations were divided into two clusters; notably, nearly all populations from Jiangxi Province—except ZX—grouped into one cluster. Because three to five individuals were sampled from all populations investigated in the present study, we must increase the number of individuals sampled to verify the results of this study in the future. Nonetheless, the findings of the present study will assist in developing conservation strategies and in the genetic improvement of C. camphora.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/10/11/1019/s1, Table S1: Location information and SSR data of the 180 sampled C. camphora individuals; Table S2: Allele frequency distribution across 180 C. camphora; Table S3: Pairwise genetic differentiation index (FST) among the 41 populations of C. camphora.
Author Contributions
Conceptualization: Y.Z. and F.Y.; Methodology: Y.Z. and A.Y.; Software: A.Y., Z.L., and Z.W.; Validation: Y.L., T.L., and L.L.; Funding acquisition: Y.Z.; Investigation: Y.Z., A.Y., and Y.L.; Resources: L.L., H.Z., and Z.W.; Project administration: Y.Z. and F.Y.; Writing—original draft: Y.Z.; Writing—review and editing: Y.Z., F.Y., M.X., and A.Y.
Funding
This research was funded by the National Natural Science Foundation of China (no. 31860079), the Forestry Science and Technology Innovation Special of Jiangxi (201701), and the Postdoctoral Science Foundation of Jiangxi Academy of Sciences.
Acknowledgments
We are grateful to Bowei Yu for his assistance in map-making. We also appreciate Li’an Xu for his help in revising the manuscript and his constructive advice.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Chaw, S.M.; Liu, Y.C.; Wu, Y.W.; Wang, H.Y.; Lin, C.I.; Wu, C.S.; Ke, H.M.; Chang, L.Y.; Hsu, C.Y.; Yang, H.T.; et al. Stout camphor tree genome fills gaps in understanding of flowering plant genome evolution. Nat. Plants 2019, 5, 63–73. [Google Scholar] [CrossRef]
- Joy, P.; Maridass, M. Inter Species Relationship of Cinnamomum Species Using RAPD Marker Analysis. Ethnobot. Leafl. 2008, 12, 476–480. [Google Scholar]
- Editorial Board of Flora of the Trees in China. In Flora of the Trees in China; Science Press: Beijing, China, 1982.
- Chen, Z.; Wu, B.; Li, J.; Zhao, T.; Zhou, X.; Zhang, Y. Germination of the seeds and growth of the seedlings of Cinnamomum camphora (L.) Presl. Plant Species Biol. 2004, 19, 55–58. [Google Scholar] [CrossRef]
- Doh, E.J.; Kim, J.H.; Oh, S.E.; Lee, G. Identification and monitoring of Korean medicines derived from Cinnamomum spp. by using ITS and DNA marker. Genes Genom. 2017, 39, 101–109. [Google Scholar] [CrossRef]
- Panetta, F.D. Seedling emergence and seed longevity of the tree weeds Celtis sinensis and Cinnamomum camphora. Weed Res. 2010, 41, 83–95. [Google Scholar] [CrossRef]
- Firth, D.J. Camphor laurel (Cinnamomum camphora)—A new weed in north-eastern New South Wales. Aust. Weeds 1981, 2, 26–28. [Google Scholar]
- Firth, D.J. Camphor laurel: Important tree weed in the north-east. Agric. Gaz. N. S. W. 1979, 90, 14–15. [Google Scholar]
- Daubenmire, R. The Magnolia grandiflora-Quercus virginiana Forest of Florida. Am. Midl. Nat. 1990, 123, 331–347. [Google Scholar] [CrossRef]
- Kameyama, Y.; Nakajima, H. Environmental conditions for seed germination and seedling growth of Cinnamomum camphora (Lauraceae): The possibility of regeneration in an abandoned deciduous broad-leaved forest, eastern Japan. J. For. Res. 2018, 23, 190–194. [Google Scholar] [CrossRef]
- Zhou, Y.; Yan, W. Conservation and applications of camphor tree (Cinnamomum camphora) in China: Ethnobotany and genetic resources. Genet. Resour. Crop Evol. 2015, 63, 1049–1061. [Google Scholar] [CrossRef]
- Singh, R.; Jawaid, T. Cinnamomum camphora (Kapur): Review. Pharmacogn. J. 2012, 4, 1–5. [Google Scholar] [CrossRef]
- Lee, H.J.; Hyun, E.-A.; Yoon, W.J.; Kim, B.H.; Rhee, M.H.; Kang, H.K.; Cho, J.Y.; Yoo, E.S. In vitro anti-inflammatory and anti-oxidative effects of Cinnamomum camphora extracts. J. Ethnopharmacol. 2006, 103, 208–216. [Google Scholar] [CrossRef]
- Li, Y.; Fu, C.; Yang, W.; Wang, X.; Feng, D.; Wang, X.; Ren, D.; Lou, H.; Shen, T. Investigation of constituents from Cinnamomum camphora (L.) J. Presl and evaluation of their anti-inflammatory properties in lipopolysaccharide stimulated RAW 264.7 macrophages. J. Ethnopharmacol. 2018, 221, 37–47. [Google Scholar] [CrossRef]
- Li, Z.R.; Chen, S.X. Jiangxi Camphor Tree; Jiangxi Science and Technology Press: Nanchang, China, 2015. [Google Scholar]
- Kameyama, Y.; Furumichi, J.; Li, J.; Tseng, Y.-H. Natural genetic differentiation and human-mediated gene flow: The spatiotemporal tendency observed in a long-lived Cinnamomum camphora (Lauraceae) tree. Tree Genet. Genomes 2017, 13, 38. [Google Scholar] [CrossRef]
- Wang, S.; Xie, Y. China Species Red List; Higer Education Press: Beijing, China, 2004. [Google Scholar]
- Yao, X.; Ren, H.; Wang, K.; Wu, K.; Gao, S. A Preliminary Study on Genetic Variance of Bud Germination at the Seedling Stage of Cinnamomum camphora (L.) presl. Acta Agric. Univ. Jiangxiensis 2001, 23, 537–543. [Google Scholar]
- Li, Z.; Zhong, Y.; Yu, F.; Xu, M. Novel SSR marker development and genetic diversity analysis of Cinnamomum camphora based on transcriptome sequencing. Plant Genet. Resour. Charact. Util. 2018, 16, 568–571. [Google Scholar] [CrossRef]
- Kameyama, Y. Development of microsatellite markers for Cinnamomum camphora (Lauraceae). Am. J. Bot. 2012, 99, e1–e3. [Google Scholar] [CrossRef]
- Liu, F.; Hong, Z.; Xu, D.; Jia, H.; Zhang, N.; Liu, X.; Yang, Z.; Lu, M. Genetic Diversity of the Endangered Dalbergia odorifera Revealed by SSR Markers. Forests 2019, 10, 225. [Google Scholar] [CrossRef]
- Tan, J.; Zhao, Z.-G.; Guo, J.-J.; Wang, C.-S.; Zeng, J. Genetic Diversity and Population Genetic Structure of Erythrophleum fordii Oliv., an Endangered Rosewood Species in South China. Forests 2018, 9, 636. [Google Scholar] [CrossRef]
- Thriveni, H.N.; Sumangala, R.C.; Shivaprakash, K.N.; Ravikanth, G.; Vasudeva, R.; Ramesh Babu, H.N. Genetic structure and diversity of Coscinium fenestratum: A critically endangered liana of Western Ghats, India. Plant Syst. Evol. 2013, 300, 403–413. [Google Scholar] [CrossRef]
- Chen, Y.-Y.; Bao, Z.-X.; Qu, Y.; Li, W.; Li, Z.-Z. Genetic diversity and population structure of the medicinal orchid Gastrodia elata revealed by microsatellite analysis. Biochem. Syst. Ecol. 2014, 54, 182–189. [Google Scholar] [CrossRef]
- Müller, M.; Cuervo-Alarcon, L.; Gailing, O.; Rajendra, K.C.; Chhetri, M.; Seifert, S.; Arend, M.; Krutovsky, K.; Finkeldey, R. Genetic Variation of European Beech Populations and Their Progeny from Northeast Germany to Southwest Switzerland. Forests 2018, 9, 469. [Google Scholar] [CrossRef]
- Powell, W.; Morgante, M.; Andre, C.; Hanafey, M.; Vogel, J.; Tingey, S.; Rafalski, A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 1996, 2, 225–238. [Google Scholar] [CrossRef]
- Stojnić, S.; Avramidou, E.V.; Fussi, B.; Westergren, M.; Orlović, S.; Matović, B.; Trudić, B.; Kraigher, H.; Aravanopoulos, F.A.; Konnert, M. Assessment of Genetic Diversity and Population Genetic Structure of Norway Spruce (Picea abies (L.) Karsten) at Its Southern Lineage in Europe. Implications for Conservation of Forest Genetic Resources. Forests 2019, 10, 258. [Google Scholar]
- Chen, C.; Zheng, Y.; Zhong, Y.; Wu, Y.; Li, Z.; Xu, L.A.; Xu, M. Transcriptome analysis and identification of genes related to terpenoid biosynthesis in Cinnamomum camphora. BMC Genom. 2018, 19, 550. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.C.; Boyle, T.B.J.; Ye, Z.H.; Mao, J.X.; Yeh, C.; Timothy, B.; Mao, X. POPGENE (Version 1.31), the User-Friendly Shareware for Population Genetic Analysis. In Molecular Biology and Biotechnology Center; University of Alberta: Edmonton, Canada, 1997. [Google Scholar]
- Nagy, S.; Poczai, P.; Cernák, I.; Gorji, A.M.; Hegedűs, G.; Taller, J. PICcalc: An Online Program to Calculate Polymorphic Information Content for Molecular Genetic Studies. Biochem. Genet. 2012, 50, 670–672. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.J.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef]
- Rohlf, F.J. NTSYS-pc Numerical Taxonomy and Multivariate Analysis System Solutions Manual. Am. Stat. 2000, 41, 330. [Google Scholar] [CrossRef]
- Manel, S.; Schwartz, M.K.; Luikart, G.; Taberlet, P. Landscape genetics combining landscape ecology and population genetics. Trends Ecol. Evol. 2003, 18, 189–197. [Google Scholar] [CrossRef]
- Yuan, X.-Y.; Sun, Y.-W.; Bai, X.-R.; Dang, M.; Feng, X.-J.; Zulfiqar, S.; Zhao, P. Population Structure, Genetic Diversity, and Gene Introgression of Two Closely Related Walnuts (Juglans regia and J. sigillata) in Southwestern China Revealed by EST-SSR Markers. Forests 2018, 9, 646. [Google Scholar] [CrossRef]
- Ellis, J.R.; Burke, J.M. EST-SSR as a resource for population genetic analyses. Heredity 2007, 99, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Dang, M.; Zhang, T.; Hu, Y.; Zhou, H.; Woeste, K.; Zhao, P. De Novo Assembly and Characterization of Bud, Leaf and Flowers Transcriptome from Juglans Regia L. for the Identification and Characterization of New EST-SSRs. Forests 2016, 7, 247. [Google Scholar] [CrossRef]
- Hung, K.-H.; Lin, C.-H.; Shih, H.-C.; Chiang, Y.-C.; Ju, L.-P. Development, characterization and cross-species amplification of new microsatellite primers from an endemic species Cinnamomum kanehirae (Lauraceae) in Taiwan. Conserv. Genet. Resour. 2014, 6, 911–913. [Google Scholar] [CrossRef]
- Dong, W.; Zhang, X.; Guansong, Y.; Yang, L.; Wang, Y.; Shen, S. Biological characteristics and conservation genetics of the narrowly distributed rare plant Cinnamomum chago (Lauraceae). Plant Divers. 2016, 38, 247–252. [Google Scholar] [CrossRef]
- Liu, S.; An, Y.; Li, F.; Li, S.; Liu, L.; Zhou, Q.; Zhao, S.; Wei, C. Genome-wide identification of simple sequence repeats and development of polymorphic SSR markers for genetic studies in tea plant (Camellia sinensis). Mol. Breed. 2018, 38, 59. [Google Scholar] [CrossRef]
- Väli, U.; Einarsson, A.; Waits, L.; Ellegren, H. To what extent do microsatellite markers reflect genome-wide genetic diversity in natural populations? Mol. Ecol. 2008, 17, 3808–3817. [Google Scholar] [CrossRef]
- Lammi, A.; Siikamaki, P.; Mustajarvi, K. Genetic Diversity, Population Size, and Fitness in Central and Peripheral Populations of a Rare Plant Lychnis viscaria. Conserv. Biol. 1999, 13, 1069–1078. [Google Scholar] [CrossRef]
- Li, X.; Li, M.; Hou, L.; Zhang, Z.; Pang, X.; Li, Y. De Novo Transcriptome Assembly and Population Genetic Analyses for an Endangered Chinese Endemic Acer miaotaiense (Aceraceae). Genes 2018, 9, 378. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Eguiarte, L.; Montana, C. Genetic structure and outcrossing rates in Flourensia cernua (Asteraceae) growing at different densities in the South-western Chihuahuan Desert. Ann. Bot. 2004, 94, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Cheng, Y.; Li, K.; Li, H. Integrating Phylogeographic Analysis and Geospatial Methods to Infer Historical Dispersal Routes and Glacial Refugia of Liriodendron chinense. Forests 2019, 10, 565. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W.; Sherman-Broyles, S.L. Factors influencing levels of genetic diversity in woody plant species. New For. 1992, 6, 95–124. [Google Scholar] [CrossRef]
- Zhai, S.H.; Yin, G.S.; Yang, X.H. Population Genetics of the Endangered and Wild Edible Plant Ottelia acuminata in Southwestern China Using Novel SSR Markers. Biochem. Genet. 2018, 56, 235–254. [Google Scholar] [CrossRef]
- Shen, Z.; Zhang, K.; Ma, L.; Duan, J.; Ao, Y. Analysis of the genetic relationships and diversity among 11 populations of Xanthoceras sorbifolia using phenotypic and microsatellite marker data. Electron. J. Biotechnol. 2017, 26, 33–39. [Google Scholar] [CrossRef]
- Dervishi, A.; Jakše, J.; Ismaili, H.; Javornik, B.; Štajner, N. Comparative assessment of genetic diversity in Albanian olive (Olea europaea L.) using SSRs from anonymous and transcribed genomic regions. Tree Genet. Genomes 2018, 14, 53. [Google Scholar] [CrossRef]
- Boaventura-Novaes, C.R.D.; Novaes, E.; Mota, E.E.S.; Telles, M.P.C.; Coelho, A.S.G.; Chaves, L.J. Genetic drift and uniform selection shape evolution of most traits in Eugenia dysenterica DC. (Myrtaceae). Tree Genet. Genomes 2018, 14, 76. [Google Scholar] [CrossRef]
- Long, X.; Weng, Y.; Liu, S.; Hao, Z.; Sheng, Y.; Guan, L.; Shi, J.; Chen, J. Genetic Diversity and Differentiation of Relict Plant Liriodendron Populations Based on 29 Novel EST-SSR Markers. Forests 2019, 10, 334. [Google Scholar] [CrossRef]
- Guo, H.-Y.; Wang, Z.-L.; Huang, Z.; Chen, Z.; Yang, H.-B.; Kang, X.-Y. Genetic Diversity and Population Structure of Alnus cremastogyne as Revealed by Microsatellite Markers. Forests 2019, 10, 278. [Google Scholar] [CrossRef]
- Quintana, J.; Contreras, A.; Merino, I.; Vinuesa, A.; Orozco, G.; Ovalle, F.; Gomez, L. Genetic characterization of chestnut (Castanea sativa Mill.) orchards and traditional nut varieties in El Bierzo, a glacial refuge and major cultivation site in northwestern Spain. Tree Genet. Genomes 2014, 11, 826. [Google Scholar] [CrossRef][Green Version]
- Wright, S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Hartvig, I.; So, T.; Changtragoon, S.; Tran, H.T.; Bouamanivong, S.; Theilade, I.; Kjaer, E.D.; Nielsen, L.R. Population genetic structure of the endemic rosewoods Dalbergia cochinchinensis and D. oliveri at a regional scale reflects the Indochinese landscape and life-history traits. Ecol. Evol. 2018, 8, 530–545. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. Evolution in mendelian populations. Genetics 1931, 16, 97–159. [Google Scholar] [PubMed]
- Albaladejo, R.G.; Carrillo, L.F.; Aparicio, A.; Fernández-Manjarrés, J.F.; González-Varo, J.P. Population genetic structure in Myrtus communis L. in a chronically fragmented landscape in the Mediterranean: Can gene flow counteract habitat perturbation? Plant Biol. 2009, 11, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Forest, F.; Grenyer, R.; Rouget, M.; Davies, T.J.; Cowling, R.M.; Faith, D.P.; Balmford, A.; Manning, J.C.; Procheş, Ş.; Bank, M.V.D.; et al. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 2007, 445, 757–760. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).