Environmental Filtering Drives Local Soil Fungal Beta Diversity More Than Dispersal Limitation in Six Forest Types along a Latitudinal Gradient in Eastern China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design and Field Sampling
2.2. Climate Data and Geographic Distance
2.3. Soil Physicochemical Analysis
2.4. Amplification, Illumina Sequencing and Bioinformatics
2.5. Statistical Analyses
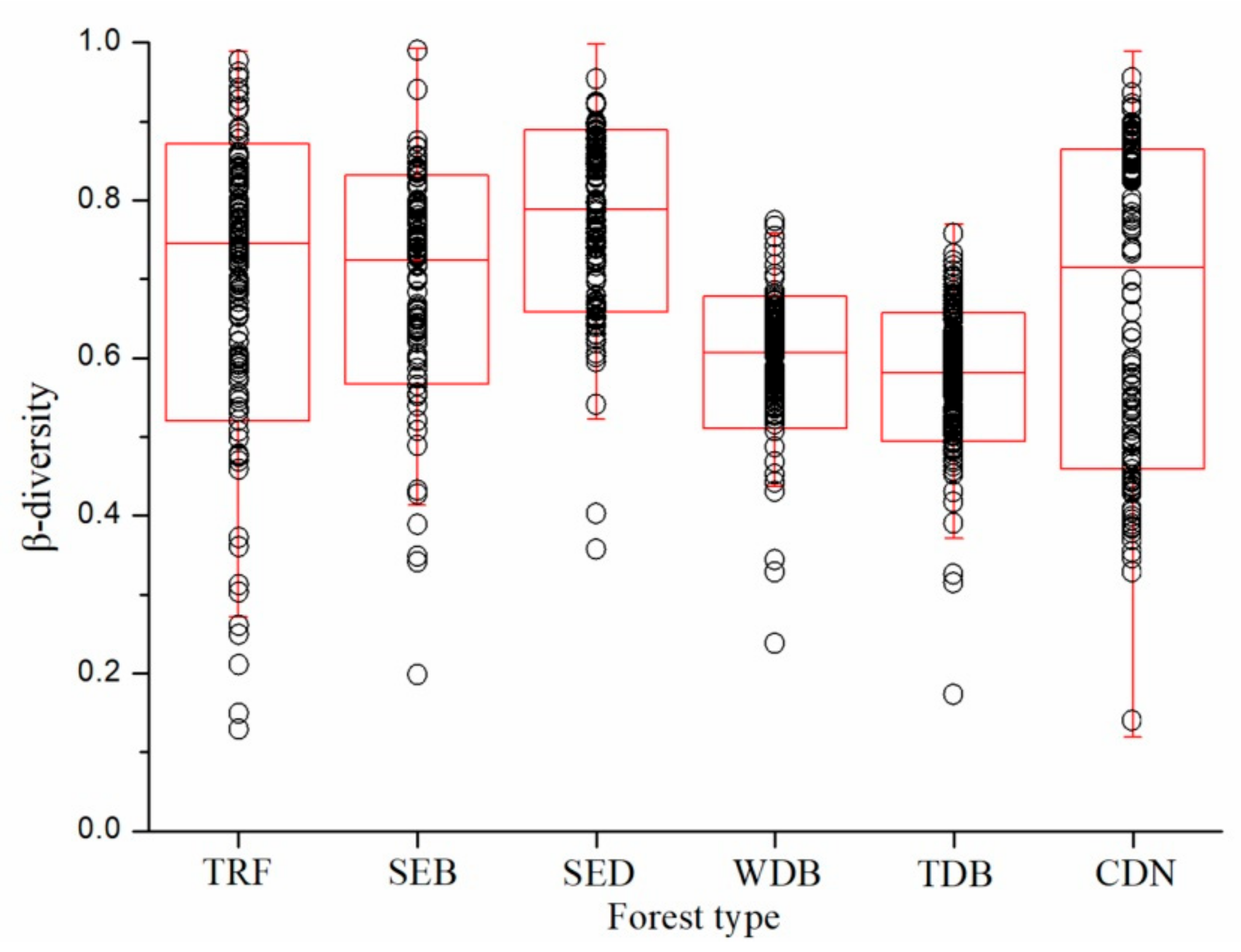
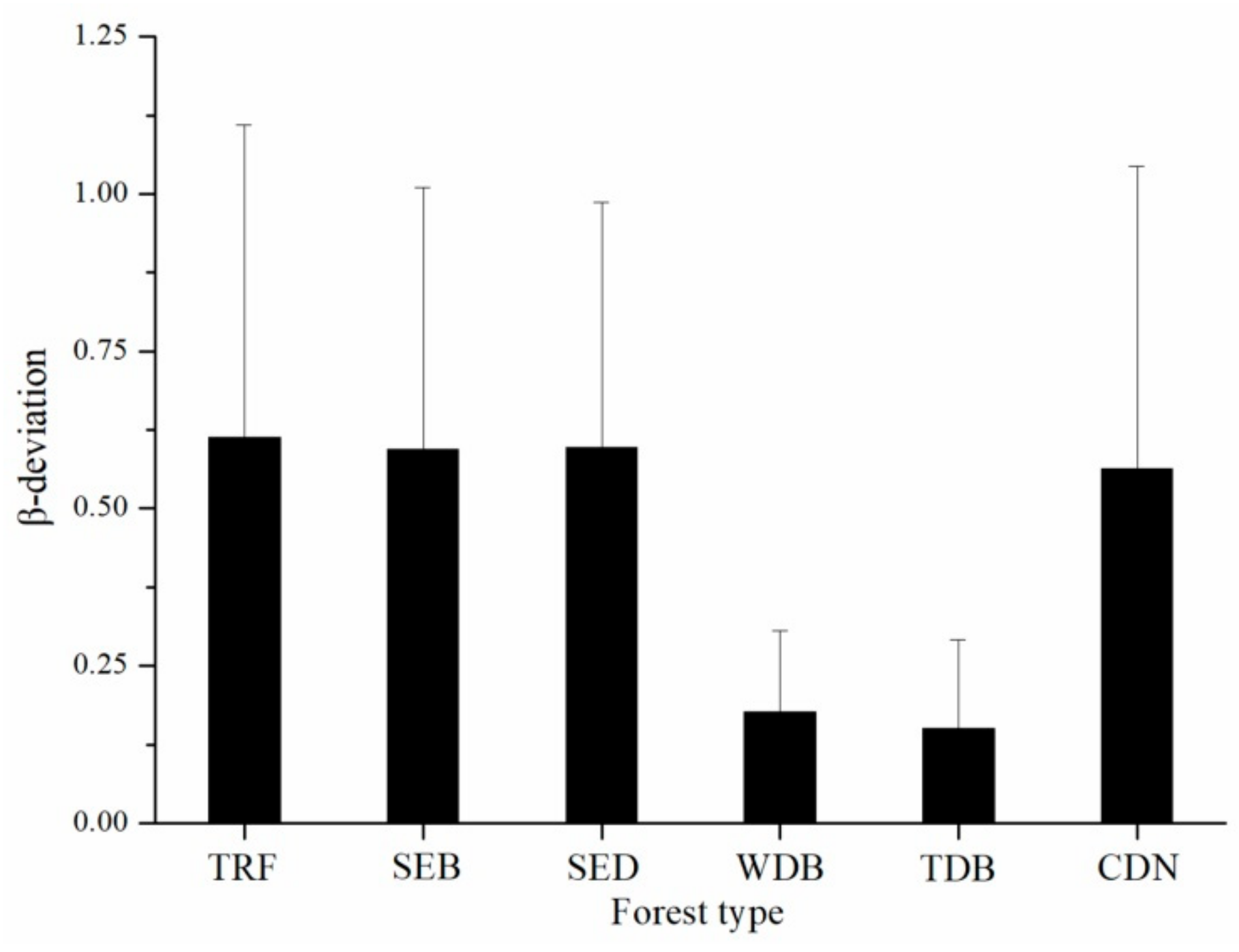
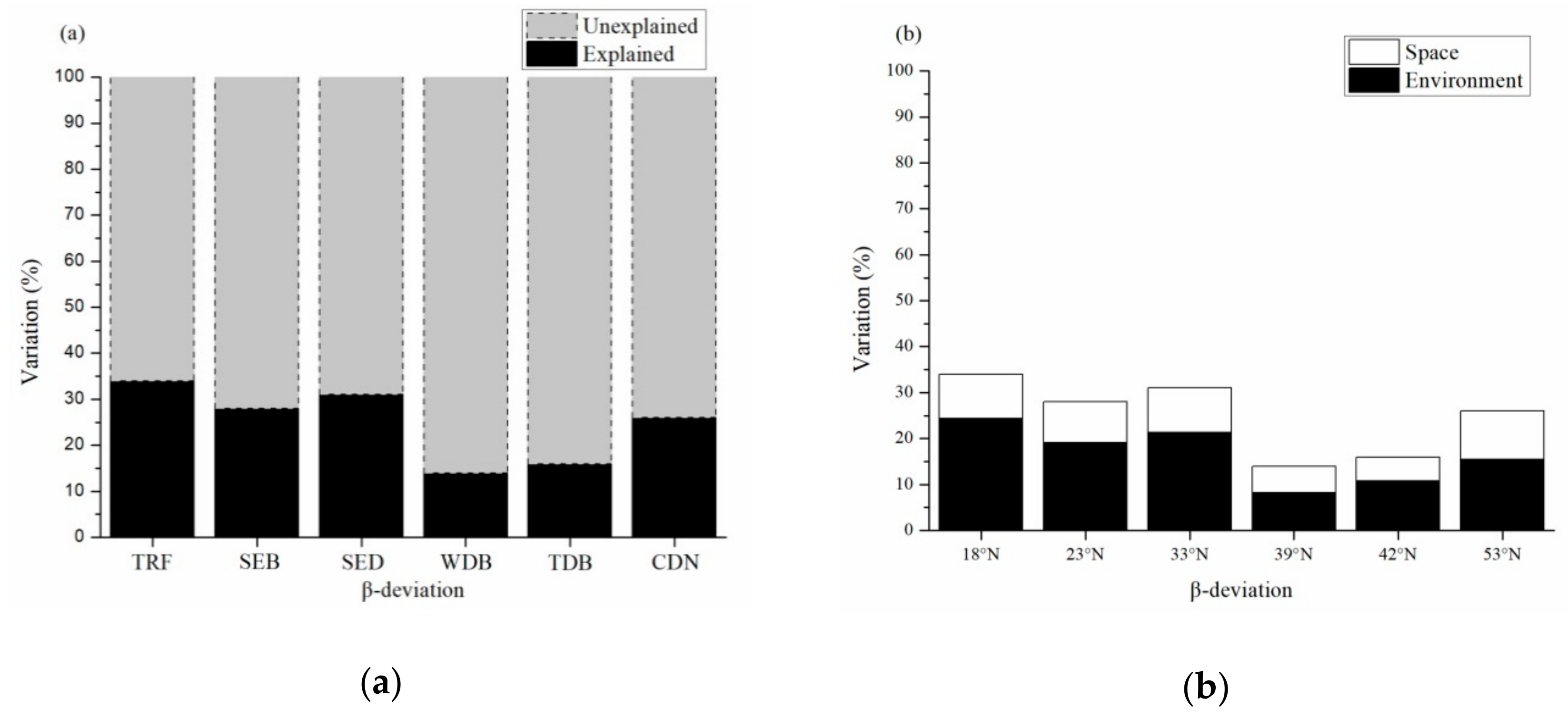
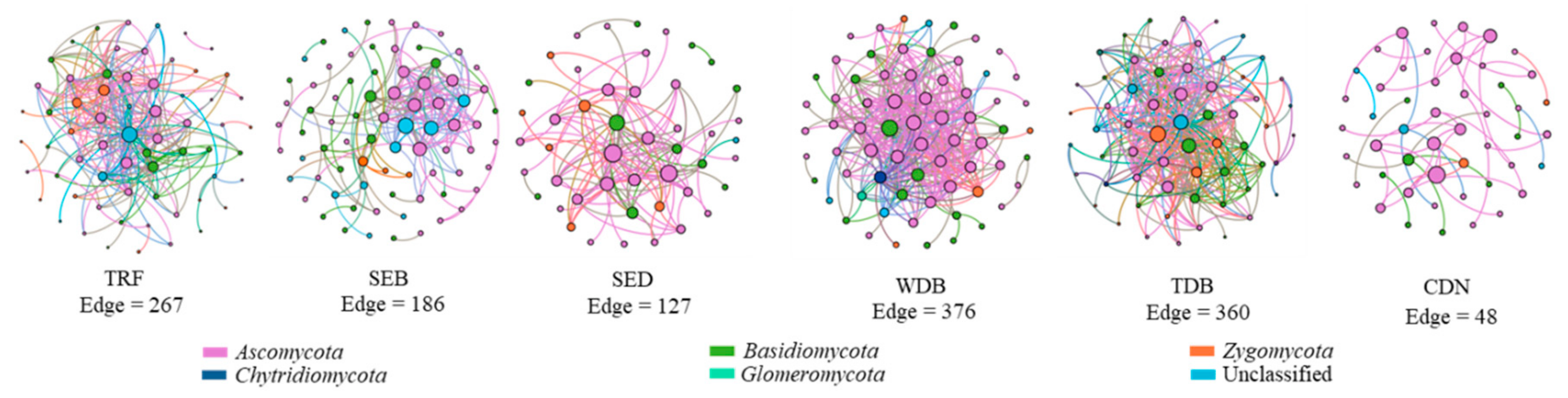
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hanson, C.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Martiny, J.B.H. Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat. Rev. Genet. 2012, 10, 497. [Google Scholar] [CrossRef] [PubMed]
- Ricklefs, R.E. A comprehensive framework for global patterns in biodiversity. Ecol. Lett. 2004, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Kraft, N.J.B.; Comita, L.S.; Chase, J.M.; Sanders, N.J.; Swenson, N.G.; Crist, T.O.; Stegen, J.C.; Vellend, M.; Boyle, B.; Anderson, M.J.; et al. Disentangling the drivers of β diversity along latitudinal and elevational gradients. Science 2011, 333, 1755–1758. [Google Scholar] [CrossRef] [PubMed]
- Glassman, S.I.; Wang, I.J.; Bruns, T.D. Environmental filtering by pH and soil nutrients drives community assembly in fungi at fine spatial scales. Mol. Ecol. 2017, 26, 6960–6973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kivlin, S.N.; Winston, G.C.; Goulden, M.L.; Treseder, K.K. Environmental filtering affects soil fungal community composition more than dispersal limitation at regional scales. Fungal Ecol. 2014, 12, 14–25. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.; Tian, J.; Bai, C.; Xiang, M.; Sun, J.; Liu, X. The biogeography of fungal communities in wetland sediments along the Changjiang River and other sites in China. ISME J. 2013, 7, 1299–1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cline, L.C.; Zak, D.R. Dispersal limitation structures fungal community assembly in a long-term glacial chronosequence. Environ. Microbiol. 2014, 16, 1538–1548. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.R.; Karunaratne, S.; Campbell, C.D.; Yao, H.; Robinson, L.; Singh, B.K. Deterministic processes vary during community assembly for ecologically dissimilar taxa. Nat. Commun. 2015, 6, 8444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Condit, R.; Pitman, N.; Leigh, E.G.; Chave, J.; Terborgh, J.; Foster, R.B.; Núñez, P.; Aguilar, S.; Valencia, R.; Villa, G.; et al. Beta-Diversity in Tropical Forest Trees. Science 2002, 295, 666–669. [Google Scholar] [CrossRef] [Green Version]
- Anderson, M.J.; Crist, T.O.; Chase, J.M.; Vellend, M.; Inouye, B.D.; Freestone, A.L.; Sanders, N.J.; Cornell, H.V.; Comita, L.S.; Davies, K.F. Navigating the multiple meanings of β diversity: A roadmap for the practicing ecologist. Ecol. Lett. 2011, 14, 19–28. [Google Scholar] [CrossRef]
- Myers, J.A.; Chase, J.M.; Jiménez, I.; Jørgensen, P.M.; Araujo-Murakami, A.; Paniagua-Zambrana, N.; Seidel, R. Beta-diversity in temperate and tropical forests reflects dissimilar mechanisms of community assembly. Ecol. Lett. 2013, 16, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Bell, T. Experimental tests of the bacterial distance—Decay relationship. ISME J. 2010, 4, 1357. [Google Scholar] [CrossRef]
- Tilman, D. Niche tradeoffs, neutrality, and community structure: A stochastic theory of resource competition, invasion, and community assembly. Proc. Natl. Acad. Sci. USA 2004, 101, 10854–10861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P.; et al. Patterns and Processes of Microbial Community Assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maestre, F.T.; Delgado-Baquerizo, M.; Jeffries, T.C.; Eldridge, D.J.; Ochoa, V.; Gozalo, B.; Quero, J.L.; García-Gómez, M.; Gallardo, A.; Ulrich, W.; et al. Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc. Natl. Acad. Sci. USA 2015, 112, 15684–15689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prober, S.M.; Leff, J.W.; Bates, S.T.; Borer, E.T.; Firn, J.; Harpole, W.S.; Lind, E.M.; Seabloom, E.W.; Adler, P.B.; Bakker, J.D. Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecol. Lett. 2015, 18, 85–95. [Google Scholar] [CrossRef]
- Shen, C.; Xiong, J.; Zhang, H.; Feng, Y.; Lin, X.; Li, X.; Liang, W.; Chu, H. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 2013, 57, 204–211. [Google Scholar] [CrossRef]
- Tripathi, B.M.; Stegen, J.C.; Kim, M.; Dong, K.; Adams, J.M.; Lee, Y.K. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 2018, 12, 1072–1083. [Google Scholar] [CrossRef]
- Hubbell, S.P. The Unified Neutral Theory of Biodiversity and Biogeography (MPB–32); Princeton University Press: Princeton, NJ, USA, 2001. [Google Scholar]
- Vanschoenwinkel, B.; Buschke, F.; Brendonck, L. Disturbance regime alters the impact of dispersal on alpha and beta diversity in a natural metacommunity. Ecology 2013, 94, 2547–2557. [Google Scholar] [CrossRef]
- Wang, X.B.; Lü, X.T.; Yao, J.; Wang, Z.W.; Deng, Y.; Cheng, W.X.; Zhou, J.Z.; Han, X.G. Habitat–specific patterns and drivers of bacterial β–diversity in China’s drylands. ISME J. 2017, 11, 1345. [Google Scholar] [CrossRef]
- Chytrý, M.; Lososová, Z.; Horsák, M.; Uher, B.; Čejka, T.; Danihelka, J.; Fajmon, K.; Hájek, O.; Juřičková, L.; Kintrová, K. Dispersal limitation is stronger in communities of microorganisms than macroorganisms across Central European cities. J. Biogeogr. 2012, 39, 1101–1111. [Google Scholar] [CrossRef]
- Bell, G. Neutral macroecology. Science 2001, 293, 2413–2418. [Google Scholar] [CrossRef]
- Violle, C.; Nemergut, D.R.; Pu, Z.; Jiang, L. Phylogenetic limiting similarity and competitive exclusion. Ecol. Lett. 2011, 14, 782–787. [Google Scholar] [CrossRef] [PubMed]
- De Cáceres, M.; Legendre, P.; Valencia, R.; Cao, M.; Chang, L.W.; Chuyong, G.; Condit, R.; Hao, Z.; Hsieh, C.F.; Hubbell, S.; et al. The variation of tree beta diversity across a global network of forest plots. Glob. Ecol. Biogeogr. 2012, 21, 1191–1202. [Google Scholar] [CrossRef]
- Wang, J.; Shen, J.; Wu, Y.; Tu, C.; Soininen, J.; Stegen, J.C.; He, J.Z.; Liu, X.; Zhang, L.; Zhang, E. Phylogenetic beta diversity in bacterial assemblages across ecosystems: Deterministic versus stochastic processes. ISME J. 2013, 7, 1310–1321. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Van Nostrand, J.D.; Deng, Y.; Lü, X.; Wang, C.; Zhou, J.; Han, X. Scale–dependent effects of climate and geographic distance on bacterial diversity patterns across northern China’s grasslands. FEMS Microbiol. Ecol. 2015, 91, fiv133. [Google Scholar] [CrossRef] [PubMed]
- Püttker, T.; de Arruda Bueno, A.; Prado, P.I.; Pardini, R. Ecological filtering or random extinction? Beta-diversity patterns and the importance of niche-based and neutral processes following habitat loss. Oikos 2015, 124, 206–215. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Trivedi, P.; Osanai, Y.; Liu, Y.R.; Hamonts, K.; Jeffries, T.C.; Singh, B.K. Carbon content and climate variability drive global soil bacterial diversity patterns. Ecol. Monogr. 2016, 86, 373–390. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Knight, R. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. USA 2007, 104, 11436–11440. [Google Scholar] [CrossRef] [Green Version]
- Peay, K.G.; Garbelotto, M.; Bruns, T.D. Evidence of dispersal limitation in soil microorganisms: Isolation reduces species richness on mycorrhizal tree islands. Ecology 2010, 91, 3631–3640. [Google Scholar] [CrossRef]
- Shi, Y.; Li, Y.; Xiang, X.; Sun, R.; Yang, T.; He, D.; Zhang, K.; Ni, Y.; Zhu, Y.G.; Adams, J.M.; et al. Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China Plain. Microbiome 2018, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Veresoglou, S.D.; Tedersoo, L.; Xu, T.; Ge, T.; Liu, L.; Chen, Y.; Hao, Z.; Su, Y.; Rillig, M.C.; et al. Contrasting latitudinal diversity and co-occurrence patterns of soil fungi and plants in forest ecosystems. Soil Biol. Biochem. 2019, 131, 100–110. [Google Scholar] [CrossRef]
- Zeilinger, S.; Gupta, V.K.; Dahms, T.E.S.; Silva, R.N.; Singh, H.B.; Upadhyay, R.S.; Gomes, E.V.; Tsui, C.K.M.; Nayak, S.C. Friends or foes? Emerging insights from fungal interactions with plants. FEMS Microbiol. Rev. 2015, 40, 182–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X. Vegetation and Geographical Patterns in China; Geological Publishing House: Beijing, China, 2007; pp. 1–40. [Google Scholar]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, X.; Zang, R.; Fu, S.; Ai, X.; Yao, L.; Ding, Y.; Huang, J.; Lu, X. Functional recovery of a subtropical evergreen-deciduous broadleaved mixed forest following clear cutting in central China. Sci. Rep. 2018, 8, 16458. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, S.; Huang, Y.; Fu, S.; Wang, J.; Ming, A.; Li, X.; Yao, M.; Li, H. Tree species mixture inhibits soil organic carbon mineralization accompanied by decreased r-selected bacteria. Plant Soil 2018, 431, 203–216. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, Y.; Li, X.; Zhang, Y. Successional changes of fungal communities along the biocrust development stages. Biol. Fertil. Soils 2018, 54, 285–294. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Curd, E.E.; Martiny, J.B.H.; Li, H.; Smith, T.B. Bacterial diversity is positively correlated with soil heterogeneity. Ecosphere 2018, 9, e02079. [Google Scholar] [CrossRef]
- Borcard, D.; Legendre, P.; Avois-Jacquet, C.; Tuomisto, H. Dissecting the spatial structure of ecological data at multiple scales. Ecology 2004, 85, 1826–1832. [Google Scholar] [CrossRef]
- Griffith, D.A.; Peres-Neto, P.R. Spatial modeling in ecology: The flexibility of eigenfunction spatial analyses. Ecology 2006, 87, 2603–2613. [Google Scholar] [CrossRef]
- Blanchet, F.G.; Legendre, P.; Borcard, D. Forward selection of explanatory variables. Ecology 2008, 89, 2623–2632. [Google Scholar] [CrossRef] [PubMed]
- Peres-Neto, P.R.; Legendre, P.; Dray, S.; Borcard, D. Variation partitioning of species data matrices: Estimation and comparison of fractions. Ecology 2006, 87, 2614–2625. [Google Scholar] [CrossRef]
- Lichstein, J.W. Multiple regression on distance matrices: A multivariate spatial analysis tool. Plant Ecol. 2007, 188, 117–131. [Google Scholar] [CrossRef]
- Friedman, J.; Alm, E.J. Inferring Correlation Networks from Genomic Survey Data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. In Proceedings of the Third international AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Dini-Andreote, F.; Silva, M.D.; Triadó-Margarit, X.; Casamayor, E.O.; Van elsas, J.D.; Salles, J.F. Dynamics of bacterial community succession in a salt marsh chronosequence: Evidences for temporal niche partitioning. ISME J. 2014, 8, 1989–2001. [Google Scholar] [CrossRef]
- Veresoglou, S.D.; Liu, L.; Xu, T.; Rillig, M.C.; Wang, M.; Wang, J.; Chen, Y.; Hu, Y.; Hao, Z.; Chen, B. Biogeographical constraints in Glomeromycotinan distribution across forest habitats in China. J. Ecol. 2019, 107, 684–695. [Google Scholar] [CrossRef]
- Martiny, J.B.H.; Bohannan, B.J.; Brown, J.H.; Colwell, R.K.; Fuhrman, J.A.; Green, J.L.; Horner–Devine, M.C.; Kane, M.; Krumins, J.A.; Kuske, C.R. Microbial biogeography: Putting microorganisms on the map. Nat. Rev. Microbiol. 2006, 4, 102. [Google Scholar] [CrossRef]
- Chesson, P. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 2000, 31, 343–366. [Google Scholar] [CrossRef]
- Xue, Y.; Chen, H.; Liu, M.; Huang, B.; Yang, J.R.; Yang, J. Distinct patterns and processes of abundant and rare eukaryotic plankton communities following a reservoir cyanobacterial bloom. ISME J. 2018, 12, 2263–2277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, S.; Powell, J.R.; Drigo, B.; Cairney, J.W.; Anderson, I.C. The role of stochasticity differs in the assembly of soil- and root-associated fungal communities. Soil Biol. Biochem. 2015, 80, 18–25. [Google Scholar] [CrossRef]
- Graham, C.H.; Fine, P.V.A. Phylogenetic beta diversity: Linking ecological and evolutionary processes across space in time. Ecol. Lett. 2008, 11, 1265–1277. [Google Scholar] [CrossRef]
- Caruso, T.; Chan, Y.; Lacap, D.C.; Lau, M.C.Y.; McKay, C.P.; Pointing, S.B. Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME J. 2011, 5, 1406–1413. [Google Scholar] [CrossRef] [PubMed]
- Kraus, T.E.C.; Dahlgren, R.A.; Zasoski, R.J. Tannins in nutrient dynamics of forest ecosystems—A review. Plant Soil 2003, 256, 41–66. [Google Scholar] [CrossRef]
- Courty, P.E.; Franc, A.; Garbaye, J. Temporal and functional pattern of secreted enzyme activities in an ectomycorrhizal community. Soil Biol. Biochem. 2010, 42, 2022–2025. [Google Scholar] [CrossRef]
- Courty, P.E.; Pritsch, K.; Schloter, M.; Hartmann, A.; Garbaye, J. Activity profiling of ectomycorrhiza communities in two forest soils using multiple enzymatic tests. New Phytol. 2005, 167, 309–319. [Google Scholar] [CrossRef]
- Grosso, F.; Bååth, E.; De Nicola, F. Bacterial and fungal growth on different plant litter in Mediterranean soils: Effects of C/N ratio and soil pH. Appl. Soil Ecol. 2016, 108, 1–7. [Google Scholar] [CrossRef]
- Nicolas, P.B.; Richard, C.; Samuel, D.; Christophe, M.; Mélanie, L.; Claudy, J.; Hamid Reza, S.; Laure, G.; Dominique, A.; Lionel, R. Validation and application of a PCR primer set to quantify fungal communities in the soil environment by real–time quantitative PCR. PLoS ONE 2011, 6, e24166. [Google Scholar]
- Johannes, R.; Brookes, P.C.; Erland, B.T. Fungal and bacterial growth responses to N fertilization and pH in the 150-year ‘Park Grass’ UK grassland experiment. FEMS Microbiol. Ecol. 2015, 76, 89–99. [Google Scholar]
- Ni, Y.; Yang, T.; Zhang, K.; Shen, C.; Chu, H. Fungal Communities Along a Small-Scale Elevational Gradient in an Alpine Tundra Are Determined by Soil Carbon Nitrogen Ratios. Front. Microbiol. 2018, 9, 1815. [Google Scholar] [CrossRef] [PubMed]
- Barberán, A.; McGuire, K.L.; Wolf, J.A.; Jones, F.A.; Wright, S.J.; Turner, B.L.; Essene, A.; Hubbell, S.P.; Faircloth, B.C.; Fierer, N. Relating belowground microbial composition to the taxonomic, phylogenetic, and functional trait distributions of trees in a tropical forest. Ecol. Lett. 2015, 18, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Brookes, P.C.; Bååth, E. Investigating the mechanisms for the opposing pH relationships of fungal and bacterial growth in soil. Soil Biol. Biochem. 2010, 42, 926–934. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leprince, F.; Quiquampoix, H. Extracellular enzyme activity in soil: Effect of pH and ionic strength on the interaction with montmorillonite of two acid phosphatases secreted by the ectomycorrhizal fungus Hebeloma cylindrosporum. Eur. J. Soil Sci. 1996, 47, 511–522. [Google Scholar] [CrossRef]
- Yamanaka, T. The Effect of pH on the Growth of Saprotrophic and Ectomycorrhizal Ammonia Fungi in vitro. Mycologia 2003, 95, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.L.; Mortimer, P.E.; Slik, J.F.; Zou, X.M.; Xu, J.; Feng, W.T.; Qiao, L. Variation in forest soil fungal diversity along a latitudinal gradient. Fungal Divers. 2014, 64, 305–315. [Google Scholar] [CrossRef]
- Auguet, J.C.; Barberan, A.; Casamayor, E.O. Global ecological patterns in uncultured Archaea. ISME J. 2010, 4, 182. [Google Scholar] [CrossRef]
- Lienhard, P.; Tivet, F.; Chabanne, A.; Dequiedt, S.; Lelièvre, M.; Sayphoummie, S.; Leudphanane, B.; Prévost-Bouré, N.C.; Séguy, L.; Maron, P.A. No-till and cover crops shift soil microbial abundance and diversity in Laos tropical grasslands. Agron. Sustain. Dev. 2013, 33, 375–384. [Google Scholar] [CrossRef]
- Talbot, J.M.; Bruns, T.D.; Taylor, J.W.; Smith, D.P.; Branco, S.; Glassman, S.I.; Erlandson, S.; Vilgalys, R.; Liao, H.L.; Smith, M.E.; et al. Endemism and functional convergence across the North American soil mycobiome. Proc. Natl. Acad. Sci. USA 2014, 111, 6341–6346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, J.L.; Holmes, A.J.; Westoby, M.; Oliver, I.; Briscoe, D.; Dangerfield, M.; Gillings, M.; Beattie, A.J. Spatial scaling of microbial eukaryote diversity. Nature 2004, 432, 747–750. [Google Scholar] [CrossRef] [PubMed]
- Soininen, J.; Mcdonald, R.; Hillebrand, H. The distance decay of similarity in ecological communities. Ecography 2010, 30, 3–12. [Google Scholar] [CrossRef]
- Hanna, T.; Kalle, R.; Markku, Y.H. Dispersal, environment, and floristic variation of western Amazonian forests. Science 2003, 299, 241–244. [Google Scholar]
- Crist, T.O. The spatial distribution of termites in shortgrass steppe: A geostatistical approach. Oecologia 1998, 114, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Finlay, B.J. Global Dispersal of Free-Living Microbial Eukaryote Species. Science 2002, 296, 1061–1063. [Google Scholar] [CrossRef] [Green Version]
- Lekberg, Y.; Koide, R.T.; Rohr, J.R.; Morton, J.B.; Aldrich-Wolfe, L. Role of niche restrictions and dispersal in the composition of arbuscular mycorrhizal fungal communities. J. Ecol. 2007, 95, 95–105. [Google Scholar] [CrossRef]
- De Menezes, A.B.; Prendergast-Miller, M.T.; Richardson, A.E.; Toscas, P.; Farrell, M.; Macdonald, L.M.; Baker, G.; Wark, T.; Thrall, P.H. Network analysis reveals that bacteria and fungi form modules that correlate independently with soil parameters. Environ. Microbiol. 2015, 17, 2677–2689. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Genet. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Ma, B.; Wang, H.; Dsouza, M.; Lou, J.; He, Y.; Dai, Z.; Brookes, P.C.; Xu, J.; Gilbert, J.A. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J. 2016, 10, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Legendre, P.; Mi, X.; Ren, H.; Ma, K.; Yu, M.; Sun, I.F.; He, F. Partitioning beta diversity in a subtropical broad-leaved forest of China. Ecology 2009, 90, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Davison, J.; Öpik, M.; Daniell, T.J.; Moora, M.; Zobel, M. Arbuscular mycorrhizal fungal communities in plant roots are not random assemblages. FEMS Microbiol. Ecol. 2011, 78, 103–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Site | Forest Type | Dominant Tree Species | Location |
|---|---|---|---|
| JF | Tropical rain forest | Gironniera subaequalis Planch. | 18°44′22″ N 108°51′59″ E |
| Cryptocarya chinensis Hemsl.(Lauraceae) | |||
| DH | Subtropical evergreen broad-leaved forest | Schima superba (Theaceae) Castanea henryi (Skan) Rehd. et Wils | 23°10′03″ N 112°10′01″ E |
| BT | Subtropical evergreen-deciduous broad-leaved mixed forest | Quercus aliena .var. acuteserrata Maxim. ex Wenz. Quercus variabilis Blume | 33°29′32″ N 111°55′32″ E |
| DL | Warm-temperate deciduous broad-leaved forest | Quercus wutaishanica Mayr | 39°57′27″ N 115°25′29″ E |
| CB | Temperate needle-leaf and deciduous broad-leaved mixed forest | Pinus koraiensis Sieb. et Zucc. | 42°20′51″ N 128°8′55″ E |
| Betula platyphylla Suk | |||
| MH | Cold-temperate deciduous needle-leaf forest | Larix gmelinii (Ruprecht) Kuzeneva | 53°27′46″ N 122°20′20″ E |
| Betula platyphylla Suk |
| Environmental Variable | TRF | SEB | SED | WDB | TDB | CDN | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ρ | p | ρ | p | ρ | p | ρ | p | ρ | p | ρ | p | |
| MAP | 0.05 | n.s. | 0.04 | n.s. | 0.07 | n.s. | 0.06 | n.s. | 0.02 | n.s. | 0.06 | n.s. |
| MAT | 0.03 | n.s. | 0.07 | n.s. | 0.08 | n.s. | 0.04 | n.s. | 0.01 | n.s. | 0.04 | n.s. |
| Elevation | 0.09 | n.s. | 0.02 | n.s. | 0.05 | n.s. | 0.04 | n.s. | 0.06 | n.s. | 0.02 | n.s. |
| Slope | 0.1 | n.s. | 0.03 | n.s. | 0.02 | n.s. | 0.05 | n.s. | 0.04 | n.s. | 0.01 | n.s. |
| Soil pH | 0.22 | 0.02 | 0.08 | n.s. | 0.04 | n.s. | 0.1 | n.s. | 0.16 | 0.04 | 0.03 | n.s. |
| SWC | 0.09 | n.s. | 0.07 | n.s. | 0.12 | n.s. | 0.04 | n.s. | 0.06 | n.s. | 0.08 | n.s. |
| SOC | 0.28 | 0.001 | 0.23 | 0.02 | 0.08 | n.s. | 0.01 | n.s. | 0.02 | n.s. | 0.22 | 0.02 |
| STN | 0.25 | 0.01 | 0.08 | n.s. | 0.24 | 0.01 | 0.1 | n.s. | 0.15 | 0.04 | 0.09 | n.s. |
| SAN | 0.05 | n.s. | 0.25 | 0.01 | 0.23 | 0.01 | 0.21 | 0.02 | 0.04 | n.s. | 0.05 | n.s. |
| Soil C/N | 0.09 | n.s. | 0.14 | 0.04 | 0.2 | 0.02 | 0.09 | n.s. | 0.01 | n.s. | 0.19 | 0.02 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Zhang, X.; Fu, S.; Zhang, W. Environmental Filtering Drives Local Soil Fungal Beta Diversity More Than Dispersal Limitation in Six Forest Types along a Latitudinal Gradient in Eastern China. Forests 2019, 10, 863. https://doi.org/10.3390/f10100863
Huang Y, Zhang X, Fu S, Zhang W. Environmental Filtering Drives Local Soil Fungal Beta Diversity More Than Dispersal Limitation in Six Forest Types along a Latitudinal Gradient in Eastern China. Forests. 2019; 10(10):863. https://doi.org/10.3390/f10100863
Chicago/Turabian StyleHuang, Yongtao, Xiao Zhang, Shenglei Fu, and Weixin Zhang. 2019. "Environmental Filtering Drives Local Soil Fungal Beta Diversity More Than Dispersal Limitation in Six Forest Types along a Latitudinal Gradient in Eastern China" Forests 10, no. 10: 863. https://doi.org/10.3390/f10100863
APA StyleHuang, Y., Zhang, X., Fu, S., & Zhang, W. (2019). Environmental Filtering Drives Local Soil Fungal Beta Diversity More Than Dispersal Limitation in Six Forest Types along a Latitudinal Gradient in Eastern China. Forests, 10(10), 863. https://doi.org/10.3390/f10100863




