Pulmonary Nodule Detection from X-ray CT Images Based on Region Shape Analysis and Appearance-based Clustering
Abstract
:1. Introduction
2. Initial Detection of Nodule Candidates
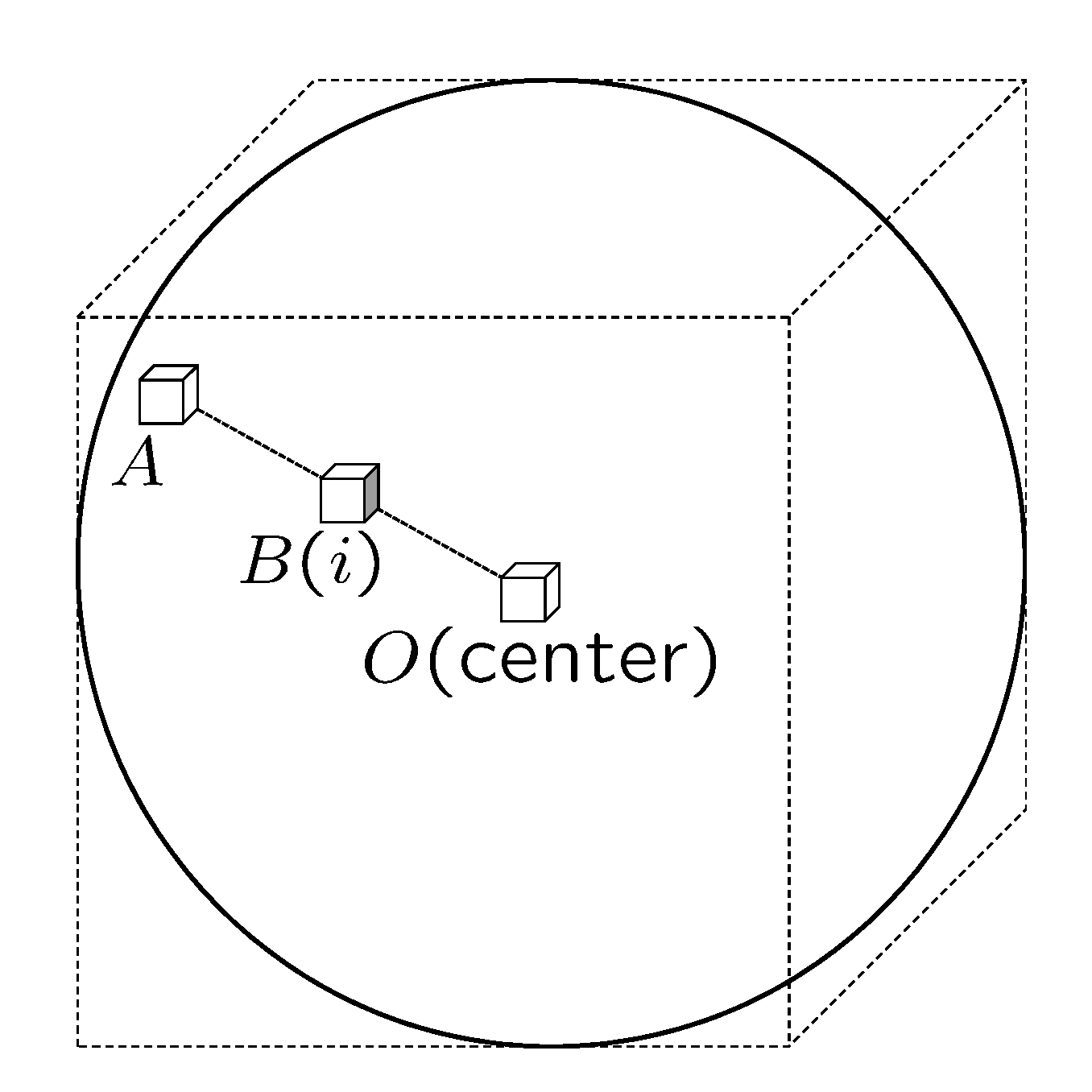
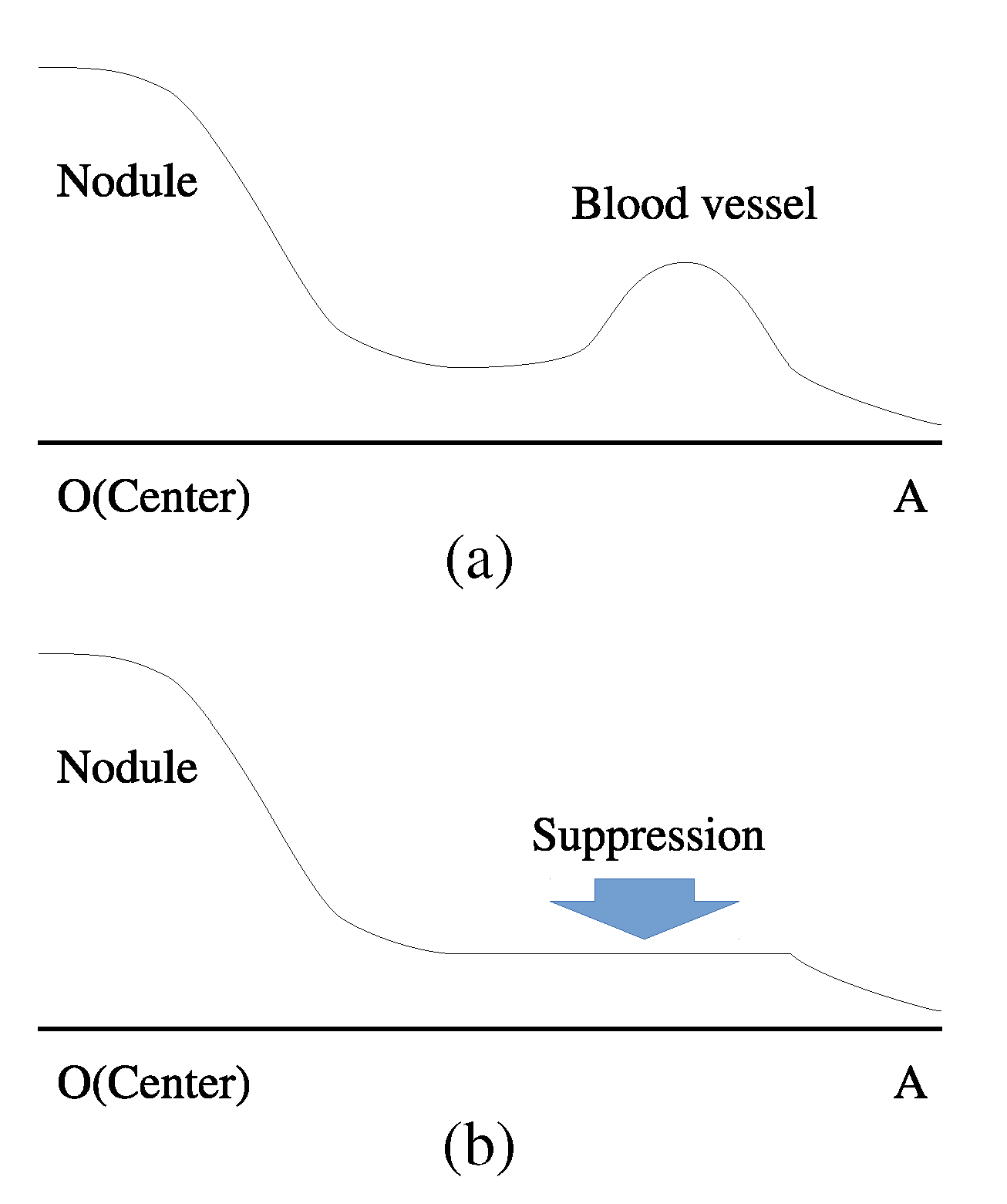
2.1. Radial Suppression Filter


2.2. Moment-of-Inertia Filter
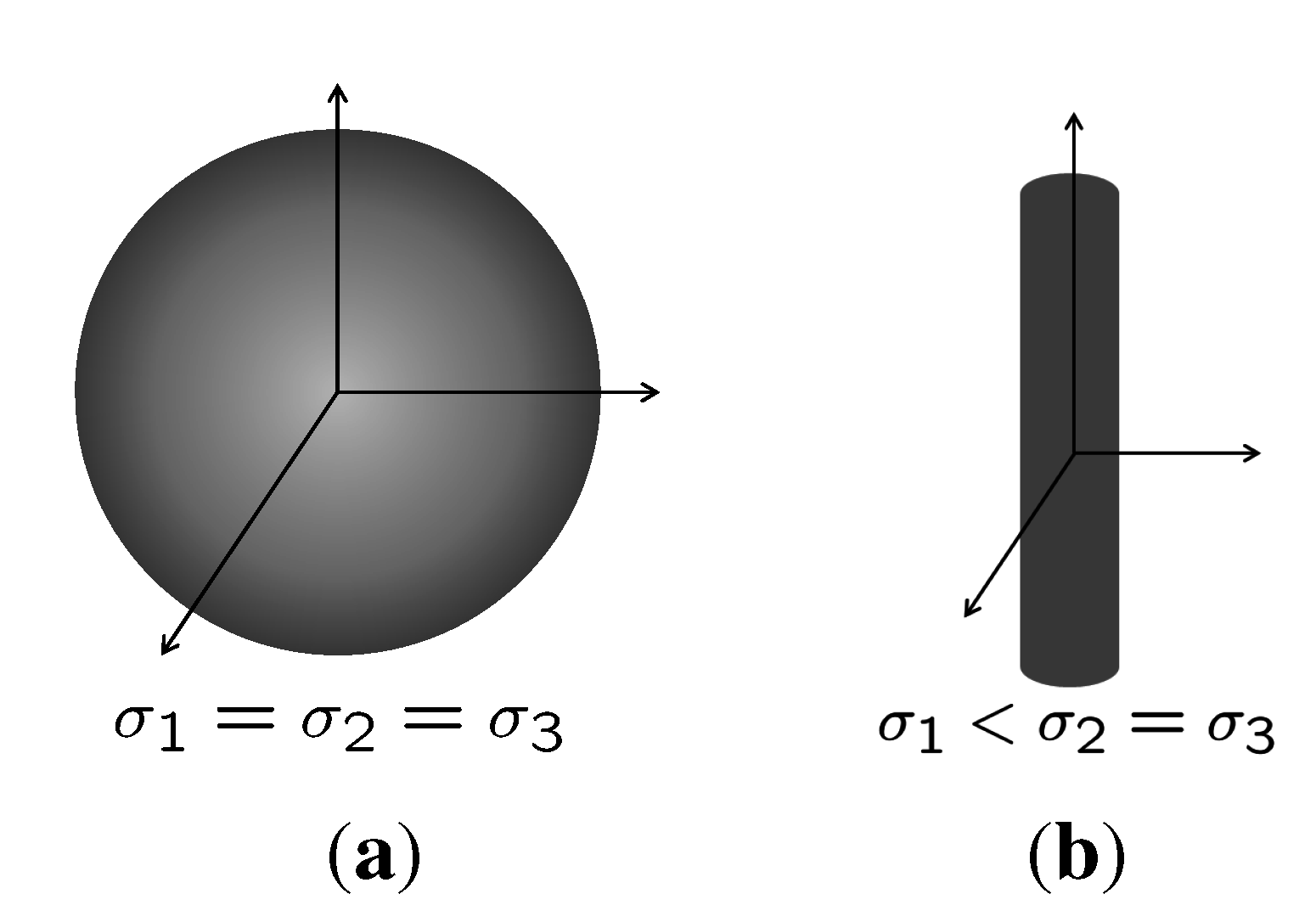
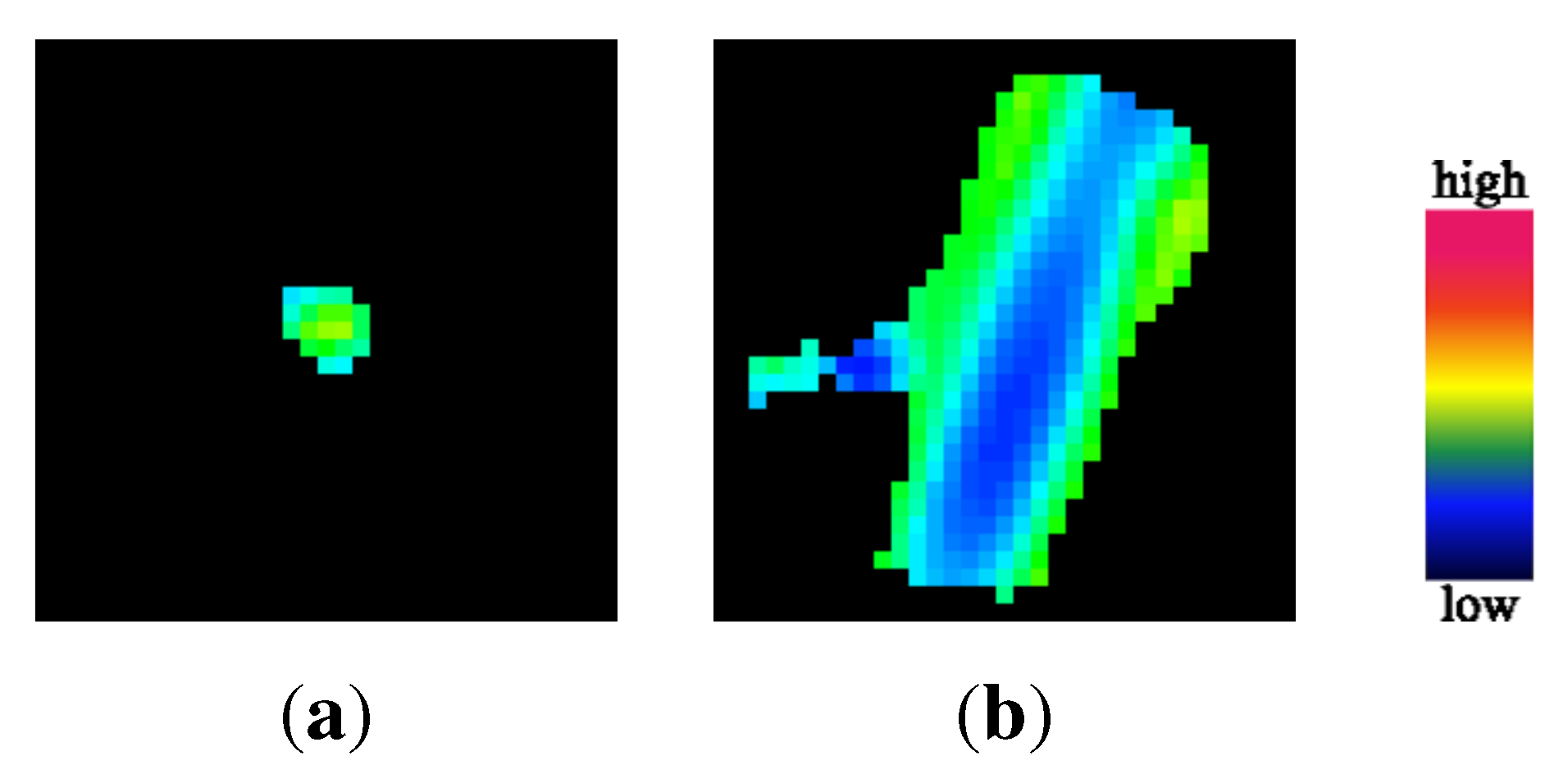
2.3. Center Displacement Filter
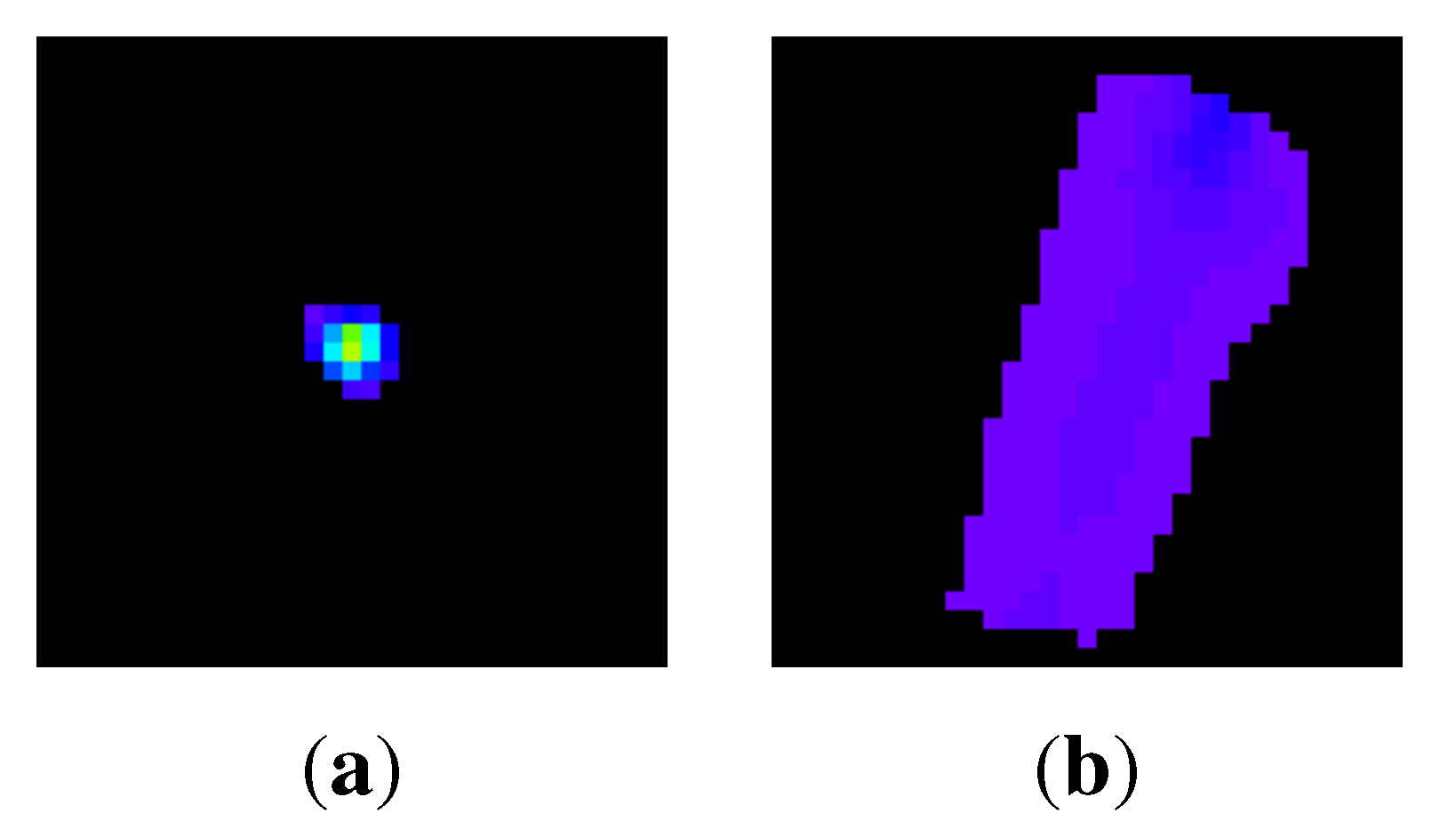
3. False positive reduction
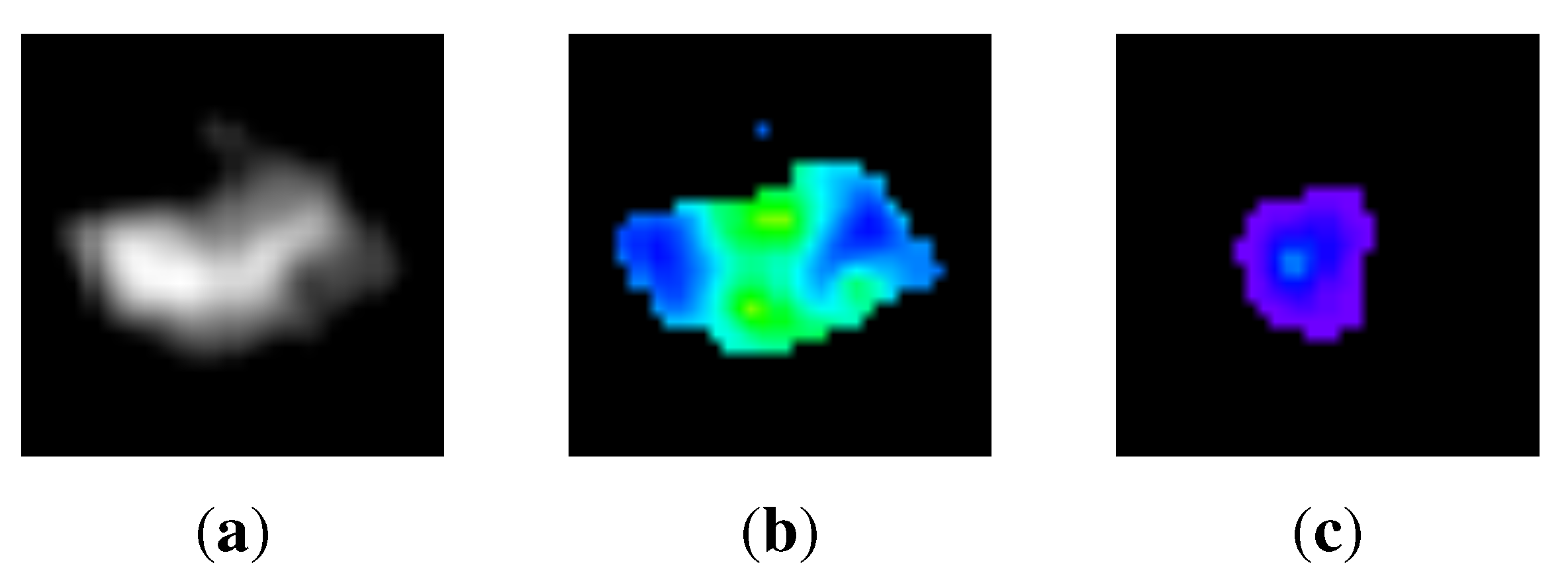
3.1. Normalization of directions of intensity distributions
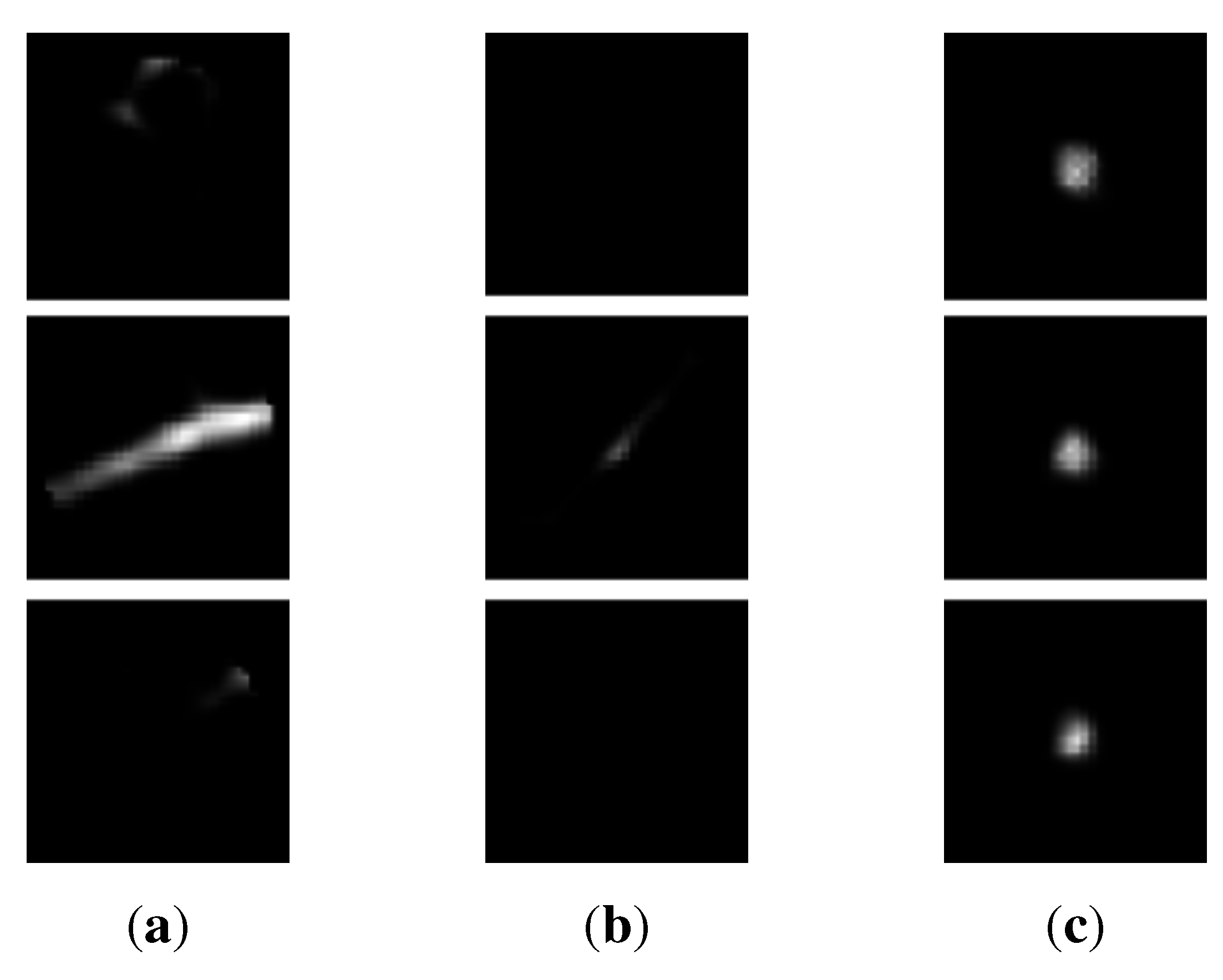
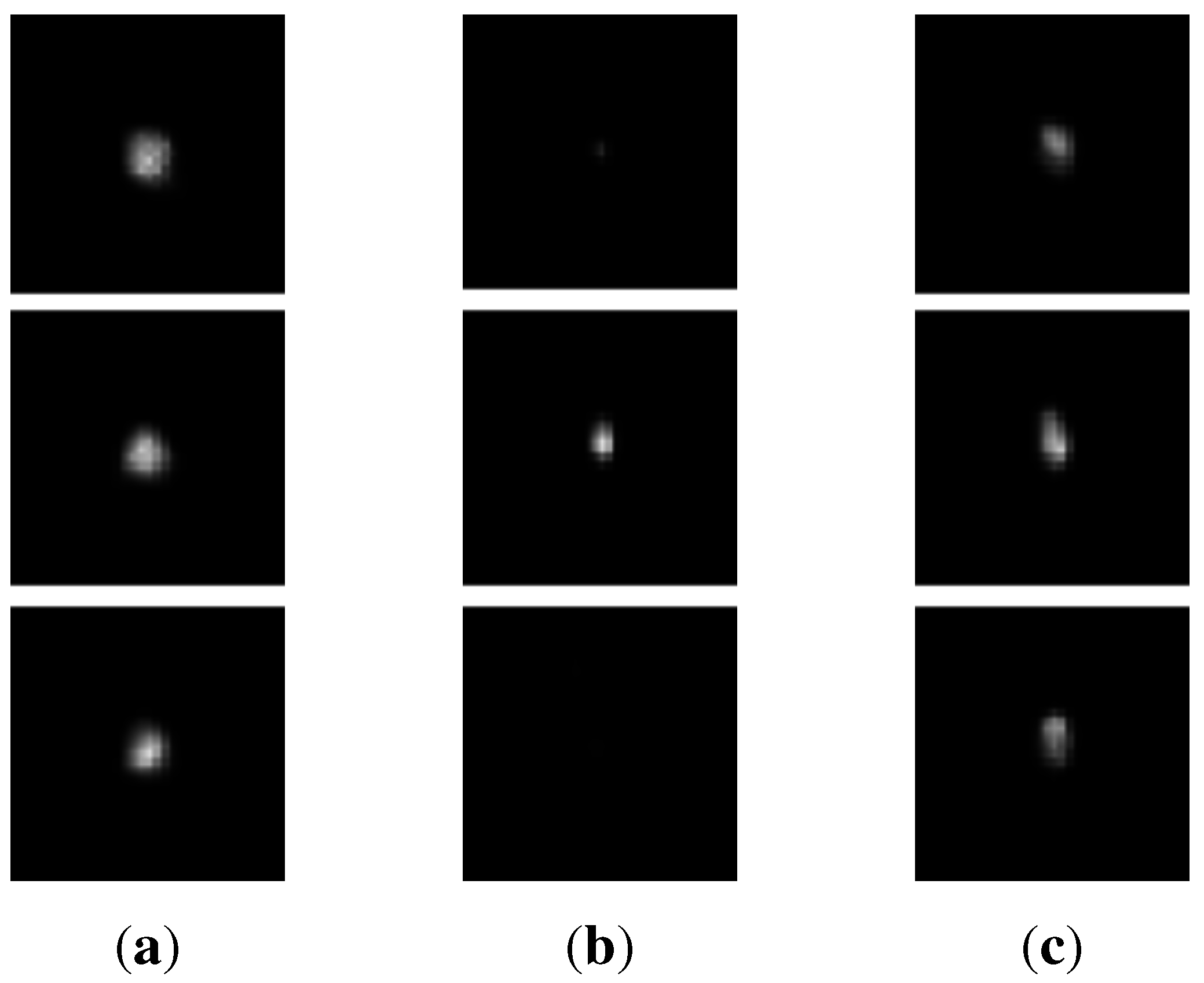
3.2. Appearance-Based k-means Clustering
3.2.1. Training Phase
3.2.2. Testing Phase
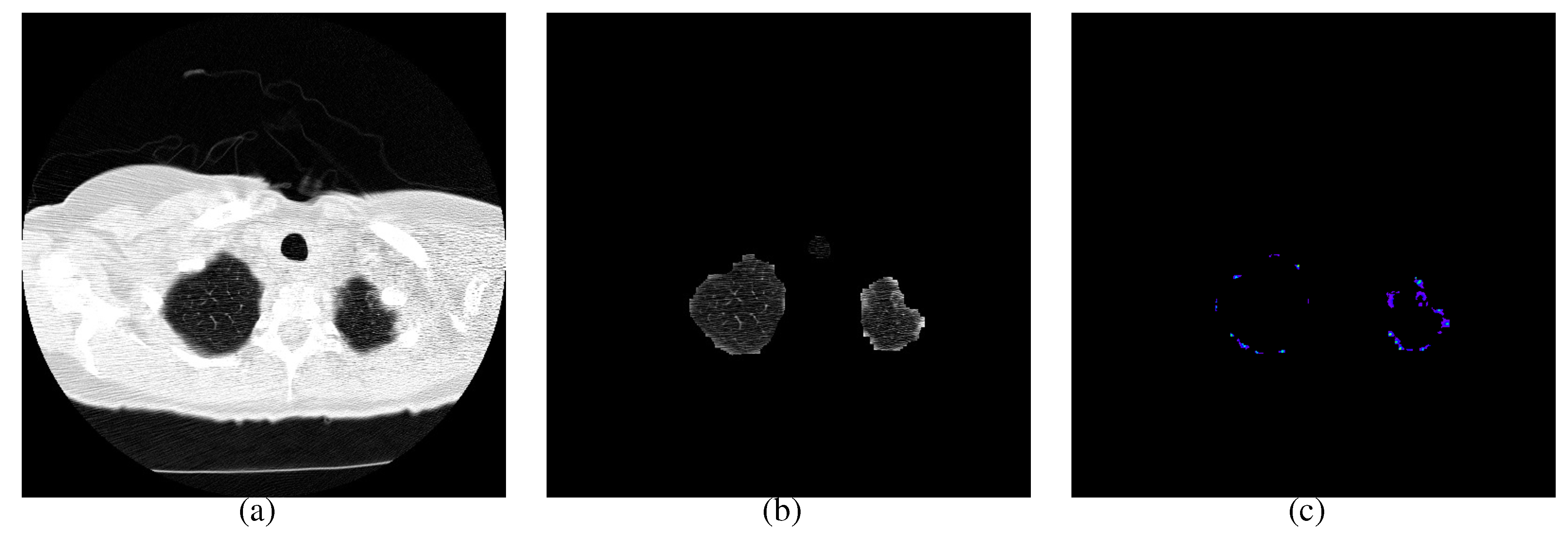
4. Experimental Results
4.1. Experimental Conditions
4.2. Initial Nodule Detection
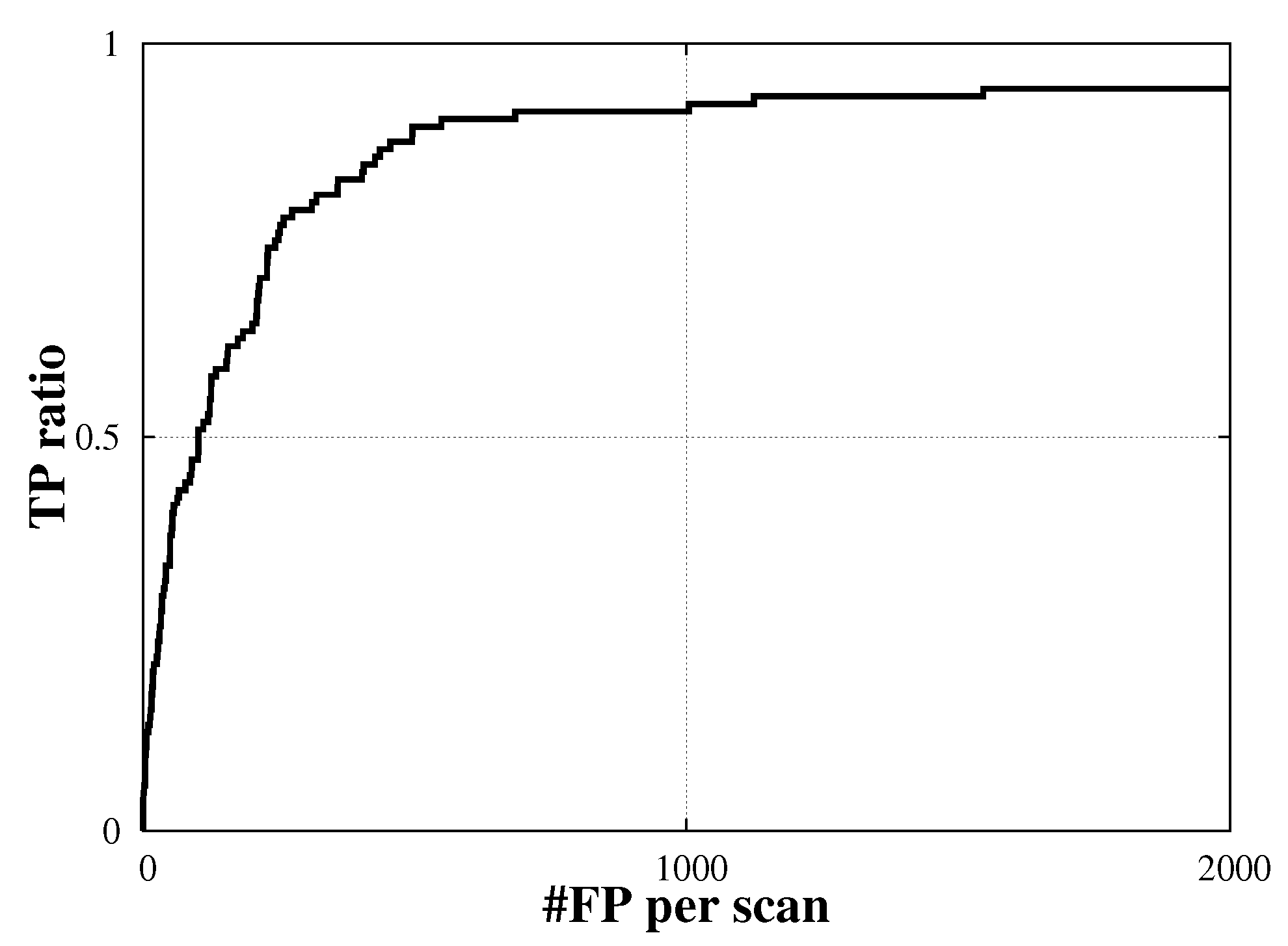
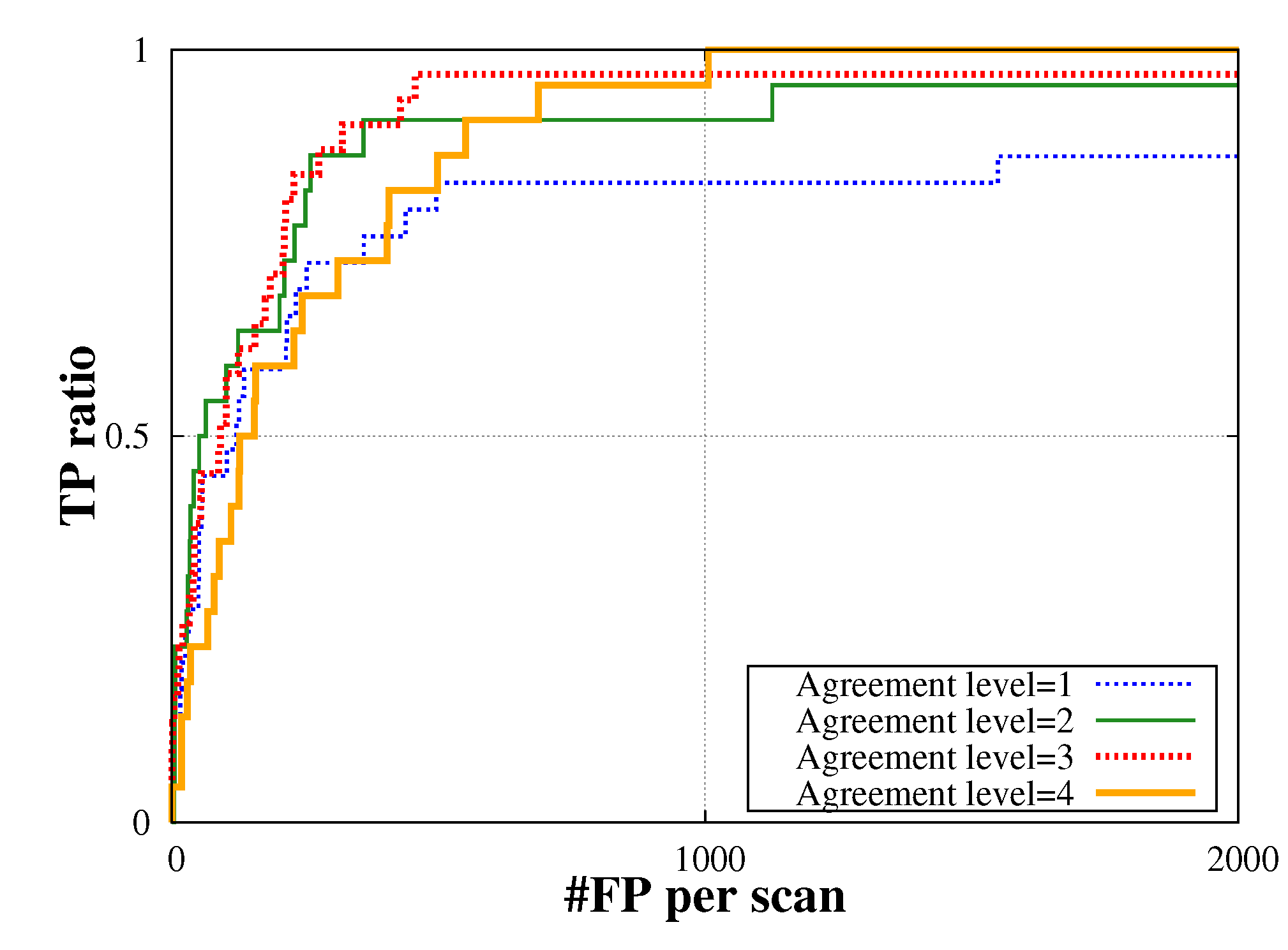
4.3. False Positive Reduction
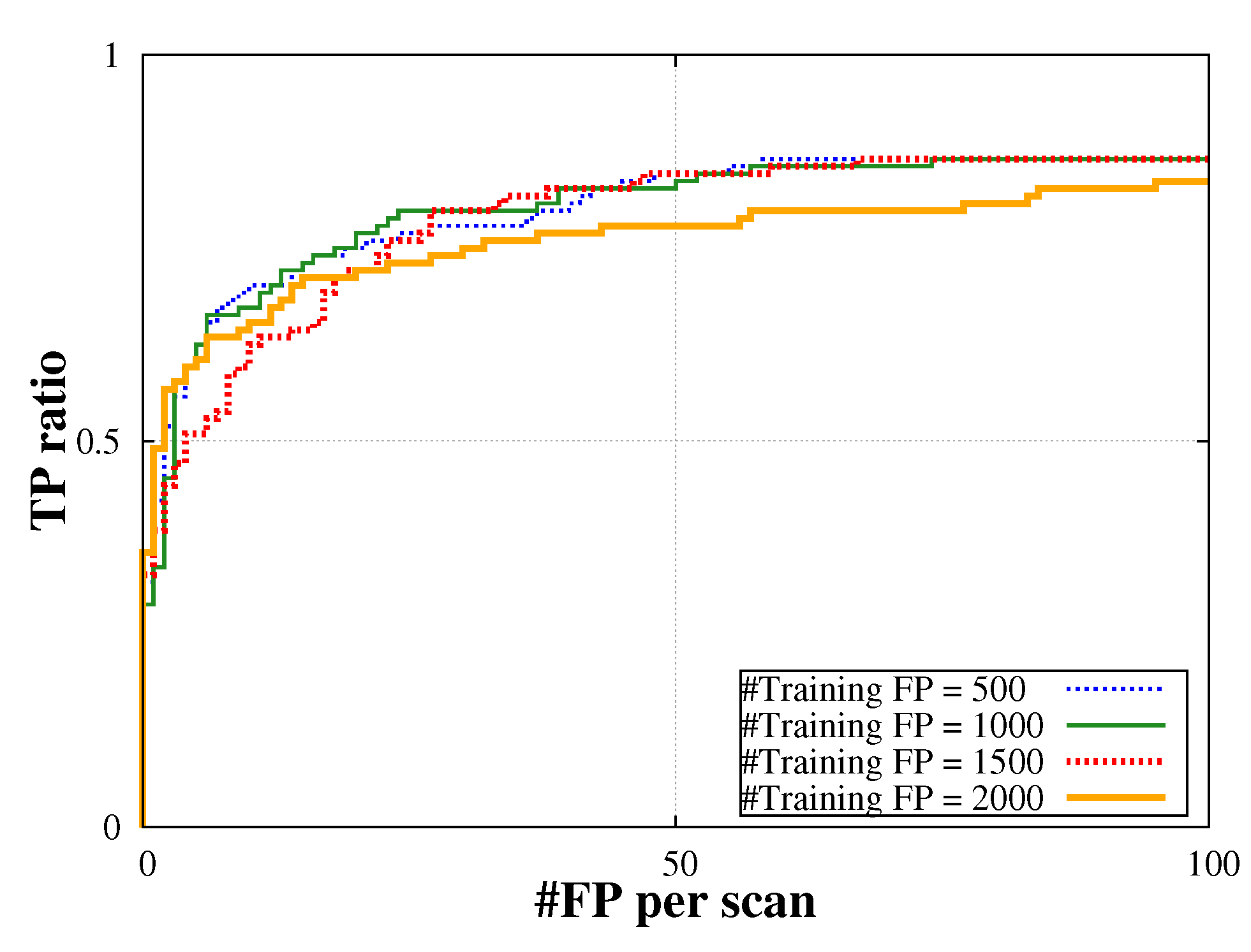
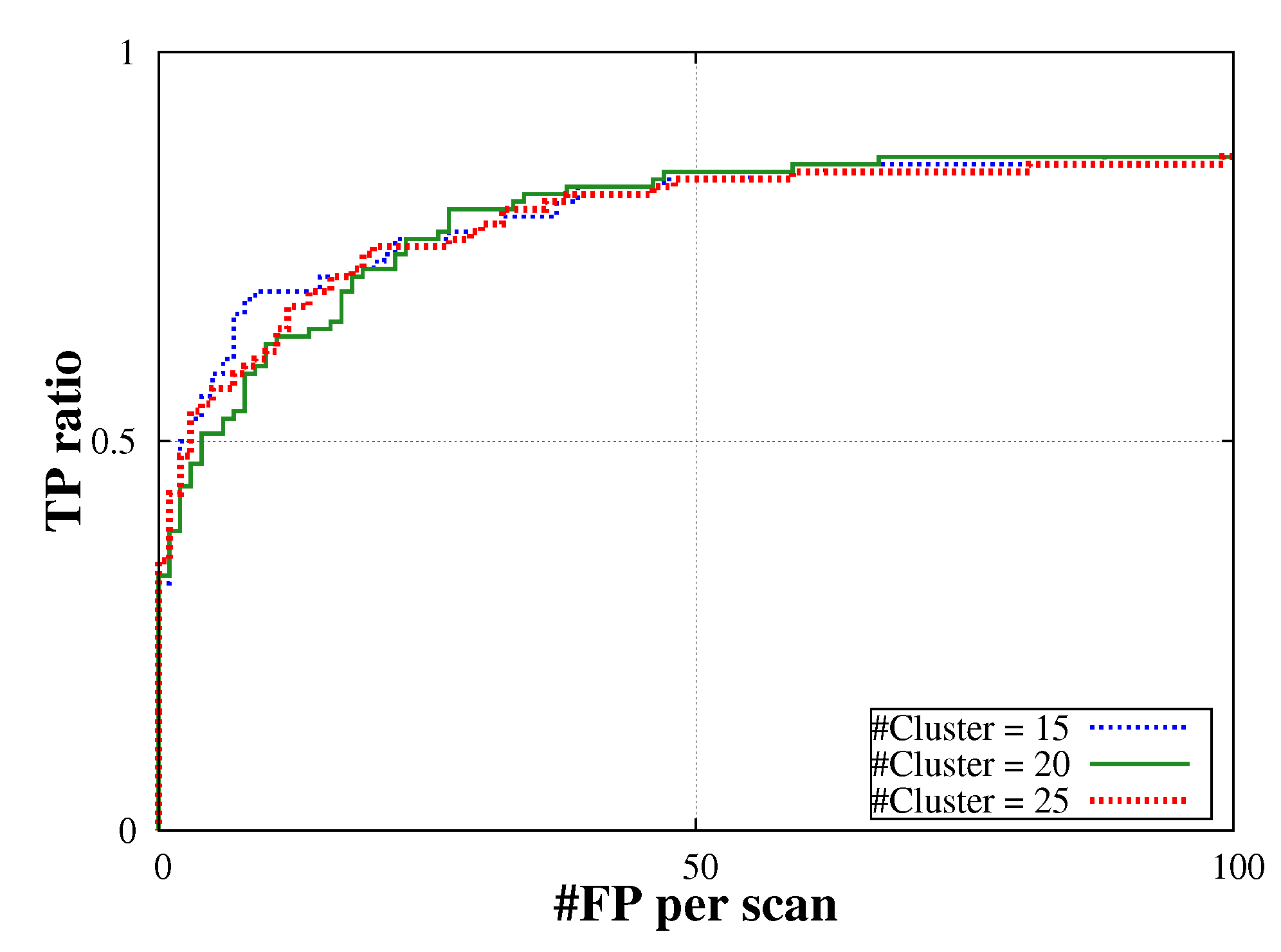
5.Discussion
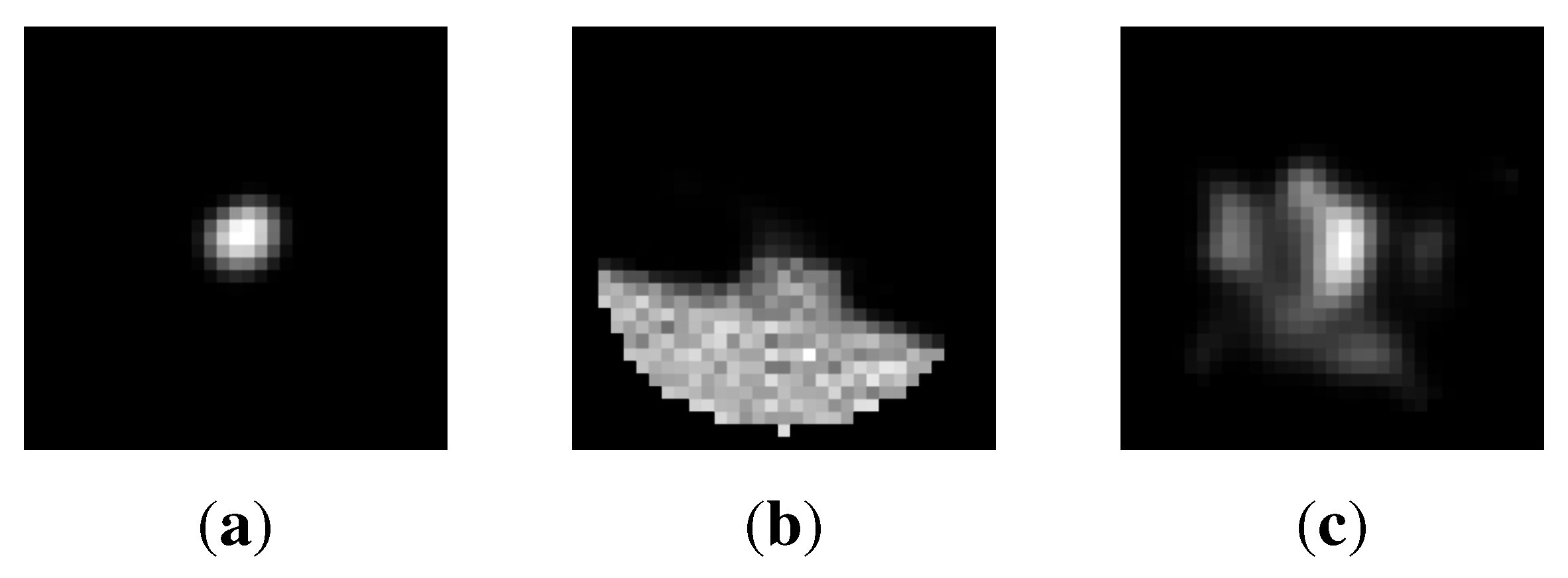
6. Conclusion
Author Contributions
Conflicts of Interest
References
- Yamamoto, S.; Tanaka, I.; Senda, M.; Tateno, Y.; Iinuma, T.; Matsumoto, T.; Matsumoto, M. Image Processing for Computer-Aided Diagnosis of Lung Cancer by CT(LSCT). Syst. Comput. Jpn. 1994, 25, 67–80. [Google Scholar] [CrossRef]
- Sone, S.; Matsumoto, T.; Honda, T.; Tsushima, K.; Takayama, F.; Hanaoka, T.; Kondo, R.; Haniuda, M. HRCT Features of Small Peripheral Lung Carcinomas Detected in a Low-dose CT Screening Program. Acad. Radiol. 2010, 17, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Van Ginneken, B. Computer-Aided Diagnosis in Thoracic Computed Tomography. Imaging Decis. 2009, 12, 11–22. [Google Scholar] [CrossRef]
- Yamamoto, S.; Matsumoto, M.; Takano, Y.; Iinuma, T.; Matsumoto, T. Quoit Filter : A New Filter Based on Mathematical Morphology to Extract the Isolated Shadow, and Its Application to Automatic Detection of Lung Cancer in X-ray CT. In Proceedings of the 13th International Conference on Pattern Recognition, 25–29 August 1996; Volume 2, pp. 3–7.
- Messay, T.; Hardie, R.C.; Rogers, S.K. A new computationally efficient CAD system for pulmonary nodule detection in CT imagery. Med. Image Anal. 2010, 14, 390–406. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Hara, T.; Fujita, H.; Itoh, S.; Ishigaki, T. Automated Detection of Pulmonary Nodules in Helical CT Images Based on an Improved Template-Matching Technique. IEEE Trans. Med. Imaging 2001, 20, 595–604. [Google Scholar] [PubMed]
- Tan, M.; Bister, M.; Cornelis, J. A novel computer-aided lung nodule detection system for CT images. Med. Phys. 2011, 38, 5630–5645. [Google Scholar] [CrossRef] [PubMed]
- Stanley, K.O.; Miikkulainen, R. Evolving Neural Networks through Augmenting Topologies. Evolut. Comput. 2002, 10, 99–127. [Google Scholar] [CrossRef] [PubMed]
- Ozekes, S.; Osman, P. Computerized Lung Nodule Detection Using 3D Feature Extraction and Learning Based Algorithms. J. Med. Syst. 2010, 34, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.L.A.; Kouzani, A.Z.; Hu, E.J. Random forest based lung nodule classification aided by clustering. Comput. Med. Imaging Graph. 2010, 34, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.J.; Choi, T.S. Genetic programming-based feature transform and classification for the automatic detection of pulmonary nodules on computed tomography images. Inf. Sci. 2012, 212, 57–78. [Google Scholar] [CrossRef]
- Ye, X.; Lin, X.; Dehmeshki, J.; Slabaugh, G.; Beddoe, G. Shape-based computer-aided detection of lung nodules in thoracic CT images. IEEE Eng. Med. Biol. Soc. 2009, 56, 1810–1820. [Google Scholar]
- Brown, M.; McNitt-Gray, M.; Goldin, J.; Suh, R.; Sayre, J.; Aberle, D. Patient-specific models for lung nodule detection and surveillance in CT images. IEEE Trans. Med. Imaging 2001, 20, 1242–1250. [Google Scholar] [CrossRef] [PubMed]
- Osman, O.; Ozekes, S.; Ucan, O.N. Lung nodule diagnosis using 3D template matching. Comput. Biol. Med. 2007, 37, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Paik, D.S.; Beaulieu, C.F.; Rubin, G.D.; Acar, B.; Jeffrey, J.; Yee, J.; Dey, J.; Napel, S. Surface normal overlap: A computer-aided detection algorithm with application to colonic polyps and lung nodules in helical CT. IEEE Trans. Med. Imaging 2004, 23, 661–675. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Cuenca, J.; Tahoces, P.; Souto, M.; Lado, M.; Remy-Jardin, M.; Remy, J.; Vidal, J.J. Application of the iris filter for automatic detection of pulmonary nodules on computed tomography images. Comput. Biol. Med. 2009, 39, 921–933. [Google Scholar] [CrossRef] [PubMed]
- Golosio, B.; Masala, G.L.; Piccioli, A.; Oliva, P.; Carpinelli, M.; Cataldo, R.; Cerello, P.; DeCarlo, F.; Falaschi, F.; Fantacci, M.E.; et al. A novel multithreshold method for nodule detection in lung CT. Med. Phys. 2009, 36, 3607–3618. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Armato, S.G., 3rd; Li, F.; Sone, S.; Doi, K. Massive training artificial neural network (MTANN) for reduction of false positives in computerized detection of lung nodules in low-dose computed tomography. Med. Phys. 2003, 30, 1602–1617. [Google Scholar] [CrossRef] [PubMed]
- Armato, S.G.; McLennan, G.; McNitt-Gray, M.F.; Meyer, C.R.; Yankelevitz, D.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; Kazerooni, E.A.; MacMahon, H.; et al. Lung image database consortium: Developing a resource for the medical imaging research community. Radiology 2004, 232, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Takizawa, H.; Nishizako, H. Lung Cancer Detection from X-ray CT Scans Using Discriminant Filters and View-based Support Vector Machine. J. Insti. Image Electron. Eng. Jpn. 2011, 40, 59–66. [Google Scholar]
- Fukano, G.; Nakamura, Y.; Takizawa, H.; Mizuno, S.; Yamamoto, S.; Doi, K.; Katsuragawa, S.; Matsumoto, T.; Tateno, Y.; Iinuma, T. Eigen Image Recognition of Pulmonary Nodules from Thoracic CT Images by Use of Subspace Method. IEICE Trans. Inf. Syst. 2005, E88-D-II, 1273–1283. [Google Scholar] [CrossRef]
- Takizawa, H.; Yamamoto, S.; Shiina, T. Recognition of Pulmonary Nodules in Thoracic CT Scans Using 3-D Deformable Object Models of Different Classes. Algorithms 2010, 3, 125–144. [Google Scholar] [CrossRef]
- Forsyth, D.A.; Ponce, J. Computer Vision: A Modern Approach; Prentice Hall: Upper Saddle River, NJ, USA, 2003. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yanagihara, T.; Takizawa, H. Pulmonary Nodule Detection from X-ray CT Images Based on Region Shape Analysis and Appearance-based Clustering. Algorithms 2015, 8, 209-223. https://doi.org/10.3390/a8020209
Yanagihara T, Takizawa H. Pulmonary Nodule Detection from X-ray CT Images Based on Region Shape Analysis and Appearance-based Clustering. Algorithms. 2015; 8(2):209-223. https://doi.org/10.3390/a8020209
Chicago/Turabian StyleYanagihara, Takanobu, and Hotaka Takizawa. 2015. "Pulmonary Nodule Detection from X-ray CT Images Based on Region Shape Analysis and Appearance-based Clustering" Algorithms 8, no. 2: 209-223. https://doi.org/10.3390/a8020209
APA StyleYanagihara, T., & Takizawa, H. (2015). Pulmonary Nodule Detection from X-ray CT Images Based on Region Shape Analysis and Appearance-based Clustering. Algorithms, 8(2), 209-223. https://doi.org/10.3390/a8020209





