A Novel Algorithm for Macromolecular Epitope Matching
Abstract
:1. Introduction
2. Results and Discussion
2.1. Description of problem and assessment of problem size
2.2. Algorithm
Rationale
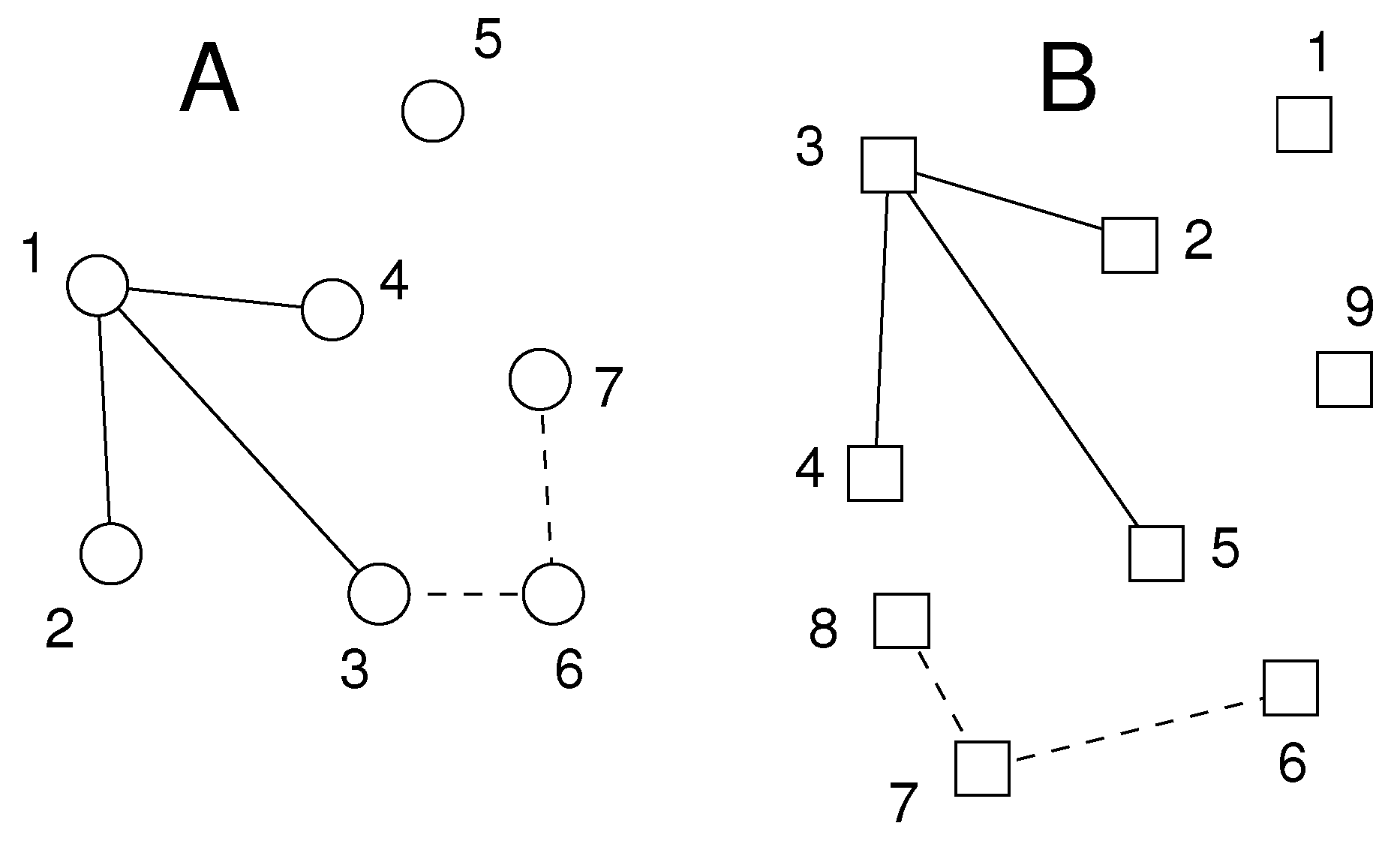

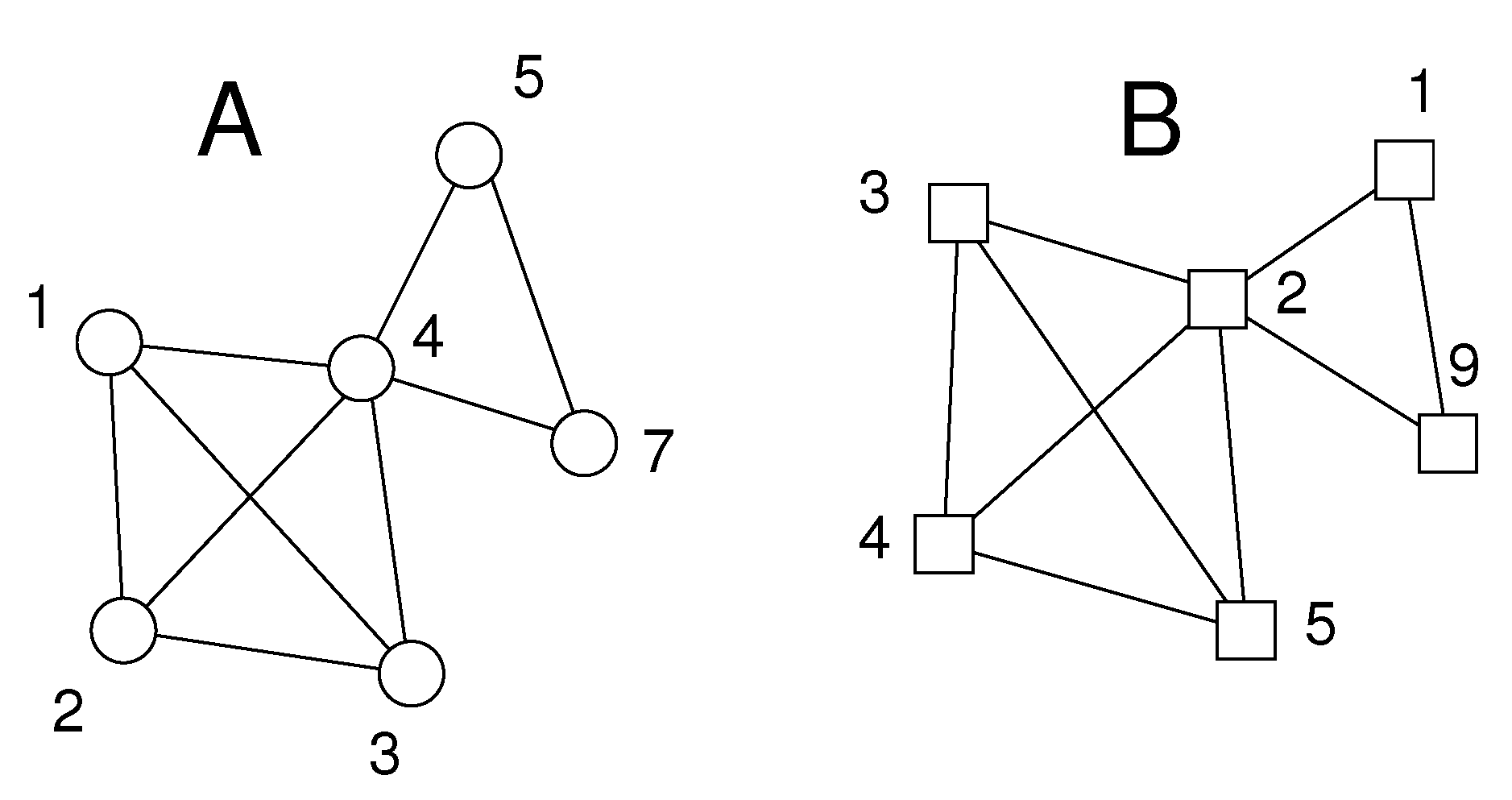
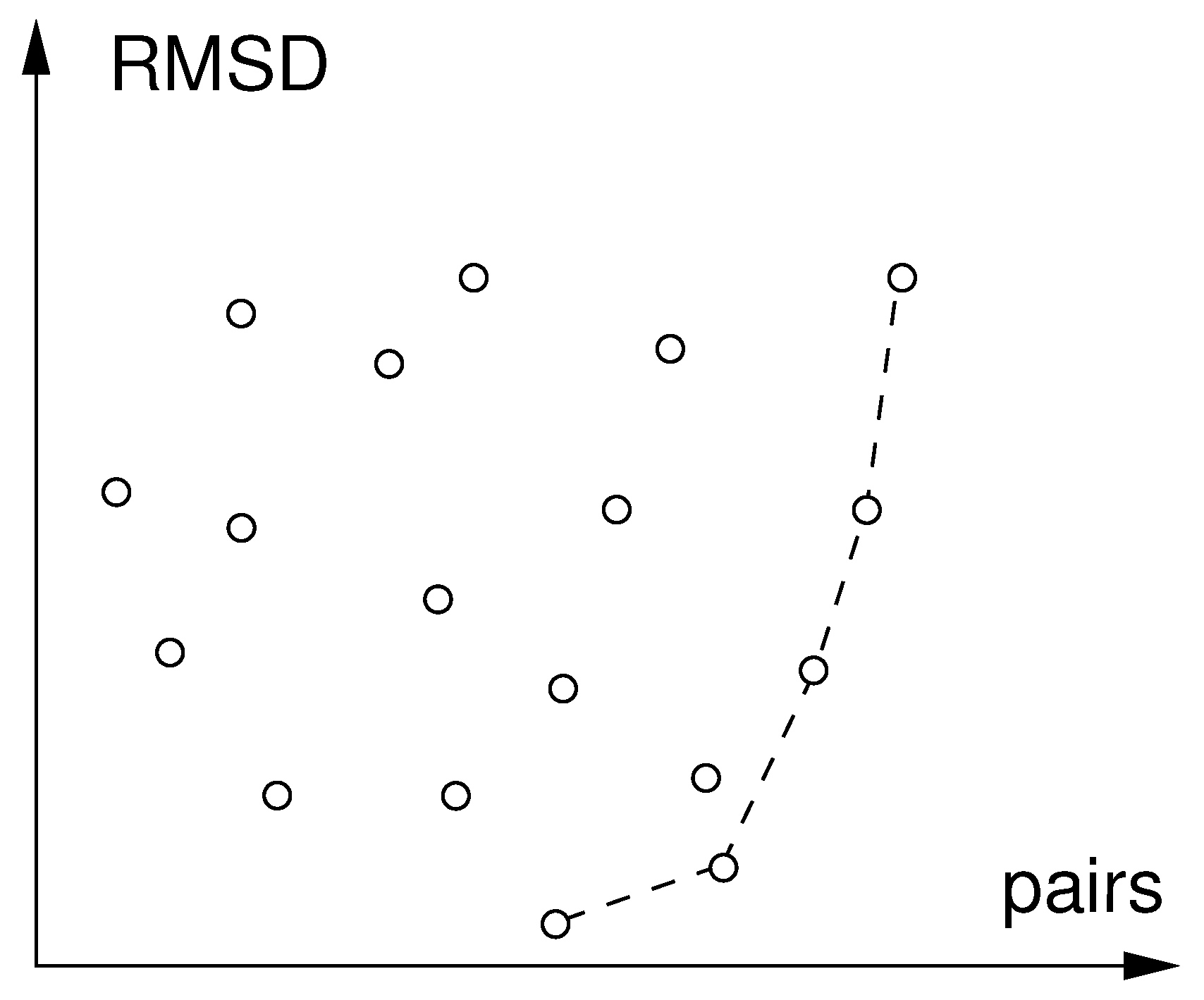
2.3. Applications
Matching two protein structures
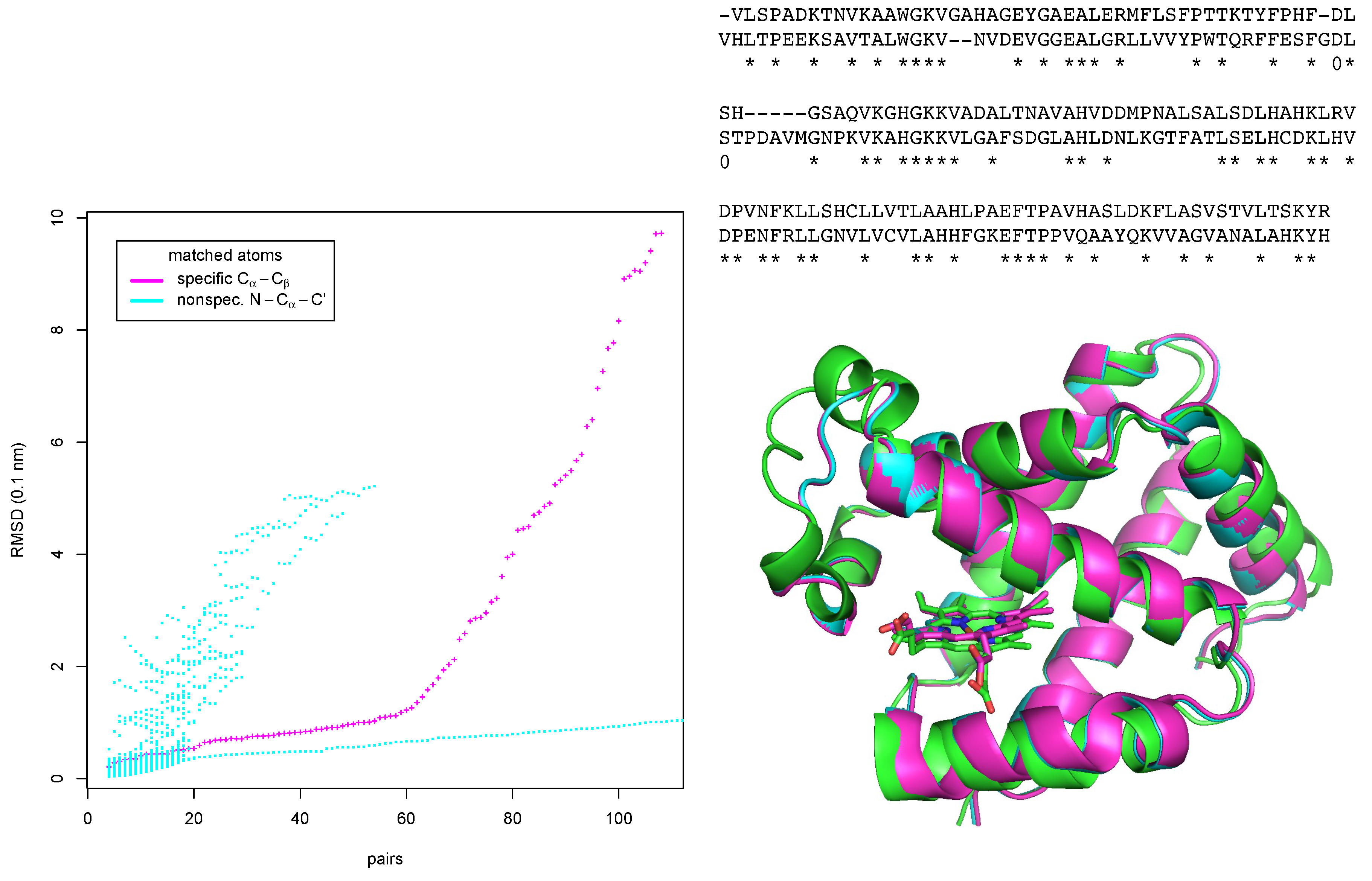
Recognition of a binding site in a homolog
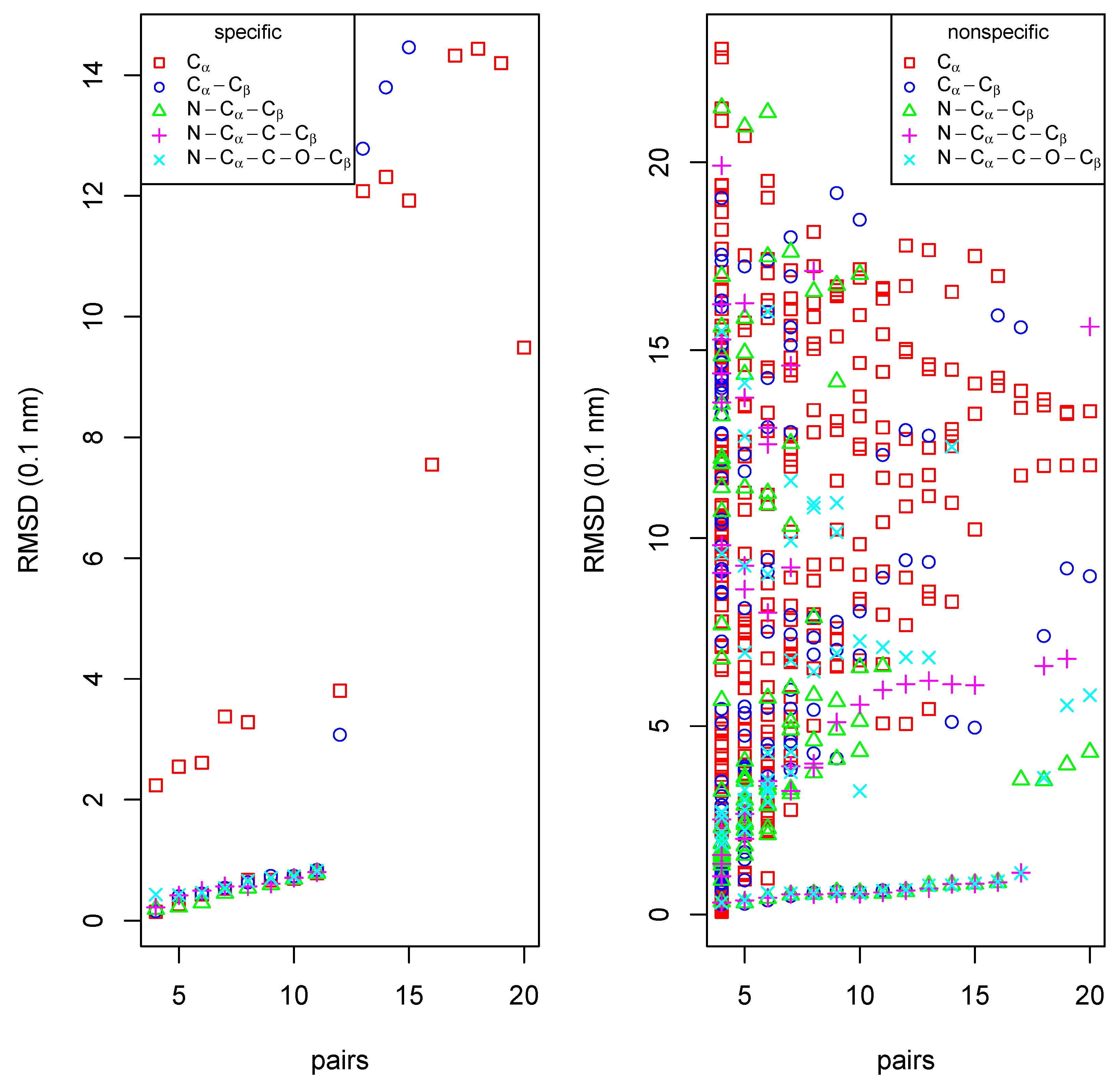
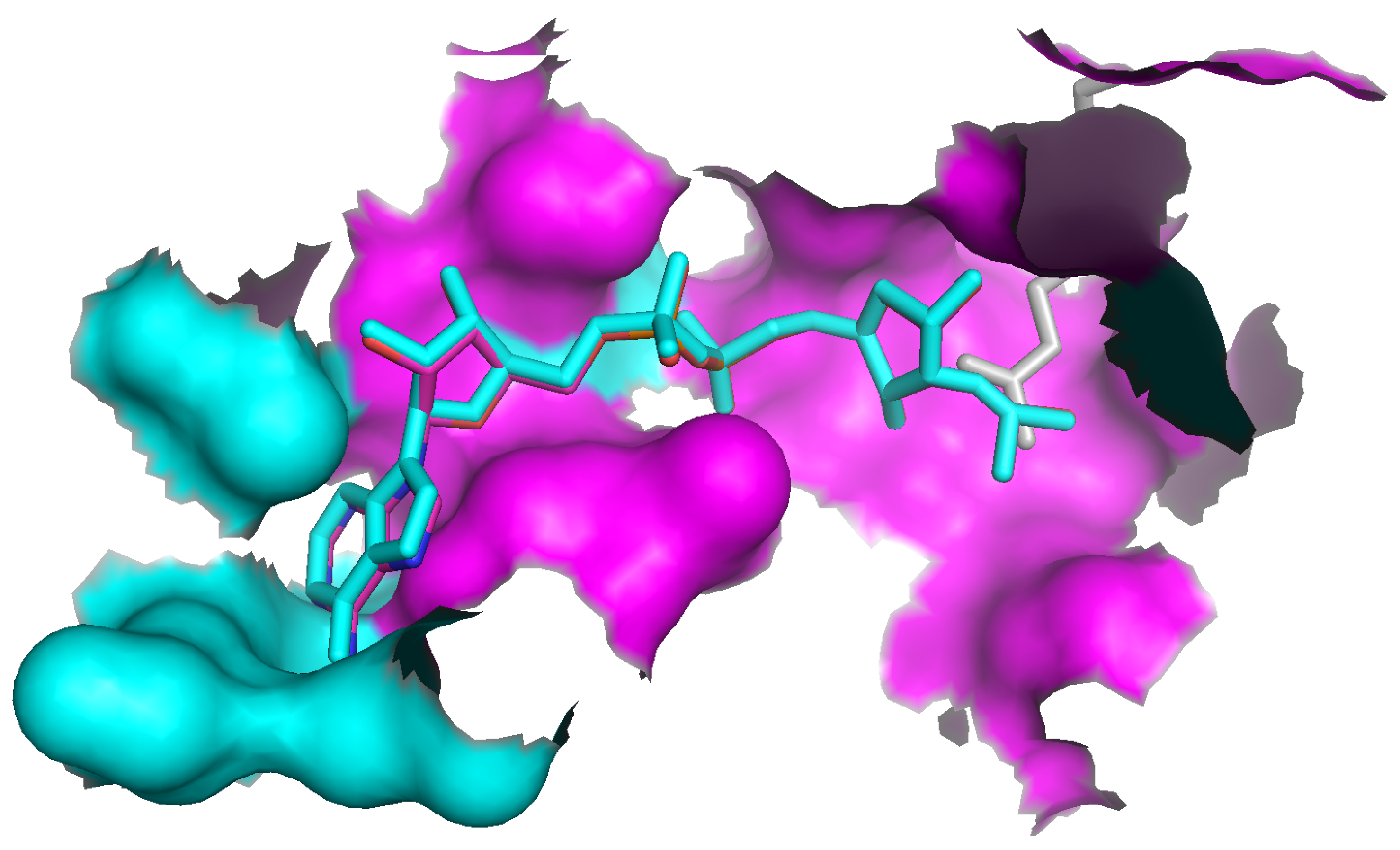
Identification of a ligand binding site in a database
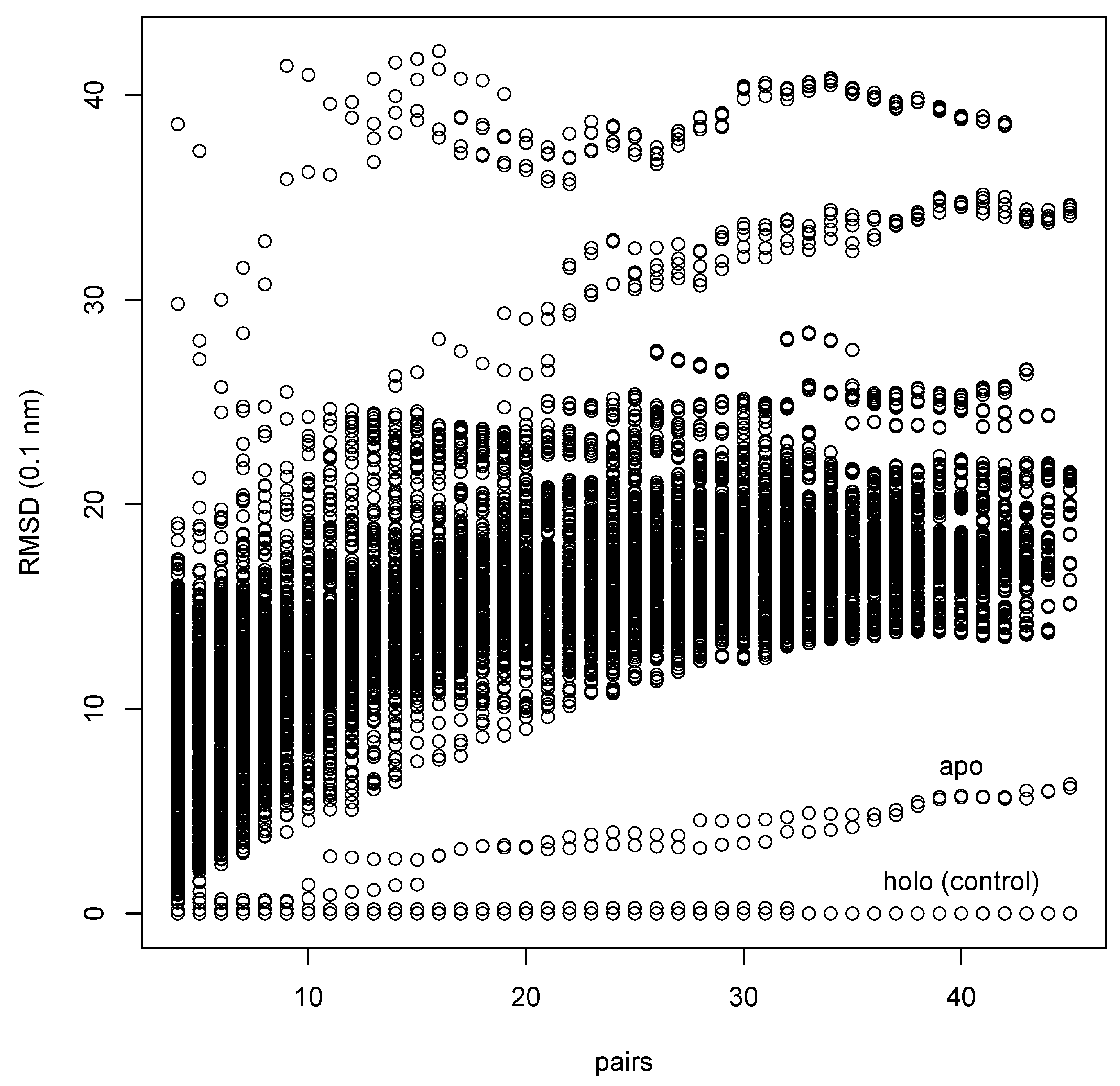
2.4. Comparative test
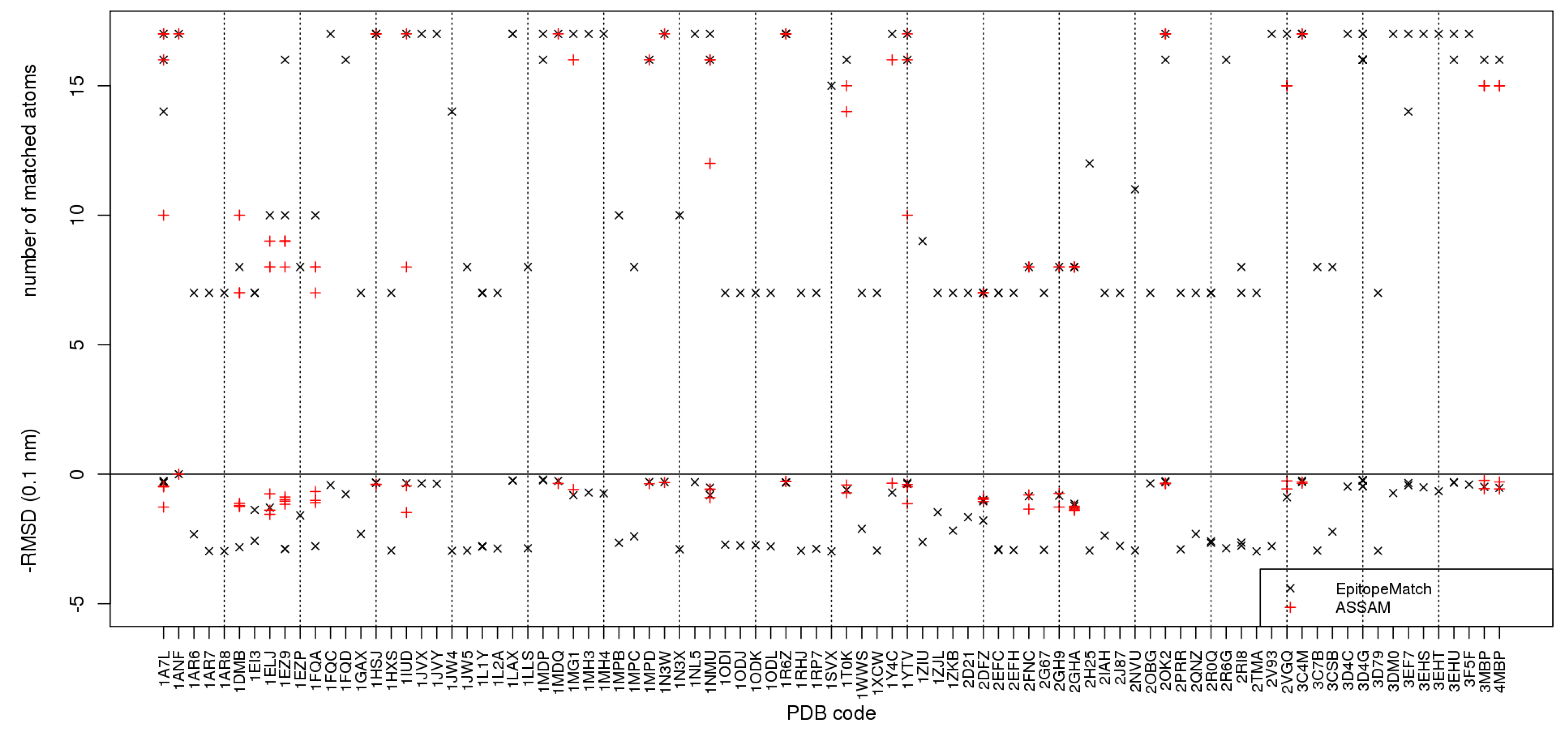
2.5. Discussion of practical performance
3. Supplementary material
References and Notes
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Gronwald, W.; Hohm, T.; Hoffmann, D. Evolutionary Pareto-optimization of stably folding peptides. BMC Bioinformatics 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Lemmen, C.; Lengauer, T. Computational Methods for the Structural Alignment of Molecules. J. Comput. Aided. Mol. Des. 2000, 14, 215–232. [Google Scholar] [CrossRef] [PubMed]
- Halperin, I.; Ma, B.; Wolfson, H.; Nussinov, R. Principles of Docking: An Overview of Search Algorithms and a Guide to Scoring Functions. Proteins 2002, 47, 409–443. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Sander, C. Protein Structure Comparison by Alignment of Distance Matrices. J. Mol. Biol. 1993, 233, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Taylor, W.R.; Orengo, C.R. Protein Structure Alignment. J. Mol. Biol. 1989, 208, 1–22. [Google Scholar] [CrossRef]
- Norel, R.; Fischer, D.; Wolfson, H.J.; Nussinov, R. Molecular Surface Recognition by a Computer vision-based Technique. Protein Eng. 1994, 7, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Raymond, J.W.; Willett, P. Maximum Common Subgraph Isomorphism Algorithms for the Matching of Chemical Structures. J. Comput.-Aided Mol. Des. 2002, 16, 521–533. [Google Scholar] [CrossRef] [PubMed]
- Artymiuk, P.J.; Poirrette, A.R.; Grindley, H.M.; Rice, D.W.; Willett, P. A Graph-theoretic Approach to the Identification of Three-dimensional Patterns of Amino Acid Side-chains in Protein Structures. J. Mol. Biol. 1994, 243, 327–344. [Google Scholar] [CrossRef] [PubMed]
- Varkony, T.; Shiloach, Y.; Smith, D. Computer-Assisted Examination of Chemical Compounds for Structural Similarities. J. Chem. Inf. Comput. Sci. 1979, 19, 104–111. [Google Scholar] [CrossRef]
- Bron, C.; Kerbosch, J. Algorithm 457: Finding All Cliques of an Undirected Graph. Comm. of the ACM 1973, 16, 575–577. [Google Scholar] [CrossRef]
- Kabsch, W. A Solution for the Best Rotation to Relate Two Sets of Vectors. Acta Cryst. 1976, 922, 922–923. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Positions-Specific Gap Penalties and Weight Matrix Choice. Nucl. Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Chai, X.; Marmorstein, R. Structure of the Yeast Hst2 Protein Deacetylase in Ternary Complex with 2’-O-Acetyl ADP Ribose and Histone Peptide. Structure 2003, 11, 1403–1411. [Google Scholar] [CrossRef] [PubMed]
- Ullmann, J.R. An Algorithm for Subgraph Isomorphism. J. Assoc. Comput. Machinery 1976, 16, 31–42. [Google Scholar] [CrossRef]
- Gunasekaran, K.; Nussinov, R. How Different are Structurally Flexible and Rigid Binding Sites? Sequence and Structural Features Discriminating Proteins that Do and Do not Undergo Conformational Change upon Ligand Binding. J. Mol. Biol. 2007, 365, 257–273. [Google Scholar] [CrossRef] [PubMed]
Appendix
| Algorithm 1 The central part of the EpitopeMatch algorithm. Several parameters are supplied by the user, including the minimum, maximum, and increment values of the distance tolerance δ1(δ1min, δ1max, δ1incr), the quotient δquot = δ2/δ1, the minimum degree ν of vertices in stars and cliques, and the structure A of the query epitope (m atoms) and B of the target protein (n atoms). | ||
| 1: | procedure StructureAlignment(δ1min, δ1max, δ1incr, δquot, ν, A, B) | |
| 2: | m ← |A|, n ← |B| | |
| 3: | δ1 ← δ1min | |
| 4: | Hmn ← m · |B| + n + 1 | ▹ Hash key matrix encodes all vertex pairs. |
| 5: | sizeMCS ← 0 | |
| 6: | stars ← 0 | ▹ Declaration of hash table for stars. |
| 7: | repeat | |
| 8: | δ1 ← δ2 · δquot | |
| 9: | DetermineMCS(δ1, δ2) | |
| 10: | δ1 ← δ1 + δ1incr | |
| 11: | until δ1 > δ1max ∨ sizeMCS == m | ▹ See Step 3 in text. |
| 12: | end procedure | |
| 13: | procedure DetermineMCS(δ1, δ2) | ▹ Determines Maximum Common Subgraphs. |
| 14: | DetermineStars(δ1) | ▹ See Algorithm 2. |
| 15: | for i ← 1, |enlargedStars| do | ▹ Corresponding to Step 2 in text. |
| 16: | star ← enlargedStarsi | |
| 17: | DetermineClique(star, δ2) | ▹ See Algorithm 3. |
| 18: | end for | |
| 19: | sizeMCS ← CombineCliques(cliques) | |
| 20: | end procedure | |
| Algorithm 2 Pseudocode of Step 1 of EpitopeMatch algorithm covering the determination of stars, i.e. the seed matches. If several condition are fulfilled, including Inequality 2 in line 9, the star graph around atoms i of A and k of B is extended by an edge to atoms j of A and l of B, and vice versa. Line 10 indicates that the star around i and k is addressed by hash key Hik. The corresponding hash table entry in stars is then also updated. | ||
| 1: | procedure DetermineStars(δ1) | |
| 2: | starsold ← stars | |
| 3: | for i ← 1, m do | ▹ Dimension m of square matrix DA |
| 4: | for j ← i + 1, m do | |
| 5: | for k ← 1, n do | ▹ Dimension n of square matrix DB |
| 6: | for l ← k + 1, n do | |
| 7: | if si == tk ∨ sj == tl then | ▹ Atom property comparison. |
| 8: | if DB,kl ≤ DA,max then | ▹ Check distance range. |
| 9: | if |DA,ij − DB,kl| ≤ δ1 then | ▹ Inequality 2. |
| 10: | extend starHik ((j, l)), stars (starHik) | |
| 11: | extend starHjl ((i, k)), stars (starHjl) | |
| 12: | end if | |
| 13: | end if | |
| 14: | end if | |
| 15: | end if | |
| 16: | end if | |
| 17: | end if | |
| 18: | end if | |
| 19: | enlargedStars ← stars : deg(stars) ≥ max (ν, deg (starsold)) | ▹ Collect all enlarged stars |
| and new stars with central node degree of ν or higher. | ||
| 20: | end procedure | |
| Algorithm 3 Determination of cliques. The non-central nodes of a previously determined star are central nodes of new stars (reducedStars) that have to be tested for fulfilment of the second distance condition (line 7). ReduceToClique then recursively reduces the set of these stars to a clique. | ||
| 1: | procedure DetermineClique(star, δ2) | |
| 2: | reducedStars | ▹ Declaration of hash table for reduced stars. |
| 3: | for m ← 1, |star| do | |
| 4: | for n ← m + 1, |star| do | |
| 5: | i, k ← starm | |
| 6: | j, l ← starn | |
| 7: | if |DA,ij − DB,kl| ≤ δ2 then | ▹ Second distance condition. |
| 8: | extend reducedStarHik (Hjl), reducedStars (reducedStarHik) | |
| 9: | extend reducedStarHjl (Hik), reducedStars (reducedStarHjl) | |
| 10: | end if | |
| 11: | end for | |
| 12: | end for | |
| 13: | if |reducedStars| > 0 then | |
| 14: | ReduceToClique(reducedStars) | |
| 15: | end for | |
| 16: | end procedure | |
| 17: | procedure ReduceToClique(reducedStars) | |
| 18: | for i ← 1, |reducedStars| do | |
| 19: | di ← deg (reducedStarsi) | ▹ Degree of the central node. |
| 20: | for j ← 1, |reducedStars| do | |
| 21: | if j ≠ i then | |
| 22: | di ← di + deg (reducedStarsij) | |
| 23: | end if | |
| 24: | end for | |
| 25: | end for | |
| 26: | if ∀i, j : di == dj then | |
| 27: | extend cliques(reducedStars) | ▹ reducedStars is a new clique. |
| 28: | else | |
| 29: | remove reducedStarsi:min(di) | |
| 30: | ReduceToClique(reducedStars) | |
| 31: | end if | |
| 32: | end procedure | |
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Jakuschev, S.; Hoffmann, D. A Novel Algorithm for Macromolecular Epitope Matching. Algorithms 2009, 2, 498-517. https://doi.org/10.3390/a2010498
Jakuschev S, Hoffmann D. A Novel Algorithm for Macromolecular Epitope Matching. Algorithms. 2009; 2(1):498-517. https://doi.org/10.3390/a2010498
Chicago/Turabian StyleJakuschev, Stanislav, and Daniel Hoffmann. 2009. "A Novel Algorithm for Macromolecular Epitope Matching" Algorithms 2, no. 1: 498-517. https://doi.org/10.3390/a2010498
APA StyleJakuschev, S., & Hoffmann, D. (2009). A Novel Algorithm for Macromolecular Epitope Matching. Algorithms, 2(1), 498-517. https://doi.org/10.3390/a2010498



