Abstract
Based on a protein-protein docking approach we have developed a procedure to verify or falsify protein-protein interactions that were proposed by other methods such as yeast-2-hybrid assays. Our method currently utilizes intermolecular energies but can be expanded to incorporate additional terms such as amino acid based pair-potentials. We show some early results that demonstrate the general applicability of our approach.
1. Introduction
Proteins are an integral component for most of the reactions taking part in the cell. An important aspect in protein research is their three-dimensional structure, which is required to understand their function in detail. The most common methods to determine these structures are X-ray crystallography and NMR spectroscopy. To date almost 47,000 protein structures have been deposited in the PDB [1]. However, cellular functions are rarely carried out by single proteins but rather by complexes of several interacting proteins and only a very small part of the deposited structures correspond to protein-protein complexes. It is currently estimated that each protein has on average nine interaction partners [2]. High-throughput methods for detecting protein interactions, like yeast-2-hybrid assays or tandem-affinity-purification mass spectrometry, produce large expected protein-protein interaction maps. These experimental approaches are supplemented by bioinformatic methods such as phylogenetic profiling, investigations of gene neighborhoods, and gene fusion analysis.
Unfortunately it is not possible to determine the structures for all of them by experimental methods since there are often limitations concerning large or transient complexes. In addition the experimental structure determination of complexes is in most cases a very time-consuming and challenging process. For that reason computational approaches such as docking algorithms that predict the structure of protein-protein complexes are needed.
During the last few years considerable effort has been put in the development and application of docking algorithms. For a recent overview on protein-protein docking readers should refer to the review by D.W. Ritchie [3]. The success of docking algorithms has consistently improved over the last years as measured by the CAPRI (Critical Assessment of PRedicted Interactions) blind docking experiment [4]. Therefore, in many cases reliable results can be obtained.
1.1. Motivation
As mentioned above, protein-protein interactions play a major role in cellular processes and both experimental and bioinformatic high-throughput methods like yeast-2-hybrid assays are widely used for obtaining interaction maps. However, since these methods are not always applicable and often contain a considerable number of false positives [5], there is a need for computational approaches to verify or falsify protein-protein interactions that were predicted by other methods. Since protein-protein interactions are critically dependent on the three-dimensional structures of the individual molecules it seems logical to use this information for judging putative protein-protein interactions. Aloy and Russell [6], for example, have suggested a method to model putative interactions on known 3D complexes to investigate the compatibility of a proposed interaction with this complex. In another approach comparative docking together with the analysis of steric clashes is used to analyze putative interactions [7]. In this method information about the interacting residues of both partner proteins is required as well.
As detailed below, we propose in this contribution a new method based on protein-protein docking where only interface information of one of the partner proteins is sufficient to assess putative interactions. Methods providing the three-dimensional structures of protein complexes from the structures of the individual molecules (docking algorithms) are readily available. With the constant increase of experimental structures deposited in the PDB the individual structures are available in many cases. Additionally, it should be possible to use at least in some cases high quality homology models as input for docking programs.
Usually docking algorithms are used to predict the complex structure of two proteins that are known to interact. In their scoring step a great amount of different possible complex structures are compared to select those that are near-native. That means discrimination between native and non native interactions. Similarly it should be possible to extend this analysis to protein pairs where it is not known a priori whether they interact in nature or not. That means in other words to perform docking runs with different proteins, even those that do not interact or are not known to do so and finally, after the interpretation of the structures, get as a result whether two proteins are suggested to build complexes in nature or not.
This is actually a computational method to verify proposed protein-protein interactions. In this contribution we will investigate the general applicability of the suggested approach.
2. Results and Discussion
2.1. Quality Check for Docking
It is obvious that it is important for our approach that docking of two truly interacting proteins really lead to results that are close to the native structure of the complex. As a test case we used the Barnase-Barstar complex (PDBids: Barnase: 1RGH B, Barstar: 1A19 B, complex: 1AY7 A:B). Barnase excreted from the bacterium Bacillus amyloliquefaciens is a protein of 110 amino acids that possesses ribonulease activity. It forms a tight complex with its inhibitor Barstar. The above input structures were used in a standard HADDOCK [8] run that resulted in 200 final complex structures, which then were analyzed in view of a correlation between RMSD to the native structure and the score calculated by HADDOCK. A short description of the HADDOCK docking algorithm can be found in the methods section.
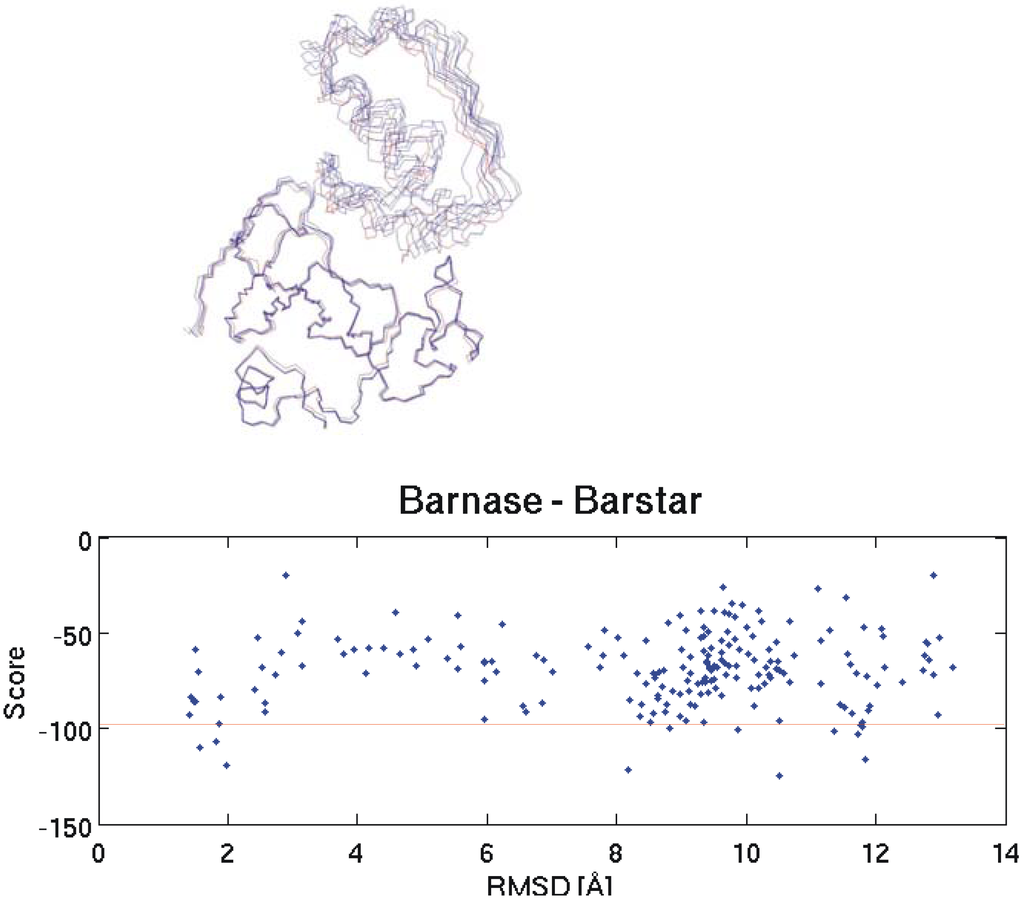
Figure 1.
On the top the backbone of the native Barnase-Barstar complex structure (red) is compared to four near native solutions obtained by docking. The graph on the bottom shows the correlation between RMSD to the native structure and HADDOCK score for all 200 docking results. Pair-wise RMSD calculations are based on the coordinates of the Cα atoms. A description of the score calculation can be found in the experimental section. The horizontal red line separates the 10 best structures according to the calculated scores.
Figure 1.
On the top the backbone of the native Barnase-Barstar complex structure (red) is compared to four near native solutions obtained by docking. The graph on the bottom shows the correlation between RMSD to the native structure and HADDOCK score for all 200 docking results. Pair-wise RMSD calculations are based on the coordinates of the Cα atoms. A description of the score calculation can be found in the experimental section. The horizontal red line separates the 10 best structures according to the calculated scores.
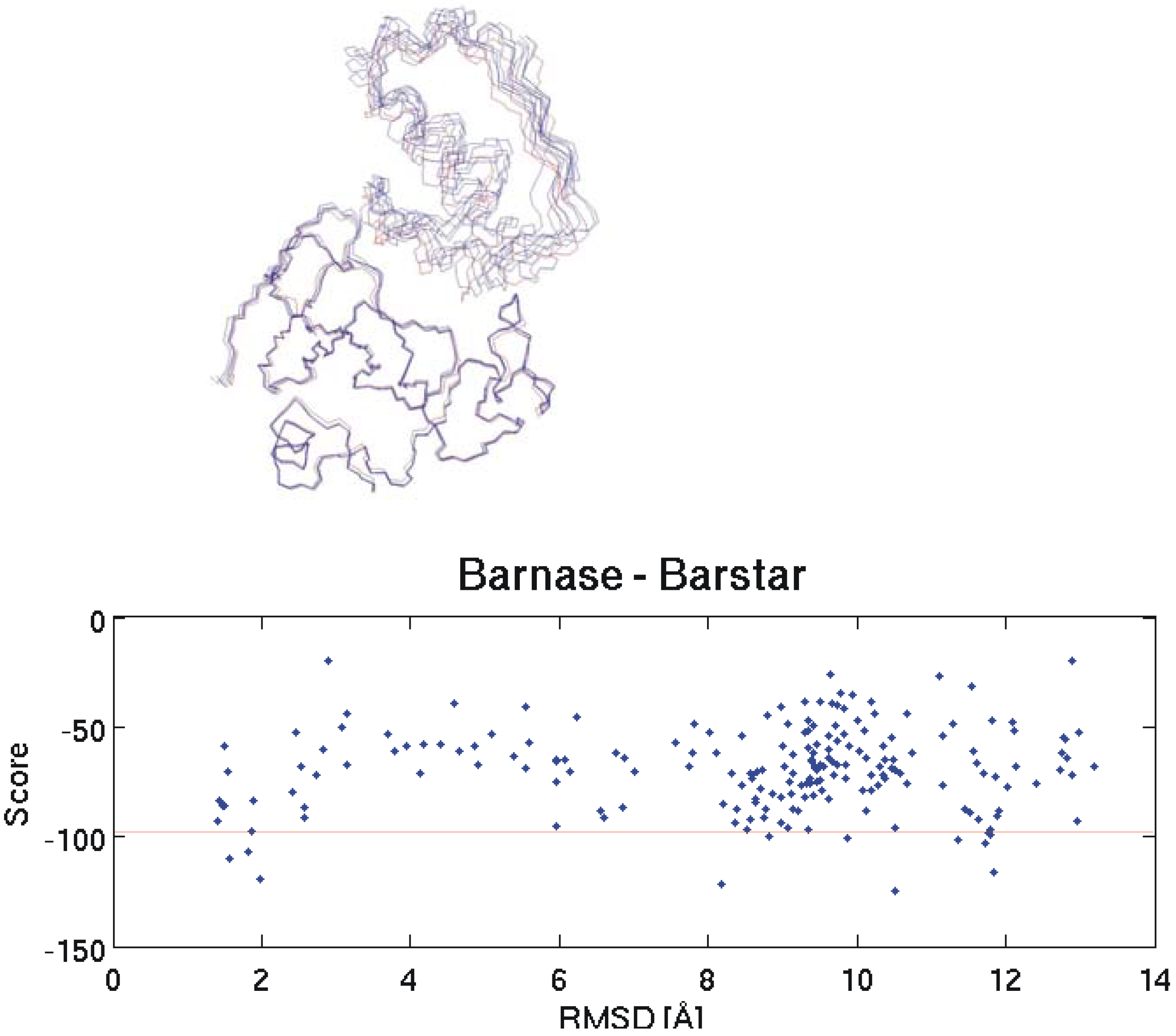
In the first attempt we did not use the facility of HADDOCK to define the interacting residues since we were interested in investigating whether the correct orientation of the molecules can be found by the algorithm without additional data. Unfortunately, this approach was unsuccessful and thus we did a second test with slightly more information where one side of the interface was defined: here for Barnase the interacting residues were incorporated as ambiguous interaction restraints (these are residues number 27, 59, 60, 83, 87 and 102) and for Barstar no interface information was included. One thousand complex structures were obtained by rigid body docking and 200 of these were further refined by semi-flexible simulated annealing in torsion angle space.
All other parameters were set to default values. This time there were several near-native structures among the 10 top ranked docking results. Ranking was based on the HADDOCK score calculation. On the top part of Figure 1 the backbones of four selected structures obtained by docking are overlaid to the native structure. It is obvious that this time near native structures could be obtained.
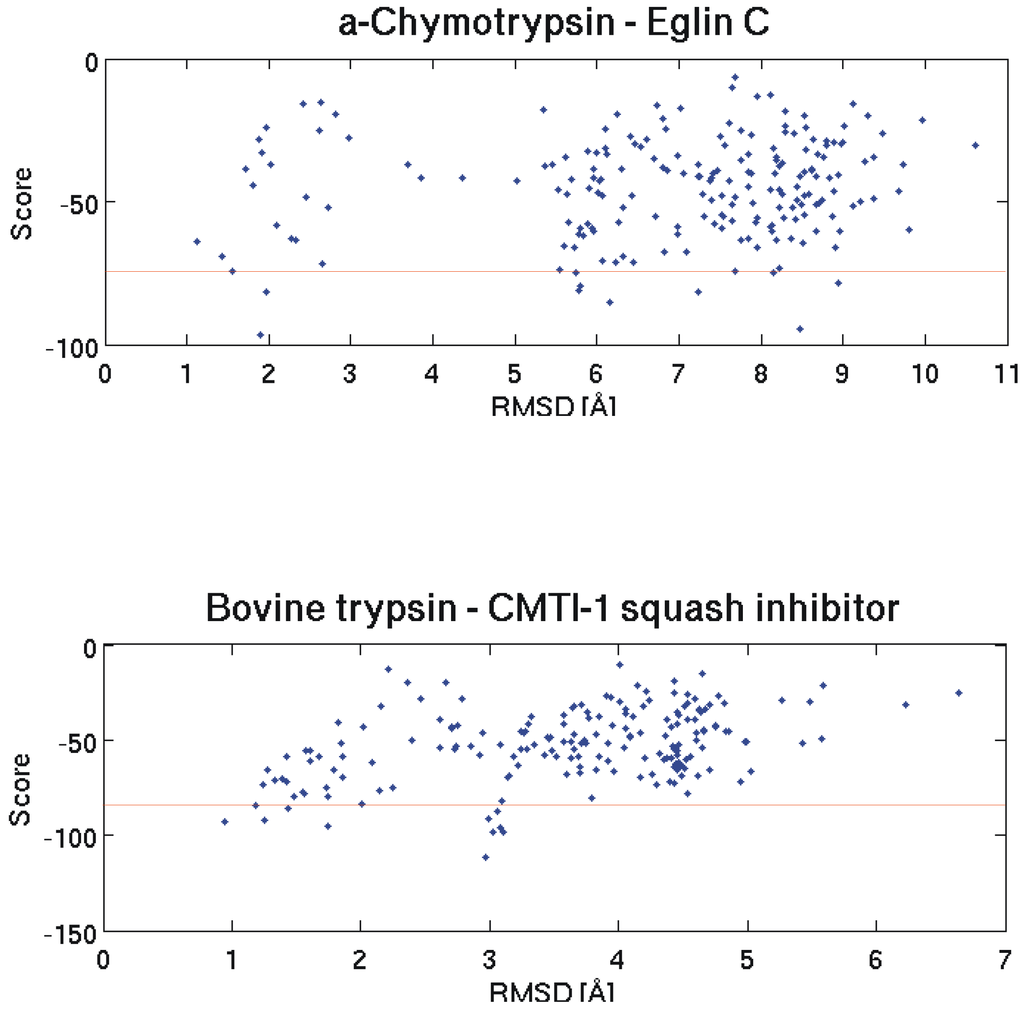
Figure 2.
The two graphs show the correlation between RMSD to the native structure and Haddock score for all 200 docking results for the α- Chymotrypsin - Eglin C and Bovine trypsin - CMTI - 1 squash inhibitor test cases. Pair-wise RMSD calculations are based on the coordinates of the C atoms. A description of the score calculation can be found in the experimental section. The horizontal red lines separate the 10 best structures according to the calculated scores.
Figure 2.
The two graphs show the correlation between RMSD to the native structure and Haddock score for all 200 docking results for the α- Chymotrypsin - Eglin C and Bovine trypsin - CMTI - 1 squash inhibitor test cases. Pair-wise RMSD calculations are based on the coordinates of the C atoms. A description of the score calculation can be found in the experimental section. The horizontal red lines separate the 10 best structures according to the calculated scores.
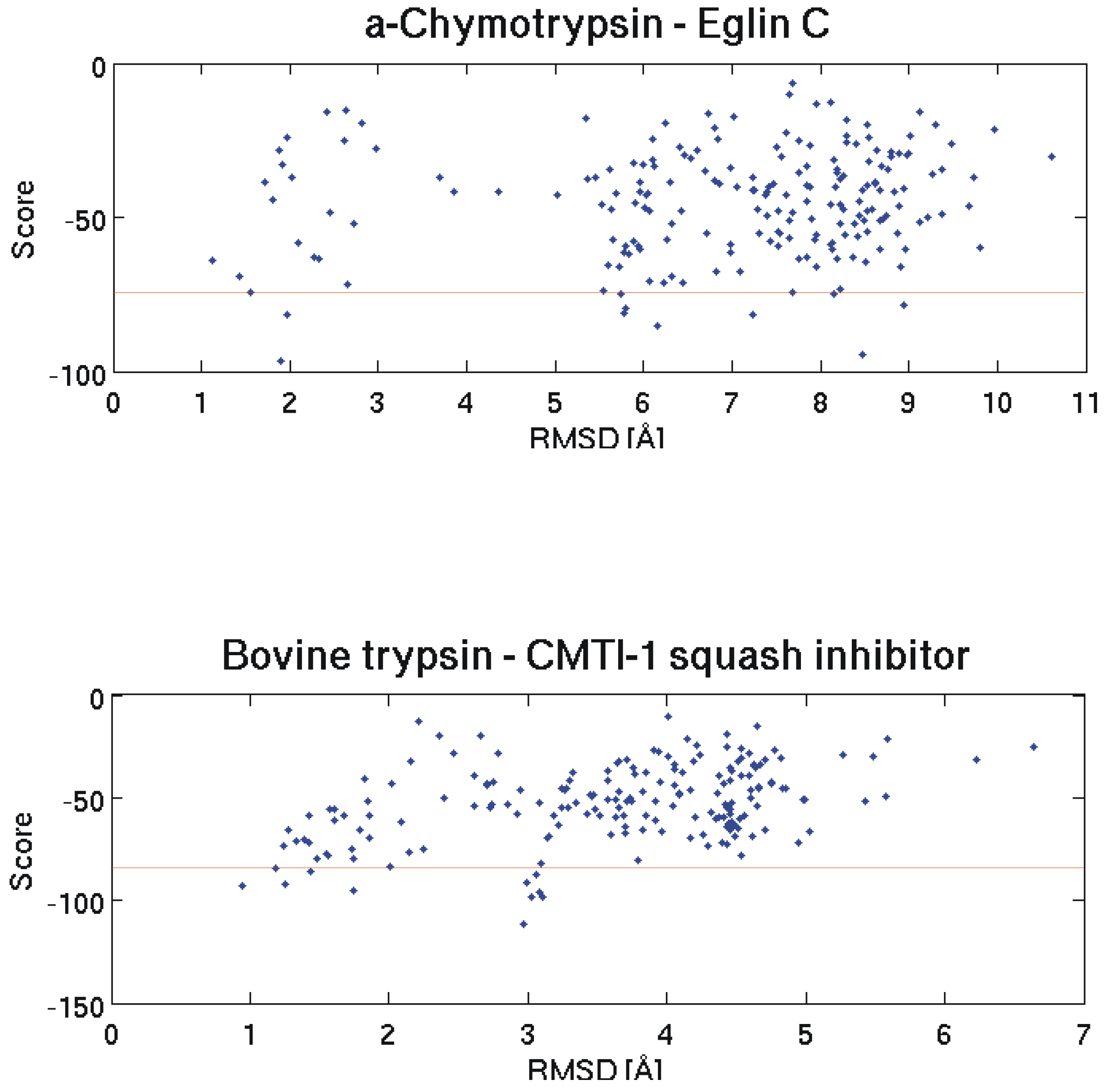
This shows that it is possible to get the right conformation of a protein-protein complex even if only one interaction side is defined. However, the challenge remains to find the correct solutions among the proposed results.
2.2. Discrimination with Interaction Energy
In the next step we were interested in investigating whether it is possible to select the native protein-protein interaction from a set of possible solutions. For this purpose we analyzed enzyme-inhibitor interactions for which the corresponding structures of the free proteins as well as the structures of the native complexes were available [9]. For each test case several putative inhibitors were docked to the known interaction site of one given enzyme employing again the HADDOCK docking algorithm. As detailed in the methods section average interaction energies, which are the sum of van der Waals energies and electrostatic energies between intermolecular atom-pairs were calculated for the 10 finally selected complex structures of each docking run.

Table 1.
Comparison of intermolecular interaction energies of native (shaded in gray) and corresponding non-native complexes. aThe energy is always the average of ten complexes that were top ranked from the docking algorithm. bThe interaction energy of the complex top ranked by the docking algorithm is shown. cResults are ranked according to the average interaction energies provided in the third column.
| Receptor | Ligand | aEint = EvdW + Eelec [kJ/mol] | bEint = EvdW + Eelec [kJ/mol] | cRank |
|---|---|---|---|---|
| Barnase | Soybean trypsin inhibitor | -670.0 | -1173.9 | 2 |
| Barnase | Ovomucoid 3rd domain | -575.0 | -481.3 | 3 |
| Barnase | Eglin C | -510.6 | -546.6 | 4 |
| Barnase | Pancreatic secretory trypsin inhibitor | -504.7 | -910.7 | 5 |
| Barnase | APPI | -481.3 | -301.3 | 6 |
| α-Chymotrypsin | Barstar | -505.5 | -341.5 | 2 |
| α-Chymotrypsin | APPI | -445.7 | -307.6 | 3 |
| α-Chymotrypsin | Soybean trypsin inhibitor | -364.9 | -469.1 | 4 |
| α-Chymotrypsin | Pancreatic secretory trypsin inhibitor | -306.3 | -397.2 | 5 |
| Bovine trypsin | Glycosylase inhibitor | -761.3 | -876.3 | 1 |
| Bovine trypsin | RAGI inhibitor | -492.2 | -565.0 | 3 |
| Bovine trypsin | Soybean trypsin inhibitor | -436.5 | -513.5 | 4 |
| Bovine trypsin | Streptomyces subtilisin inhibitor | -412.6 | -509.7 | 5 |
| Bovine trypsin | Amicyanin | -323.9 | -352.4 | 6 |
We investigated the potential of these interaction energies to discriminate between native and non-native interactions. In the following we will show three typical example test cases. The first one is again Barnase and its interactions with a set of putative inhibitors. We docked Barstar (the native inhibitor), soybean trypsin inhibitor, APPI, Ovomucoid 3rd domain and Pancratic secretory trypsin inhibitor (inhibitors that do not interact with Barnase) to Barnase. As can be seen on the bottom part of Figure 1 and on the top and bottom part of Figure 2 the structure with the lowest HADDOCK score is not necessarily the best one (smallest RMSD to the native structure). Therefore, to include at least some near native structures in the analysis, we always calculated the mean interaction energies for the 10 selected structures of every docking run and compared these mean values to each other. The results for Barnase and two similar tests with α - Chymotrypsin and Bovine trypsin can be seen in Table 1. For reasons of comparison the interaction energy obtained for the top ranked structure of each docking run is displayed in the second last column of the table. Note that these structures do not necessarily possess the best interaction energy since the ranking within a docking run is based on calculated scores and not solely on interaction energies.
The above results show clearly that in many cases it is possible to select the correct binder from a set of putative interaction partners. This is the case for Barnase where the interaction with the natural interaction partner Barstar shows the lowest (best) average interaction energy of -913.2 kJ/mol. The same is true for the interaction of α-Chymotrypsin where also the native complex with Eglin C possesses the lowest average interaction energy of -552.8 kJ/mol.
For the third test case shown, the correct interaction of Bovine trypsin with the CMTI-1 squash inhibitor is only the second best solution with an average interaction energy of -588.4 kJ/mol, whereas here the lowest interaction energy of -761.3 kJ/mol was obtained for the non-native interaction with the glycosylase inhibitor. These results demonstrate the potential of our approach. However, it is also apparent that further improvements in the scoring of the possible solutions in addition to the use of these simple interaction energies are required. For example simple scoring functions that neglect factors such as entropic contributions often reward large binding interfaces and therefore tend to favor larger binding partners.
2.3. Conclusions and Outlook
We could show that for pair-wise interactions the interface of only one of the two proteins needs to be known to obtain realistic docking results. Also docking is a useful tool to discriminate native interactions from non-native ones. We could show that in principle it is possible to select the native interaction partner among a set of non-native ones. Currently we are working on the combination of the van der Waals and electrostatic energies with pair-wise amino acid scores in a probability based scoring scheme to obtain improved predictions whether a hypothetical complex can be supposed to exist in nature or not.
3. Experimental Section
3.1. Docking
The hypothesis underlying docking predictions is that the native complex structure is the state with the lowest free energy available to the system. There are several different approaches on how to develop docking algorithms but the common, basic idea is to first do a sampling of possible conformations followed by a scoring of these conformations. Scoring means to analyze the putative complex structures generated in the first step with regard to chemical, physical and knowledge based aspects. Selecting suitable scoring terms and weighting them in an appropriate way is one of the great challenges in docking. The aim is to rank all putative structures in a way that most of the native-like structures are found in the top part of the ranked output.
In our work we are using HADDOCK2.0, which was developed by Dominguez et al. [8]. There are two main reasons for this choice: first, HADDOCK introduces the possibility to drive the sampling step by known data about the contact regions of the interacting proteins. These data can for example originate from nuclear magnetic resonance chemical shift perturbation data [10] or from mutagenesis experiments. This information is introduced as ambiguous interaction restraints (AIRs). By this it is possible to define on all docking partners the residues, which are supposed to be in the interface region of the complex. The docking is then driven by a force that pulls the defined regions together. This mechanism scales down the search space considerably and makes it possible to get reasonable results in a passable time. The other main advantage of HADDOCK is that main and side-chain flexibility can be incorporated into the docking process. In this process a three step docking procedure is used. In the first step a rigid body energy minimization is performed followed by semi-flexible simulated annealing in torsion angle space. In the last stage an optional refinement in explicit solvent, e.g. water, can be done. Since in the current application many different complex structures had to be computed this quite time consuming step was omitted. The ranking of the complex structures is based on a score calculated from EvdW, Eelec, EAIR, BSA, and Edesolv, where BSA is the buried surface area and Edesolv describes the desolvation energy. From each docking run the 10 best structures in terms of this score were selected for further analysis. As mentioned before docking runs were performed for native and non native interactions. In the next step the selected complex structures of each docking run were used to discriminate between the native and non native complexes. To base the decisions on something that resembles real physical energies the interaction energies Eint = EvdW + Eelec were calculated for the interface of each of the selected complex structures. From these individual energies the average interaction energy was obtained for the 10 selected structures of each docking run and these average interaction energies were used to discriminate between native and non native interactions.
Acknowledgements
This work was supported by the Bavarian Genomic Network BayGene. The authors thank Rainer Merkl for helpful discussions.
References
- Westbrook, J.; Feng, Z.; Chen, L.; Yang, H.; Berman, H.M. The Protein Data Bank and structural genomics. Nucl. Acid. Res. 2003, 31, 489–491. [Google Scholar] [CrossRef]
- Aloy, P.; Russell, R.B. Ten thousand interactions for the molecular biologist. Nat. Biotechnol. 2004, 22, 1317–1321. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, D.W. Recent progress and future directions in protein-protein docking. Curr. Protein Pept. Sci. 2008, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Vajda, S. Classification of protein complexes based on docking difficulty. Proteins 2005, 60, 176–180. [Google Scholar] [CrossRef] [PubMed]
- von Mering, C.; Krause, R.; Snel, B.; Cornell, M.; Oliver, S.G.; Fields, S.; Bork, P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature 2002, 417, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Aloy, P.; Russell, R.B. Interrogating protein interaction networks through structural biology. Proc. Nat. Acad. Sci. USA. 2002, 99, 5896–5901. [Google Scholar] [CrossRef] [PubMed]
- Cockell, S.J.; Oliva, B.; Jackson, R.M. Structure-based evaluation of in silico predictions of protein-protein interactions using comparative docking. Bioinformatics 2007, 23, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A Protein-Protein Docking Approach Based on Biochemical or Biophysical Information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Mintseris, J.; Wiehe, K.; Pierce, B.; Anderson, R.; Chen, R.; Janin, J.; Weng, Z. Protein-protein docking benchmark 2.0: An update. Proteins 2005, 60, 214–216. [Google Scholar]
- Schumann, F.H.; Riepl, H.; Maurer, T.; Gronwald, W.; Neidig, K.P.; Kalbitzer, H.R. Combined chemical shift changes and amino acid specific chemical shift mapping of protein-protein interactions. J. Biomol. NMR 2007, 39, 275–289. [Google Scholar] [CrossRef] [PubMed]
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
