Modeling How the Different Parts of the Immune System Fight Viruses
Abstract
1. Introduction
2. Materials and Methods
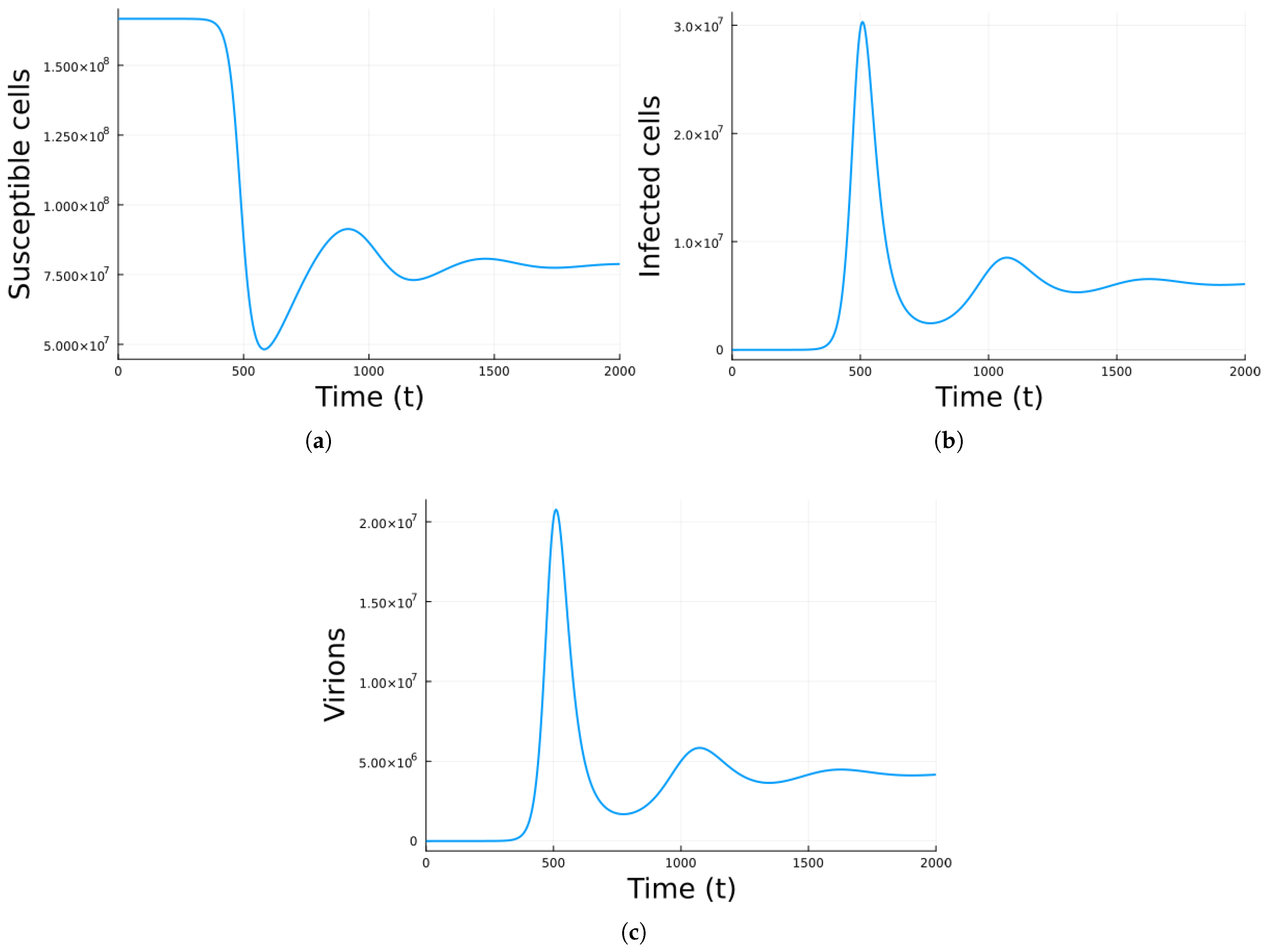
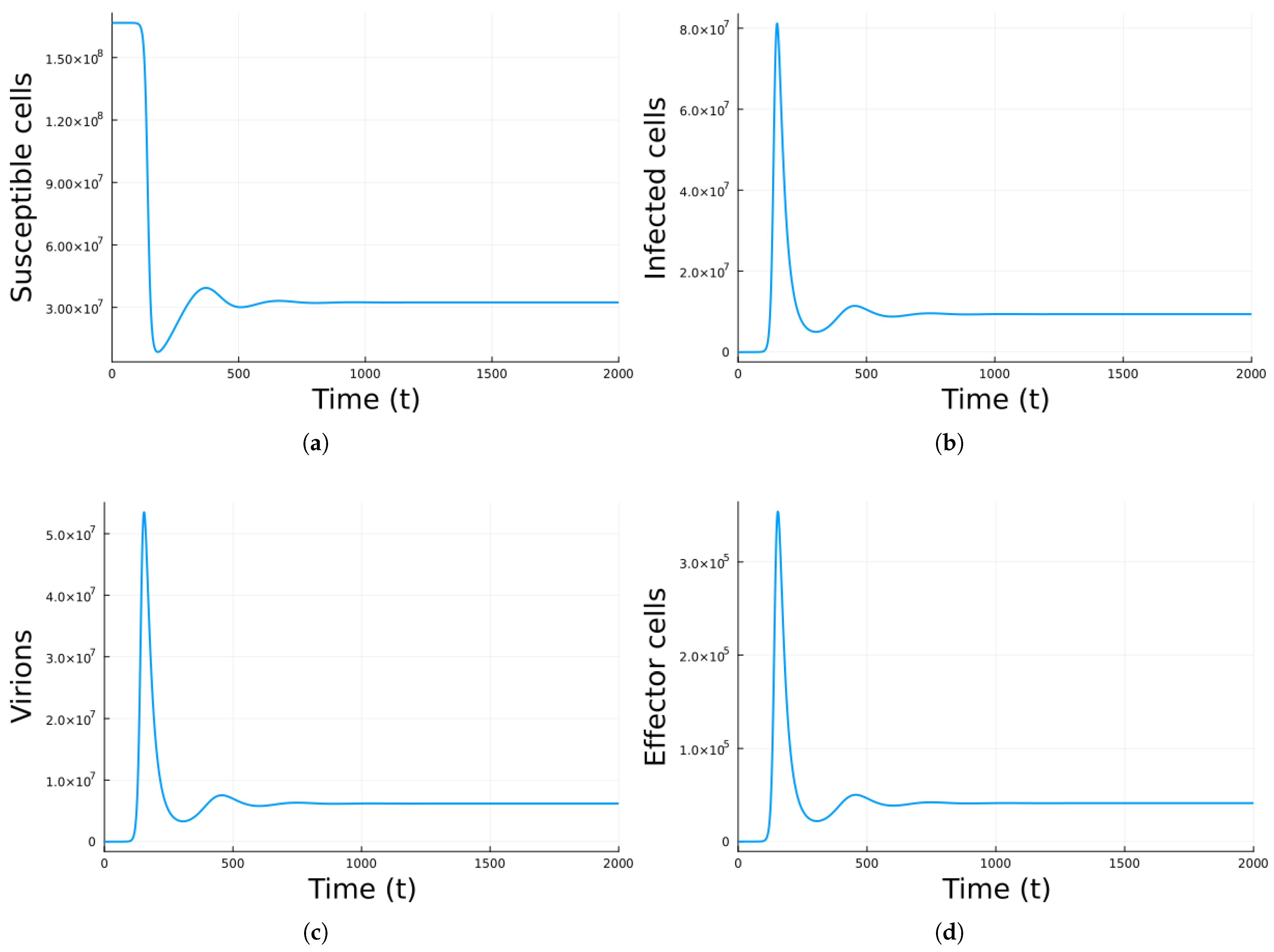
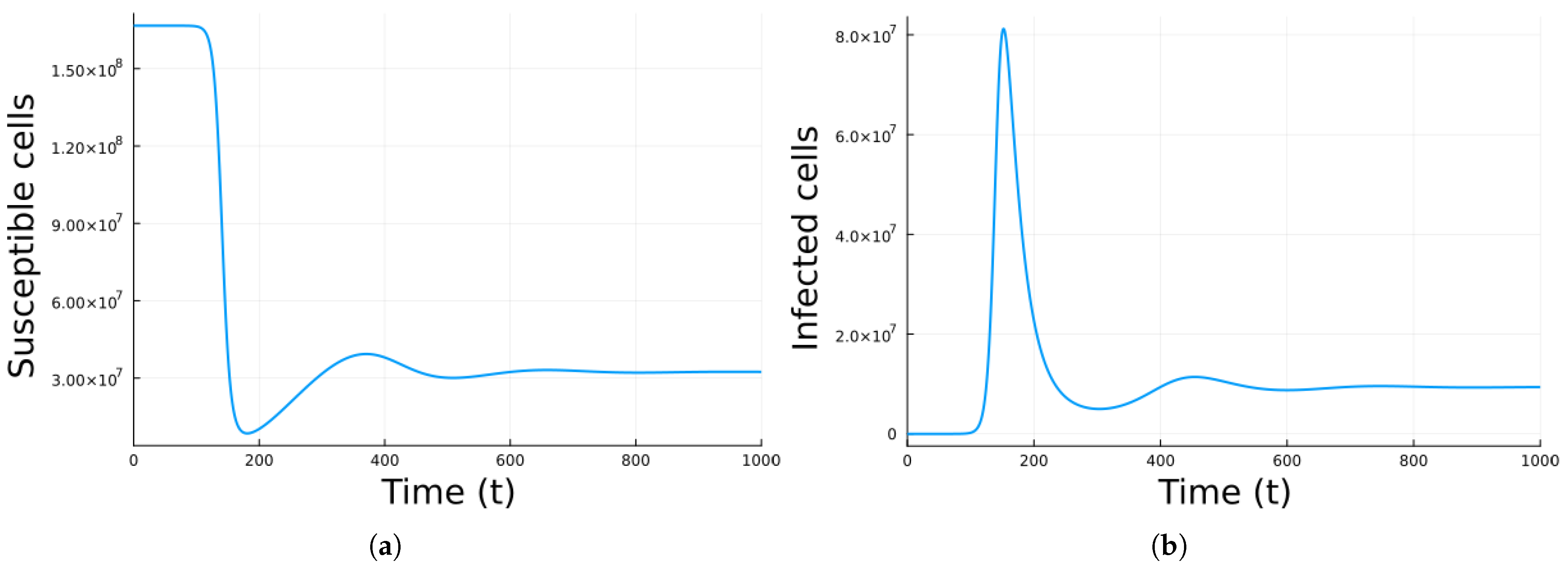
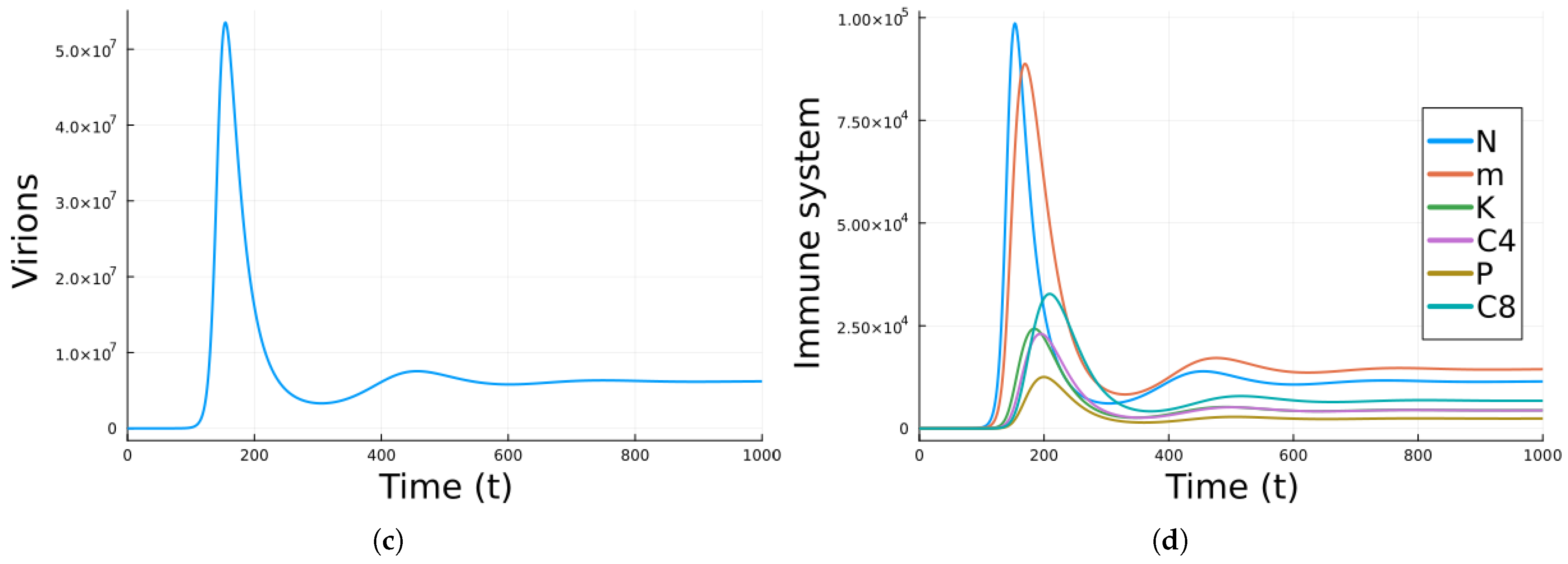
3. Results
4. Discussion
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ODE | Ordinary differential equation |
| DFE | Disease free equilibrium |
| CE | Chronic equilibrium |
| COVID-19 | Coronavirus disease 2019 |
| SARS | Severe acute respiratory syndrome coronavirus 2 |
| AIDS | Acquired immunodeficiency syndrome |
| RNA | Ribonucleic acid |
| DNA | Deoxyribonucleic acid |
| E-FAST | Extended Fourier amplitude sensitivity testing |
References
- Clark, M.A.; Douglas, M.; Choi, J. Biology 2e; Openstax: Houston, TX, USA, 2018. [Google Scholar]
- Taveira, N. Antivirals and Vaccines. Int. J. Mol. Sci. 2023, 24, 10315. [Google Scholar] [CrossRef]
- Nowak, M.A.; Bonhoeffer, S.; Hill, A.M.; Boehme, R.; Thomas, H.C.; McDade, H. Viral Dynamics in Hepatitis B Virus Infection. Proc. Natl. Acad. Sci. USA 1996, 93, 4398–4402. [Google Scholar] [CrossRef]
- Perelson, A.S.; Nelson, P.W. Mathematical Analysis of HIV-1 Dynamics in Vivo. SIAM Rev. 1999, 41, 3–44. [Google Scholar] [CrossRef]
- Baccam, P.; Beauchemin, C.; Macken, C.A.; Hayden, F.G.; Perelson, A.S. Kinetics of influenza A virus infection in humans. J. Virol. 2006, 80, 7590–7599. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M.A.; Bangham, C.R.M. Population Dynamics of Immune Responses to Persistent Viruses. Science 1996, 272, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Hattaf, K.; Yousfi, N. Dynamics of SARS-CoV-2 Infection Model with Two Modes of Transmission and Immune Response. Math. Biosci. Eng. 2020, 17, 5326–5340. [Google Scholar] [CrossRef]
- Murase, A.; Sasaki, T.; Kajiwara, T. Stability Analysis of Pathogen-Immune Interaction Dynamics. J. Math. Biol. 2005, 51, 247–267. [Google Scholar] [CrossRef] [PubMed]
- Kajiwara, T.; Sasaki, T. Global Stability of Pathogen-Immune Dynamics with Absorption. J. Biol. Dyn. 2010, 4, 258–269. [Google Scholar] [CrossRef]
- Du, S.Q.; Yuan, W. Mathematical modeling of interaction between innate and adaptive immune responses in COVID-19 and implications for viral pathogenesis. J. Med. Virol. 2020, 92, 1615–1628. [Google Scholar] [CrossRef]
- Bocharov, G.A.; Romanyukha, A.A. Mathematical model of antiviral immune response III. Influenza A virus infection. J. Theor. Biol. 1994, 167, 323–360. [Google Scholar] [CrossRef]
- Cao, P.; Wang, Z.; Yan, A.W.C.; McVernon, J.; Xu, J.; Heffernan, J.M.; Kedzierska, K.; McCaw, J.M. On the Role of CD8+ T Cells in Determining Recovery Time from Influenza Virus Infection. Front. Immunol. 2016, 7, 611. [Google Scholar] [CrossRef]
- Gómez, M.C.; Yang, H.M. Mathematical Model of the Immune Response to Dengue Virus. J. Appl. Math. Comput. 2020, 63, 455–478. [Google Scholar] [CrossRef]
- Culshaw, R.V.; Ruan, S.; Webb, G. A mathematical model of cell-to-cell spread of HIV-1 that includes a time delay. J. Math. Biol. 2003, 46, 425–444. [Google Scholar] [CrossRef] [PubMed]
- Pourbashash, H.; Pilyugin, S.S.; De Leenheer, P.; McCluskey, C. Global analysis of within host virus models with cell-to-cell viral transmission. Discrete Contin. Dyn. Syst. Ser. B 2014, 19, 3341–3357. [Google Scholar] [CrossRef]
- Shu, H.; Wang, L.; Watmough, J. Global Stability of a Nonlinear Viral Infection Model with Infinitely Distributed Intracellular Delays and CTL Immune Responses. SIAM J. Appl. Math. 2013, 73, 1280–1302. [Google Scholar] [CrossRef]
- Yang, Y.; Zou, L.; Ruan, S. Global Dynamics of a Delayed Within-Host Viral Infection Model with Both Virus-to-Cell and Cell-to-Cell Transmissions. Math. Biosci. 2015, 270, 183–191. [Google Scholar] [CrossRef]
- Zhang, S.; Li, F.; Xu, X. Dynamics and control strategy for a delayed viral infection model. J. Biol. Dyn. 2022, 16, 44–63. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, X.Q. A Within-Host Virus Model with Periodic Multidrug Therapy. Bull. Math. Biol. 2013, 75, 543–563. [Google Scholar] [CrossRef]
- Canini, L.; Perelson, A.S. Viral Kinetic Modeling: State of the Art. J. Pharmacokinet. Pharmacodyn. 2014, 41, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Chimal-Eguia, J.C. Mathematical Model of Antiviral Immune Response against the COVID-19 Virus. Mathematics 2021, 9, 1356. [Google Scholar] [CrossRef]
- Xu, Z.; Song, J.; Zhang, H.; Wei, Z.; Wei, D.; Yang, G.; Demongeot, J.; Zeng, Q. A Mathematical Model Simulating the Adaptive Immune Response in Various Vaccines and Vaccination Strategies. Sci. Rep. 2024, 14, 23995. [Google Scholar] [CrossRef]
- Eftimie, R.; Gillard, J.J.; Cantrell, D.A. Mathematical Models for Immunology: Current State of the Art and Future Research Directions. Bull. Math. Biol. 2016, 78, 2091–2134. [Google Scholar] [CrossRef]
- Holling, C.S. Some characteristics of simple types of predation and parasitism1. Can. Entomol. 1959, 91, 385–398. [Google Scholar] [CrossRef]
- Cantrell, R.S.; Cosner, C. On the dynamics of predator–prey models with the Beddington–DeAngelis functional response. J. Math. Anal. Appl. 2001, 257, 206–222. [Google Scholar] [CrossRef]
- Hahl, S.K.; Kremling, A. A Comparison of Deterministic and Stochastic Modeling Approaches for Biochemical Reaction Systems: On Fixed Points, Means, and Modes. Front. Genet. 2016, 7, 157. [Google Scholar] [CrossRef]
- Chen-Charpentier, B. On population models with delays and dependence on past values. Axioms 2024, 13, 206. [Google Scholar] [CrossRef]
- Diekmann, O.; Heesterbeek, J.; Roberts, M.G. The construction of next-generation matrices for compartmental epidemic models. J. R. Soc. Interface 2010, 7, 873–885. [Google Scholar] [CrossRef]
- Van den Driessche, P. Reproduction numbers of infectious disease models. Infect. Dis. Model. 2017, 2, 288–303. [Google Scholar] [CrossRef] [PubMed]
- Hefferman, J.; Smith, R.; Wahl, L. Perspectives on the basic reproduction ratio. J. R. Soc. Interface 2005, 2, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.D. MathematicalBiology I. An Introduction; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Allen, L. An Introduction to Mathematical Biology; Pearson-Prentice Hall: London, UK, 2007. [Google Scholar]
- Chen-Charpentier, B. A model of hepatitis B viral dynamics with delays. AppliedMath 2024, 4, 182–196. [Google Scholar] [CrossRef]
- Ciupe, S.M.; Ribeiro, R.M.; Nelson, P.W.; Dusheiko, G.; Perelson, A.S. The role of cells refractory to productive infection in acute hepatitis B viral dynamics. Proc. Natl. Acad. Sci. USA 2007, 104, 5050–5055. [Google Scholar] [CrossRef]
- Kim, H.Y.; Kwon, H.D.; Jang, T.S.; Lim, J.; Lee, H.S. Mathematical modeling of triphasic viral dynamics in patients with HBeAg-positive chronic hepatitis B showing response to 24-week clevudine therapy. PLoS ONE 2012, 7, e50377. [Google Scholar] [CrossRef]
- Herbert, J.A.; Panagiotou, S. Immune Response to Viruses. In Encyclopedia of Infection and Immunity; Elsevier: Amsterdam, The Netherlands, 2022; pp. 429–444. [Google Scholar] [CrossRef]
- Meischel, T.; Villalon-Letelier, F.; Saunders, P.M.; Reading, P.C.; Londrigan, S.L. Influenza A virus interactions with macrophages: Lessons from epithelial cells. Cell. Microbiol. 2020, 22, e13170. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Wang, X.; Ho, W.Z. Roles of Macrophages in Viral Infections. Viruses 2024, 16, 1643. [Google Scholar] [CrossRef]
- Rouse, B.T.; Sehrawat, S. Immunity and immunopathology to viruses: What decides the outcome? Nat. Rev. Immunol. 2010, 10, 514–526. [Google Scholar] [CrossRef]
- Mueller, S.N.; Rouse, B.T. Immune responses to viruses. Clin. Immunol. 2009, 421–431. [Google Scholar]
- Gracia-Hernandez, M.; Sotomayor, E.M.; Villagra, A. Targeting Macrophages as a Therapeutic Option in Coronavirus Disease 2019. Front. Pharmacol. 2020, 11, 577571. [Google Scholar] [CrossRef] [PubMed]
- Chathuranga, K.; Weerawardhana, A.; Dodantenna, N.; Lee, J.S. Regulation of Antiviral Innate Immune Signaling and Viral Evasion Following Viral Genome Sensing. Exp. Mol. Med. 2021, 53, 1647–1668. [Google Scholar] [CrossRef]
- Rodrigo, M.B.; De Min, A.; Jorch, S.K.; Martin-Higueras, C.; Baumgart, A.K.; Goldyn, B.; Becker, S.; Garbi, N.; Lemmermann, N.A.; Kurts, C. Dual Fluorescence Reporter Mice for Ccl3 Transcription, Translation, and Intercellular Communication. J. Exp. Med. 2024, 221, e20231814. [Google Scholar] [CrossRef]
- Koyama, S.; Ishii, K.J.; Coban, C.; Akira, S. Innate Immune Response to Viral Infection. Cytokine 2008, 43, 336–341. [Google Scholar] [CrossRef]
- Ciupe, S.M.; Ribeiro, R.M.; Perelson, A.S. Antibody responses during hepatitis B viral infection. PLoS Comput. Biol. 2014, 10, e1003730. [Google Scholar] [CrossRef]
- Wolfram Inc. Mathematica, version 13.2; Wolfram Inc.: Champaign, IL, USA, 2022. [Google Scholar]
- Cleveland Clinic What Do Antivirals Treat? 2025. Available online: https://my.clevelandclinic.org/health/treatments/antivirals (accessed on 25 March 2025).
- Perelson, A.S. Modelling Viral and Immune System Dynamics. Nat. Rev. Immunol. 2002, 2, 28–36. [Google Scholar] [CrossRef]
- Ciupe, S.M. Modeling the dynamics of hepatitis B infection, immunity, and drug therapy. Immunol. Rev. 2018, 285, 38–54. [Google Scholar] [CrossRef]
- Marino, S.; Hogue, I.B.; Ray, C.J.; Kirschner, D.E. A Methodology For Performing Global Uncertainty And Sensitivity Analysis In Systems Biology. J. Theor. Biol. 2008, 254, 178–196. [Google Scholar] [CrossRef]
- Rackauckas, C.; Nie, Q. DifferentialEquations.jl—A performant and feature-rich ecosystem for solving differential equations in Julia. J. Open Res. Softw. 2017, 5, 15. [Google Scholar] [CrossRef]
- Press, W.H. Numerical Recipes 3rd Edition: The Art of Scientific Computing; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Virgin, H.W.; Wherry, E.J.; Ahmed, R. Redefining Chronic Viral Infection. Cell 2009, 138, 30–50. [Google Scholar] [CrossRef] [PubMed]
- Wodarz, D. Mathematical models of immune effector responses to viral infections: Virus control versus the development of pathology. J. Comput. Appl. Math. 2005, 184, 301–319. [Google Scholar] [CrossRef]
- Kolaczkowska, E.; Kubes, P. Neutrophil Recruitment and Function in Health and Inflammation. Nat. Rev. Immunol. 2013, 13, 159–175. [Google Scholar] [CrossRef]
- Willenborg, S.; Eming, S.A. Macrophages–Sensors and Effectors Coordinating Skin Damage and Repair. JDDG J. Dtsch. Dermatol. Ges. 2014, 12, 214–221. [Google Scholar] [CrossRef]
- Nayar, S.; Dasgupta, P.; Galustian, C. Extending the Lifespan and Efficacies of Immune Cells Used in Adoptive Transfer for Cancer Immunotherapies–A Review. Oncoimmunology 2015, 4, e1002720. [Google Scholar] [CrossRef]
- De Boer, R.J.; Homann, D.; Perelson, A.S. Different Dynamics of CD4+ and CD8+ T Cell Responses During and After Acute Lymphocytic Choriomeningitis Virus Infection 1. J. Immunol. 2003, 171, 3928–3935. [Google Scholar] [CrossRef] [PubMed]
- Auner, H.W.; Beham-Schmid, C.; Dillon, N.; Sabbattini, P. The Life Span of Short-Lived Plasma Cells Is Partly Determined by a Block on Activation of Apoptotic Caspases Acting in Combination with Endoplasmic Reticulum Stress. Blood 2010, 116, 3445–3455. [Google Scholar] [CrossRef]
- Sagiv, J.Y.; Voels, S.; Granot, Z. Isolation and characterization of low-vs. high-density neutrophils in cancer. In The Tumor Microenvironment: Methods and Protocols; Springer: New York, NY, USA, 2016; pp. 179–193. [Google Scholar]
- Galli, S.J.; Borregaard, N.; Wynn, T.A. Phenotypic and Functional Plasticity of Cells of Innate Immunity: Macrophages, Mast Cells and Neutrophils. Nat. Immunol. 2011, 12, 1035–1044. [Google Scholar] [CrossRef]
- Derksen, L.Y.; Tesselaar, K.; Borghans, J.A.M. Memories That Last: Dynamics of Memory T Cells throughout the Body. Immunol. Rev. 2023, 316, 38–51. [Google Scholar] [CrossRef]
- Derksen, L.Y. Memories That Last: Dynamics of Memory Cells in Mice, Dirty Mice, and Men. Ph.D. Thesis, Utrecht University, Utrecht, The Netherlands, 2024. [Google Scholar] [CrossRef]
- Arunachalam, A.B. Vaccines Induce Homeostatic Immunity, Generating Several Secondary Benefits. Vaccines 2024, 12, 396. [Google Scholar] [CrossRef]
- Saltelli, A.; Ratto, M.; Andres, T.; Campolongo, F.; Cariboni, J.; Gatelli, D.; Saisana, M.; Tarantola, S. Global Sensitivity Analysis: The Primer; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Dixit, V.K.; Rackauckas, C. GlobalSensitivity. jl: Performant and Parallel Global Sensitivity Analysis with Julia. J. Open Source Softw. 2022, 7, 4561. [Google Scholar] [CrossRef]
- Chen, W.W.; Niepel, M.; Sorger, P.K. Classic and contemporary approaches to modeling biochemical reactions. Genes Dev. 2010, 24, 1861–1875. [Google Scholar] [CrossRef]
- De Boeck, J.; Rombouts, J.; Gelens, L. A modular approach for modeling the cell cycle based on functional response curves. PLoS Comput. Biol. 2021, 17, e1009008. [Google Scholar] [CrossRef] [PubMed]
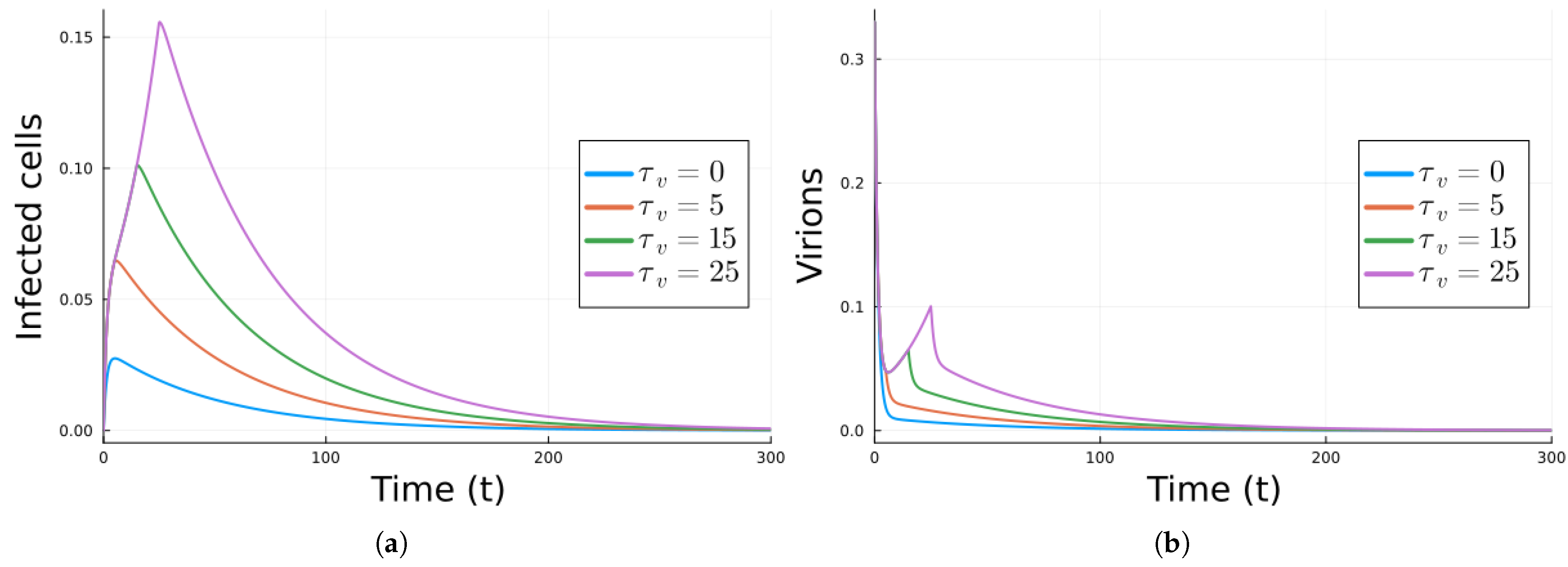
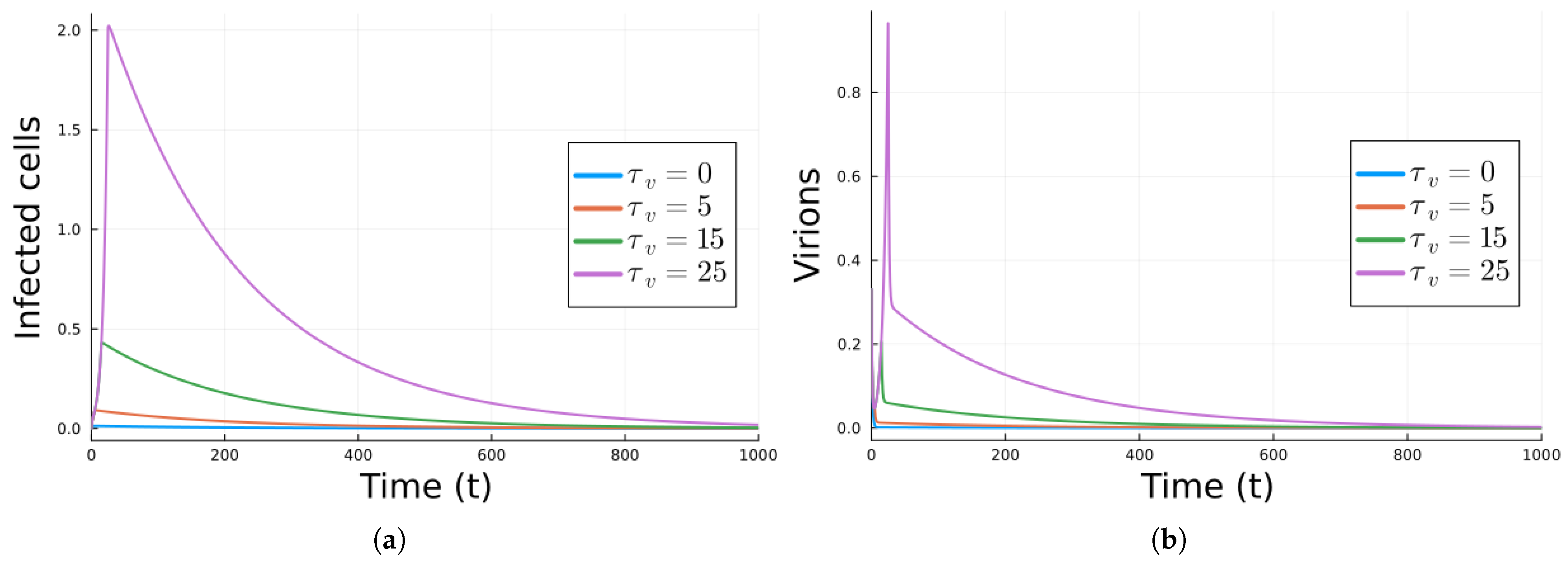
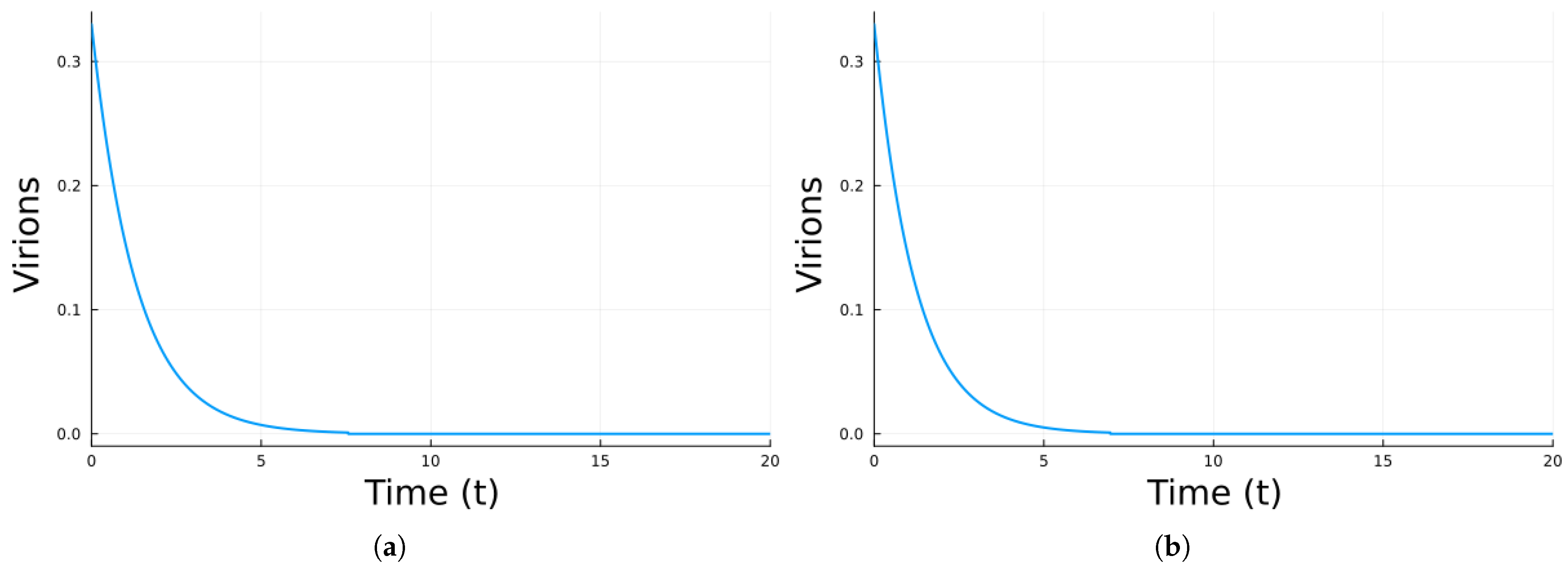
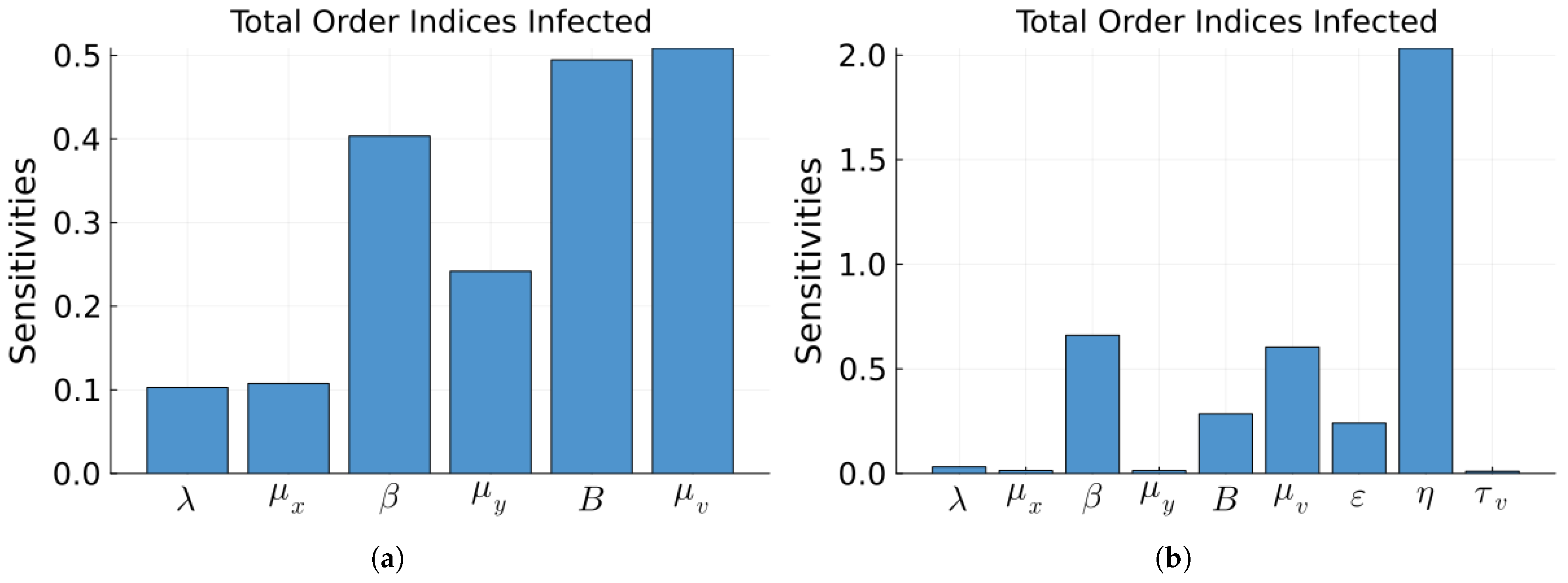
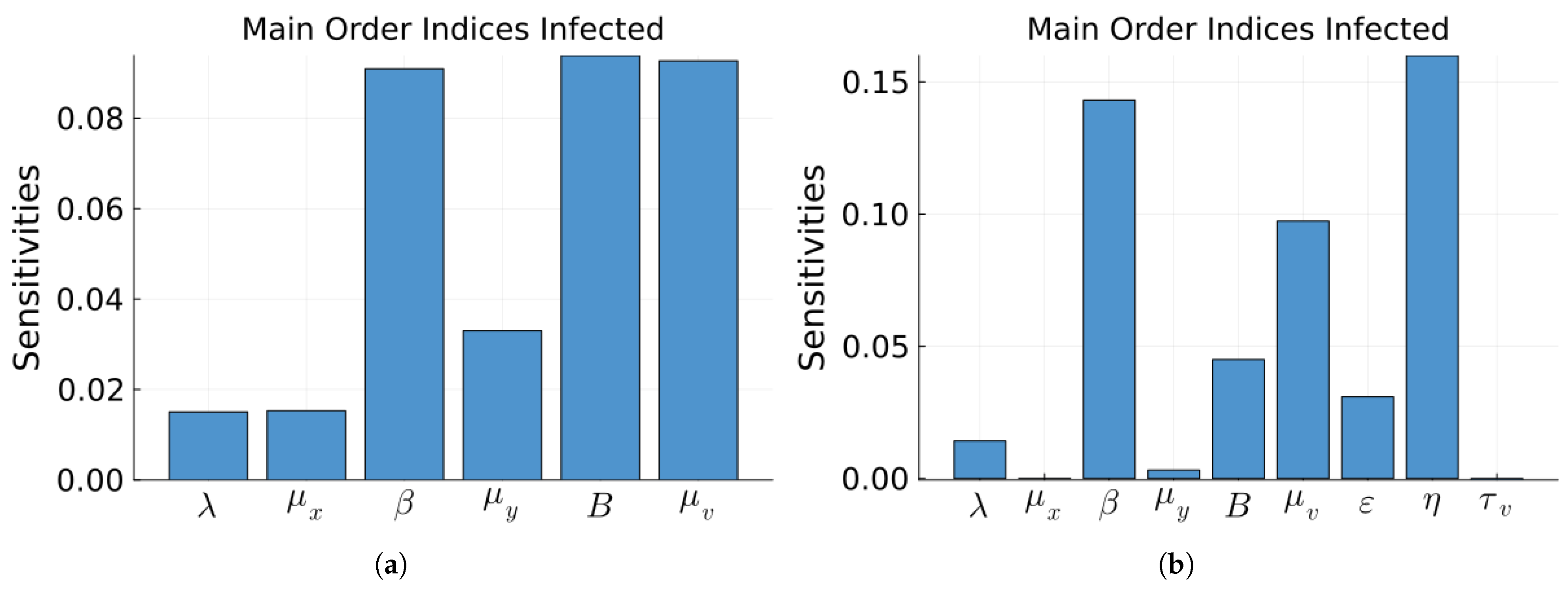
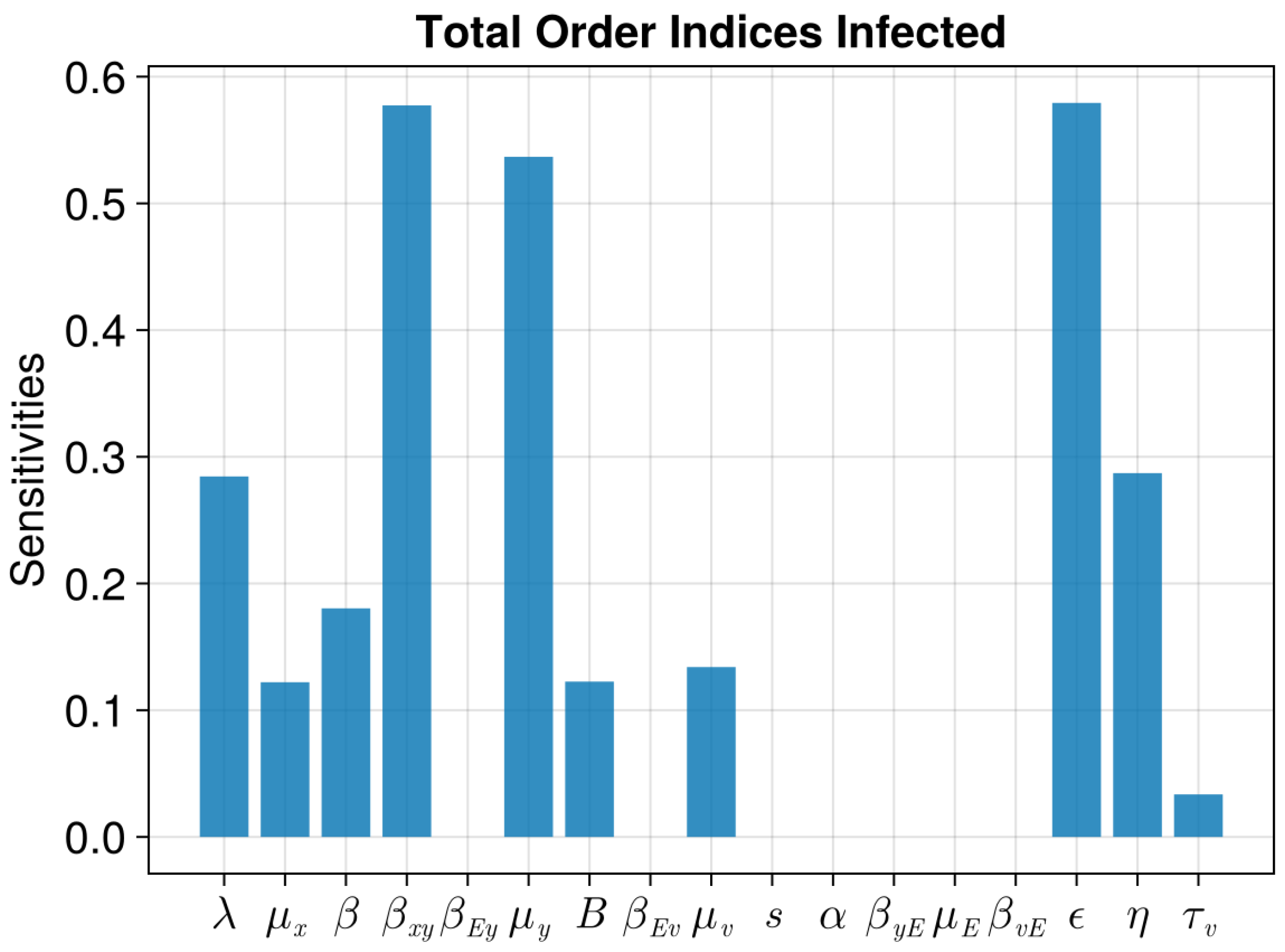
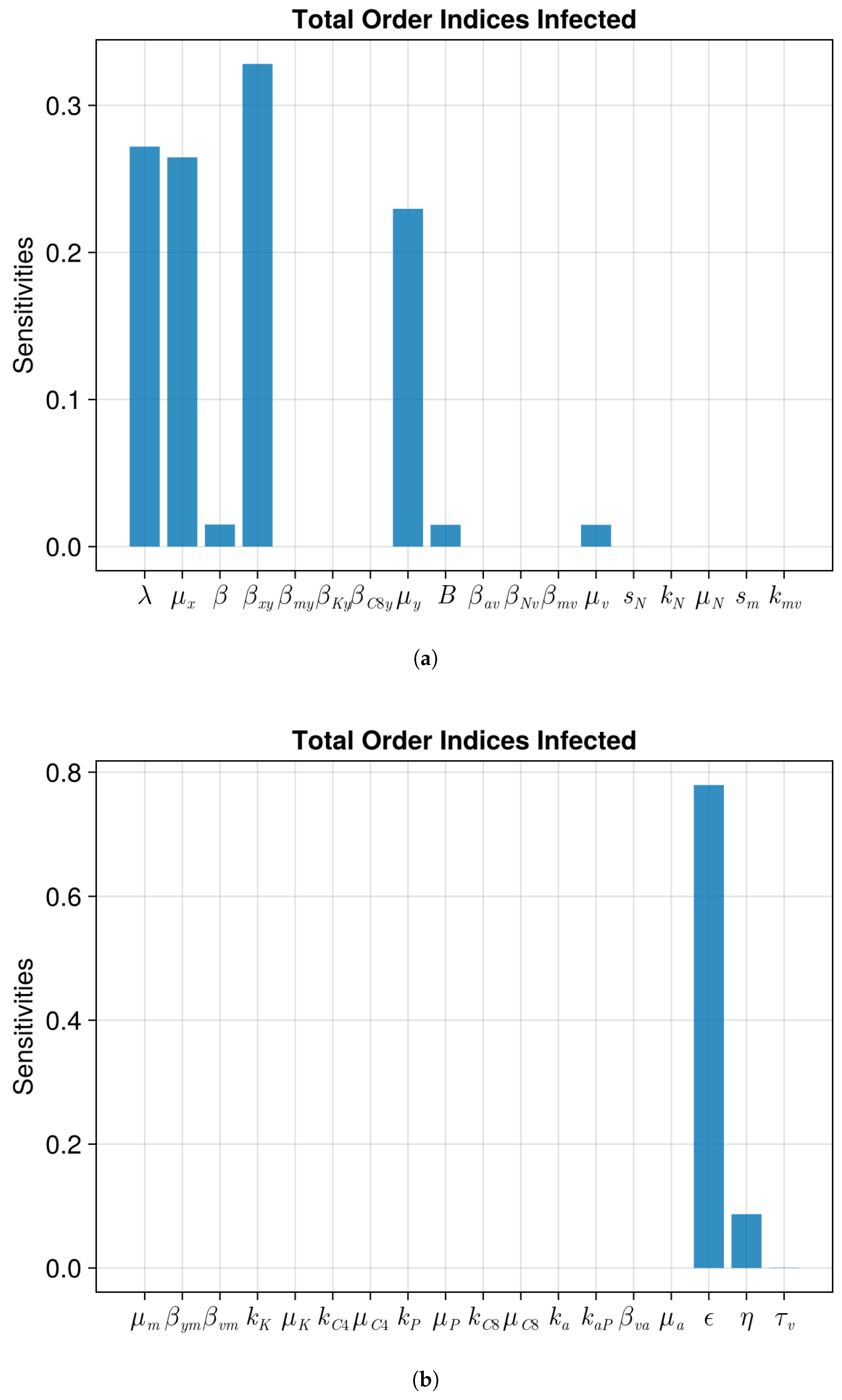
| Parameter | Value | Description |
|---|---|---|
| 5 cells/mL L/d | recruitment rate of susceptible cells | |
| 0.003/d | death rate of susceptible cells | |
| 8 mL/(virions d) | infection rate of susceptible cells by virus | |
| 0.043 L/d | death rate of infected cells | |
| B | 11.16 | number of virions produced by 1 infected cell |
| 0.7 L/d | death rate of virus |
| Parameter | Value | Description |
|---|---|---|
| 5 cells/mL L/d | recruitment rate of susceptible cells | |
| 0.003/d | death rate of susceptible cells | |
| 8 mL/(virions d) | infection rate of susceptible cells by virus | |
| 0.043/d | death rate of infected cells | |
| B | 11.16 | number of virions produced by 1 infected cell |
| 0.7 L/d | death rate of virus | |
| 4 mL/(cells d) | infection rate of susceptible cells by virus | |
| 0.6 mL/(cells d) | elimination rate of infected cells by effector cells | |
| 0.6 mL/(cells d) | removal rate of effector cells after elimination of infected cells | |
| 4 mL/(cells d) | elimination rate of virus by effector cells | |
| 4 mL/(virions d) | removal rate of y effector cells after elimination of virus | |
| s | 24 cells/mL L/d | recruitment rate of effector cells |
| 2.2 /d | recruitment rate of effector cells due to infected cells | |
| 0.5/d | death rate of effector cells |
| Parameter | Value | Description |
|---|---|---|
| 1.0 mL/(cells d) | elimination rate of infected cells by macrophages | |
| 1.0 mL/(cells d) | elimination rate of infected cells by natural killer cells | |
| 2.0 mL/(cells d) | elimination rate of infected cells by CD8+ cells | |
| 1.0 mL/ d | elimination rate of virions by antibodies | |
| 1.0 mL/(virions d) | elimination rate of antibodies due to elimination of a virion | |
| 4.0 mL/(virions d) | elimination rate of virions by neutrophils | |
| 4.0 mL/(virions d) | elimination rate of virions by macrophages | |
| 60.0 cells/mL L/(d virions) | natural recruitment rate of neutrophils | |
| 2.2 cells/mL L/d | recruitment rate of neutrophils due to virus | |
| 3.0 cells/ ml L/d | death rate of neutrophils | |
| 0 cells/mL 1/(d virions) | natural recruitment rate of macrophages | |
| 1.1 L/d | recruitment rate of macrophages due to infected cells | |
| 4.8 cells/mL L/d | death rate of macrophages | |
| 0 mL/(virions d) | elimination rate of macrophages due to eliminating virions | |
| 0 mL/(cells d) | elimination rate of macrophages due to eliminating infected cells | |
| 2.2 L/d | activation rate of natural killer cells by macrophages | |
| 7 L/d | death rate of natural killer cells | |
| 1.1 L/d | activation rate of CD4+ cells | |
| 1.1 L/d | death rate of CD4+ cells | |
| 1.1 L/d | activation rate of plasma cells | |
| 0.2 L/d | death rate of plasma cells | |
| 1.1 L/d | activation rate of CD8+ cells | |
| 7 L/d | death rate of CD8+ cells | |
| 3.6 L/mL L/d | natural recruitment rate of antibodies | |
| 2.2 L/(cells) L/d | recruitment rate of antibodies by virus | |
| 1.0 mL/(virions d) | elimination rate of antibodies by virus | |
| 0.3/d | elimination rate of antibodies |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen-Charpentier, B. Modeling How the Different Parts of the Immune System Fight Viruses. Algorithms 2025, 18, 544. https://doi.org/10.3390/a18090544
Chen-Charpentier B. Modeling How the Different Parts of the Immune System Fight Viruses. Algorithms. 2025; 18(9):544. https://doi.org/10.3390/a18090544
Chicago/Turabian StyleChen-Charpentier, Benito. 2025. "Modeling How the Different Parts of the Immune System Fight Viruses" Algorithms 18, no. 9: 544. https://doi.org/10.3390/a18090544
APA StyleChen-Charpentier, B. (2025). Modeling How the Different Parts of the Immune System Fight Viruses. Algorithms, 18(9), 544. https://doi.org/10.3390/a18090544







