CSpredR: A Multi-Site mRNA Subcellular Localization Prediction Method Based on Fusion Encoding and Hybrid Neural Networks
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
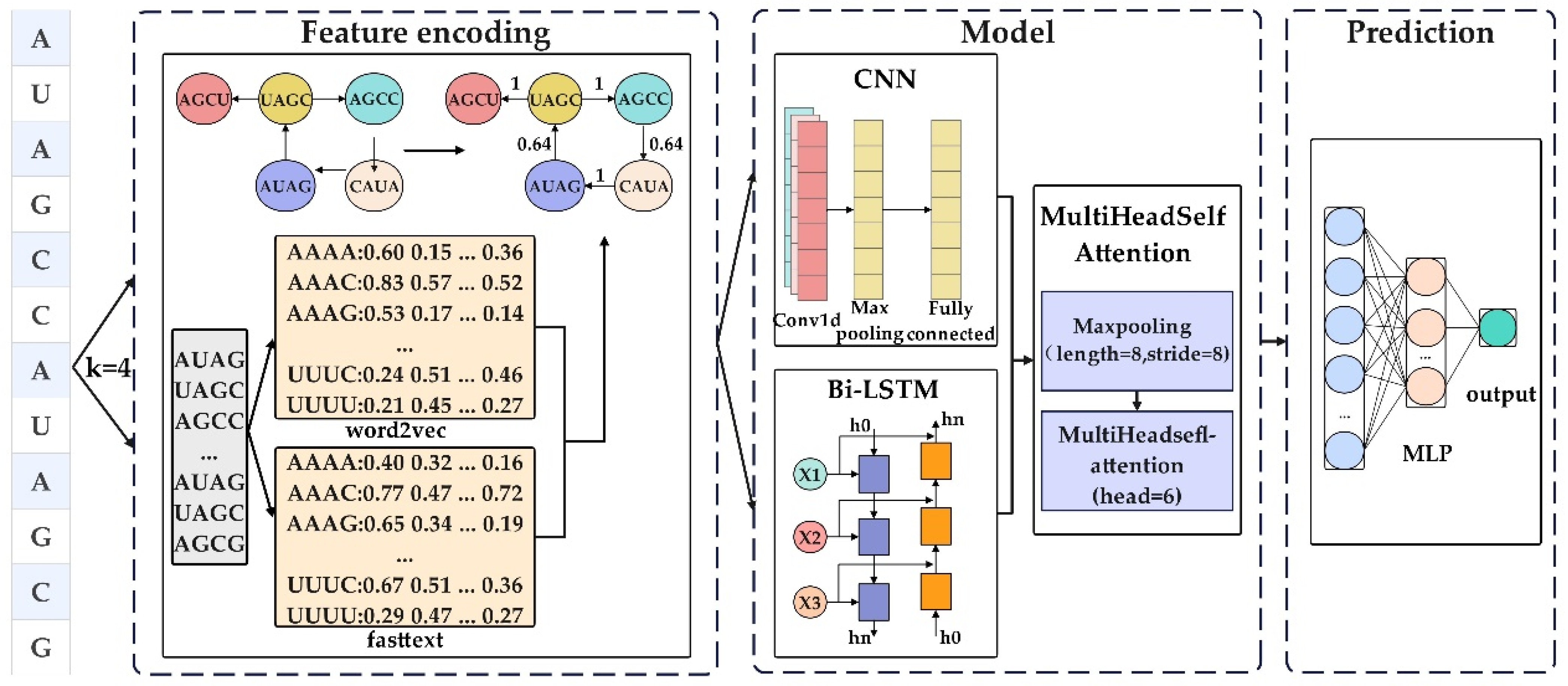
2.2. The Model Framework of CSpredR
2.3. Graph Construction
2.4. Feature Encoding
2.5. Convolutional Neural Network Method
2.6. Bidirectional Long Short-Term Memory Method
2.7. Synergistic Model: CNN and Bi-LSTM for Capturing Sequence Features
2.8. Multi-Head Attention
2.9. Performance Evaluation Metrics
2.10. Hyperparameter Optimization
3. Results and Discussion
3.1. Comparison of Different k-mer Features
3.2. Ablation Experiment
3.3. Comparison with Other Single Label Multi-Class Classification Methods
3.4. The Comparison of CSpredR with Other Prediction Models
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Di Liegro, C.M.; Schiera, G.; DI Liegro, I. Regulation of mRNA transport, localization and translation in the nervous system of mammals. Int. J. Mol. Med. 2014, 33, 747–762. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.; Garzia, A.; Tuschl, T. Simultaneous detection of the subcellular localization of RNAs and proteins in cultured cells by combined multicolor RNA-FISH and IF. Methods 2017, 118–119, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Liu-Yesucevitz, L.; Bassell, G.J.; Gitler, A.D.; Hart, A.C.; Klann, E.; Richter, J.D.; Warren, S.T.; Wolozin, B. Local rna translation at the synapse and in disease. J. Neurosci. 2011, 31, 16086–16093. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.R.; Swanson, M.S. Mechanisms of RNA-mediated Disease. J. Biol. Chem. 2009, 284, 7419–7423. [Google Scholar] [CrossRef] [PubMed]
- Chin, A.; Lécuyer, E. RNA localization: Making its way to the center stage. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2956–2970. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhu, L.; Wang, X.; Jin, H. RNA-based therapeutics: An overview and prospectus. Cell Death Dis. 2022, 13, 644. [Google Scholar] [CrossRef] [PubMed]
- Uemura, M.; Zheng, Q.; Koh, C.M.; Nelson, W.G.; Yegnasubramanian, S.; De Marzo, A.M. Overexpression of ribosomal RNA in prostate cancer is common but not linked to rDNA promoter hypomethylation. Oncogene 2011, 31, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Dolezal, J.M.; Dash, A.P.; Prochownik, E.V. Diagnostic and prognostic implications of ribosomal protein transcript expression patterns in human cancers. BMC Cancer 2018, 18, 275. [Google Scholar] [CrossRef] [PubMed]
- Sprenkle, N.T.; Sims, S.G.; Sánchez, C.L.; Meares, G.P. Endoplasmic reticulum stress and inflammation in the central nervous system. Mol. Neurodegener. 2017, 12, 1–18. [Google Scholar] [CrossRef]
- Yan, Z.; Lécuyer, E.; Blanchette, M. Prediction of mRNA subcellular localization using deep recurrent neural networks. Bioinformatics 2019, 35, i333–i342. [Google Scholar] [CrossRef] [PubMed]
- Alshubaily, I. TextCNN with attention for text classification. arXiv 2021, arXiv:2108.01921. [Google Scholar]
- Jang, B.; Kim, M.; Harerimana, G.; Kang, S.U.; Kim, J.W. Bi-LSTM model to increase accuracy in text classification: Combining Word2vec CNN and attention mechanism. Appl. Sci. 2020, 10, 5841. [Google Scholar] [CrossRef]
- Meher, P.K.; Rai, A.; Rao, A.R. mLoc-mRNA: Predicting multiple sub-cellular localization of mRNAs using random forest algorithm coupled with feature selection via elastic net. BMC Bioinform. 2021, 22, 342. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Singhal, N.; Kumar, R.; Kumar, M. mRNALoc: A novel machine-learning based in-silico tool to predict mRNA subcellular localization. Nucleic Acids Res. 2020, 48, W239–W243. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, L.; He, S.; Guo, F.; Zou, Q. SubLocEP: A novel ensemble predictor of subcellular localization of eukaryotic mRNA based on machine learning. Brief. Bioinform. 2021, 22, bbaa401. [Google Scholar] [CrossRef]
- Tang, Q.; Nie, F.; Kang, J.; Chen, W. mRNALocater: Enhance the prediction accuracy of eukaryotic mRNA subcellular localization by using model fusion strategy. Mol. Ther. 2021, 29, 2617–2623. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Shen, Z.; Liu, T.; Long, W.; Jiang, L.; Peng, S. DeepmRNALoc: ANovelPredictor of Eukaryotic mRNA Subcellular Local-ization Based on Deep Learning. Molecules 2023, 28, 2284. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Z.; Jiang, Y.; Mao, Z.; Wang, D.; Lin, H.; Xu, D. DM3Loc: Multi-label mRNA subcellular localization prediction and analysis based on multi-head self-attention mechanism. Nucleic Acids Res. 2021, 49, e46. [Google Scholar] [CrossRef]
- Bi, Y.; Li, F.; Guo, X.; Wang, Z.; Pan, T.; Guo, Y.; I Webb, G.; Yao, J.; Jia, C.; Song, J. Clarion is a multi-label problem transformation method for identifying mRNA subcellular localizations. Briefings Bioinform. 2022, 23, bbac467. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Musleh, S.; Arif, M.; Alajez, N.M.; Alam, T. Unified mRNA Subcellular Localization Predictor based on machine learning tech-niques. BMC Genom. 2024, 25, 151. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhao, B.; Yin, R.; Lu, C.; Guo, F.; Zeng, M. GraphLncLoc: Long non-coding RNA subcellular localization prediction using graph convolutional networks based on sequence to graph transformation. Brief. Bioinform. 2022, 24, bbac565. [Google Scholar] [CrossRef] [PubMed]
- Tsukiyama, S.; Hasan, M.M.; Fujii, S.; Kurata, H. LSTM-PHV: Prediction of human-virus protein-protein interactions by LSTM with word2vec. Brief. Bioinform. 2021, 22, bbab228. [Google Scholar] [CrossRef] [PubMed]
- Le, N.Q.K.; Yapp, E.K.Y.; Nagasundaram, N.; Yeh, H.-Y. Classifying Promoters by Interpreting the Hidden Information of DNA Sequences via Deep Learning and Combination of Continuous FastText N-Grams. Front. Bioeng. Biotechnol. 2019, 7, 305. [Google Scholar] [CrossRef]
- Chauhan, R.; Ghanshala, K.K.; Joshi, R.C. Convolutional neural network (CNN) for image detection and recognition. In Proceedings of the 2018 First International Conference on Secure Cyber Computing and Communication (ICSCCC), Jalandhar, India, 15–17 December 2018; pp. 278–282. [Google Scholar]
- Abdin, O.; Nim, S.; Wen, H.; Kim, P.M. PepNN: A deep attention model for the identification of peptide binding sites. Commun. Biol. 2022, 5, 503. [Google Scholar] [CrossRef]
- Hong, Z.; Zeng, X.; Wei, L.; Liu, X. Identifying enhancer–promoter interactions with neural network based on pre-trained DNA vectors and attention mechanism. Bioinformatics 2019, 36, 1037–1043. [Google Scholar] [CrossRef]
- Park, S.; Koh, Y.; Jeon, H.; Kim, H.; Yeo, Y.; Kang, J. Enhancing the interpretability of transcription factor binding site prediction using attention mechanism. Sci. Rep. 2020, 10, 13413. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Tian, S.; Gao, X.; Li, Y. mlDEEPre: Multi-functional enzyme function prediction with hierarchical multi-label deep learning. Front. Genet. 2019, 9, 714. [Google Scholar] [CrossRef]
- Ghamrawi, N.; McCallum, A. Collective multi-label classification. In Proceedings of the 14th ACM International Conference on Information and Knowledge Management, Bremen, Germany, 31 October–5 November 2005. [Google Scholar]
- Gopal, S.; Yang, Y. Multilabel classification with meta-level features. In Proceedings of the 33rd International ACM SIGIR Conference on Research and Development in Information Retrieval, Geneva, Switzerland, 19–23 July 2010. [Google Scholar]
- Bai, T.; Yan, K.; Liu, B. DAmiRLocGNet: miRNA subcellular localization prediction by combining miRNA–disease associations and graph convolutional networks. Brief. Bioinform. 2023, 24, bbad212. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Yang, Y.H.; Ding, H.; Wang, D.; Chen, W.; Lin, H. Design powerful predictor for mRNA subcellular location prediction in Homo sapiens. Brief. Bioinform. 2021, 22, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Quang, D.; Xie, X. DanQ: A hybrid convolutional and recurrent deep neural network for quantifying the function of DNA se-quences. Nucleic Acids Res. 2016, 44, e107. [Google Scholar] [CrossRef]



| Subcellular Sites | Sample Size |
|---|---|
| cytoplasm | 4016 |
| nucleus | 21,439 |
| ribosome | 8680 |
| Exosome | 31,448 |
| Nucleoplasm | 14,237 |
| chromatin | 14,328 |
| nucleolus | 11,124 |
| Cytosol | 16,312 |
| membrane | 6739 |
| Hamming Loss | One-Error | ACC | Coverage | Average Precision | Ranking Loss | |
|---|---|---|---|---|---|---|
| Word2vec | 0.219 | 0.659 | 0.625 | 6.211 | 0.650 | 0.445 |
| Fasttext | 0.274 | 0.716 | 0.588 | 6.279 | 0.648 | 0.482 |
| CSpredR | 0.182 | 0.605 | 0.657 | 6.035 | 0.675 | 0.380 |
| Word2vec | Fasttext | |
|---|---|---|
| p-value of Hamming loss | 0.03125 | 0.00484 |
| p-value of One-error | 0.03659 | 0.03496 |
| p-value of ACC | 0.03805 | 0.03778 |
| p-value of Coverage | 0.00272 | 0.00278 |
| p-value of Average precision | 0.04256 | 0.04231 |
| p-value of Ranking loss | 0.03805 | 0.04121 |
| Hamming Loss | One-Error | ACC | Coverage | Average Precision | Ranking Loss | |
|---|---|---|---|---|---|---|
| CNN | 0.322 | 0.710 | 0.437 | 7.496 | 0.496 | 0.517 |
| Bi-LSTM | 0.289 | 0.656 | 0.512 | 6.942 | 0.530 | 0.471 |
| CNN + attention | 0.276 | 0.671 | 0.525 | 6.828 | 0.579 | 0.429 |
| Bi-LSTM + attention | 0.235 | 0.632 | 0.578 | 6.440 | 0.646 | 0.402 |
| CSpredR | 0.182 | 0.605 | 0.657 | 6.035 | 0.675 | 0.380 |
| CNN | Bi-LSTM | CNN+ Attention | Bi-LSTM+ Attention | |
|---|---|---|---|---|
| p-value of Hamming loss | 0.01354 | 0.01964 | 0.01978 | 0.02169 |
| p-value of One-error | 0.03141 | 0.03783 | 0.04918 | 0.04571 |
| p-value of ACC | 0.03192 | 0.03794 | 0.04126 | 0.04780 |
| p-value of Coverage | 0.00429 | 0.01468 | 0.02779 | 0.03014 |
| p-value of Average precision | 0.03837 | 0.03994 | 0.04108 | 0.04296 |
| p-value of Ranking loss | 0.02711 | 0.02846 | 0.02971 | 0.03249 |
| Predictors | ACC | Precision | Recall | F1 Score |
|---|---|---|---|---|
| lncLocator | 0.421 | 0.374 | 0.325 | 0.289 |
| iLoc-lncRNA | 0.509 | 0.524 | 0.470 | 0.474 |
| Locate-R | 0.368 | 0.362 | 0.321 | 0.321 |
| GraphLncLoc | 0.579 | 0.736 | 0.557 | 0.584 |
| CSpredR | 0.671 | 0.755 | 0.592 | 0.643 |
| iLoc-mRNA | mRNALoc | mRNALocator | DM3Loc | Clarion | CSpredR | |
|---|---|---|---|---|---|---|
| chromatin | -- | -- | -- | -- | 81.47% | 81.50% |
| cytoplasm | -- | 54.88% | 38.90% | -- | 91.29% | 94.62% |
| Cytosol | -- | -- | -- | 57.37% | 79.77% | 83.55% |
| Exosome | -- | -- | -- | 70.00% | 92.10% | 95.46% |
| membrane | -- | -- | -- | 70.92% | 89.15% | 91.12% |
| nucleolus | -- | -- | -- | -- | 83.74% | 83.88% |
| Nucleoplasm | -- | -- | -- | -- | 80.74% | 81.20% |
| nucleus | -- | 55.18% | 57.42% | 69.52% | 79.23% | 80.06% |
| ribosome | 73.41% | -- | -- | 69.03% | 84.74% | 86.42% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Suo, W.; Wang, R. CSpredR: A Multi-Site mRNA Subcellular Localization Prediction Method Based on Fusion Encoding and Hybrid Neural Networks. Algorithms 2025, 18, 67. https://doi.org/10.3390/a18020067
Wang X, Suo W, Wang R. CSpredR: A Multi-Site mRNA Subcellular Localization Prediction Method Based on Fusion Encoding and Hybrid Neural Networks. Algorithms. 2025; 18(2):67. https://doi.org/10.3390/a18020067
Chicago/Turabian StyleWang, Xiao, Wenshuai Suo, and Rong Wang. 2025. "CSpredR: A Multi-Site mRNA Subcellular Localization Prediction Method Based on Fusion Encoding and Hybrid Neural Networks" Algorithms 18, no. 2: 67. https://doi.org/10.3390/a18020067
APA StyleWang, X., Suo, W., & Wang, R. (2025). CSpredR: A Multi-Site mRNA Subcellular Localization Prediction Method Based on Fusion Encoding and Hybrid Neural Networks. Algorithms, 18(2), 67. https://doi.org/10.3390/a18020067








