1. Introduction
Radial-basis functions have many important applications in the fields such as function interpolation [
1], meshless methods [
2], clustering classification [
3], surrogate models [
4], Autoencoder [
5], dynamic system design [
6], network event detection [
7], and modeling in energy production processes [
8], to name a few. The Gaussian-radial-basis function neural network (GRBFNN) is a neural network with one hidden layer and produces output in the form
where
is a radially symmetric unit represented by the Gaussian function such as
Herein, and are the center and width of the unit, respectively. The response of will be concentrated in the region whose a distance within 3- from the center in the i-th dimension. Therefore, one can select a group of sparsely distributed neurons with optimal centers and widths to capture the spatial characterization of a target function. This localized property of radial-basis functions has been extensively used in applications such as clustering and classification.
Although it has been shown that GRBFNNs outperform multilayer perceptrons (MLPs) in generalization [
9], tolerance to input noise [
10], and learning efficiency with a small set of data [
10], the network is not scalable for problems with high-dimensional input. This is because extensively more neurons are in need of accurate predictions, and the corresponding computations exponentially increase with the increase in dimensions. This paper aims to tackle this issue and make the network available for high-dimensional problems.
GRBFNN was proposed by Moody and Darken [
10] and Broomhhead and Lowe [
11] in the late 1980s for classification and function approximations. It was soon proven that GRBFNN is a universal approximator [
12,
13,
14] that can be arbitrarily close to a real-value function when a sufficient number of neurons is offered. The proof of universal approximability for GRBFNN can be interpreted as a process beginning with partitioning the domain of a target function into a grid, followed by using localized radial-basis functions to approximate the target function in each grid cell, then aggregating the localized functions to globally approximate the target function. It is evident that this approach is not feasible for high-dimensional problems because it will lead to the exponential growth of neurons as the number of input dimensions increases. For example, approximating a
d-variate function will require
neurons, with the domain of each dimension divided into
N segments.
To address this issue, researchers have heavily focused on selecting the optimal number of neurons as well as their centers and widths of GRBFNN such that the features of the target nonlinear map are well captured by the network. This has been mainly investigated through two strategies: (1) using supervised learning with a dynamical adjustment of neurons (e.g., numbers, centers, and widths) according to the prescribed criteria and (2) performing unsupervised-learning-based preprocessing on input to estimate the optimal placement and configuration of neurons.
For the former, Poggio and Girosi [
15] as well as Wettschereck and Dietterich [
16] applied gradient descent to train generalized-radial-basis function networks that have trainable centers. Regularization techniques [
15] were adopted to maintain the parsimonious structure of GRBFNN. Platt [
17] developed a two-layer network that dynamically allocates localized Gaussian neurons to the positions where the output pattern is not well represented. Chen et al. [
18] adopted an orthogonal least square (OLS) method and introduced a procedure that iteratively selects the optimal centers that minimize the error reduction ratio until the desired accuracy is achieved. Huang et al. [
19] proposed a growing and pruning strategy to dynamically add/remove neurons based on their contributions to learning accuracy.
The latter has been more popular because it decouples the placement of neurons and the computation of weights, reducing the complexity of program as well as computational load. Moody and Darken [
10] used the k-means clustering method [
3] to determine the centers that minimize the Euclidean distance between the training set and centers, followed by the calculation of a uniform width by averaging the distance to the nearest-neighbor of all units. Carvalho and Brizzotti [
20] investigated different clustering methods such as the iterative optimization (IO) technique, depth-first search (DF), and the combination of IO and DF for target recognition by RBFNNs. Niros and Tsekouras [
21] proposed a hierarchical fuzzy clustering method to estimate the number of neurons and trainable variables.
The optimization of widths has been of great interest more recently. Yao et al. [
22] numerically observed that the optimal widths of the radial basis function are affected by the spatial distribution of training data and the nonlinearity of approximated functions. With this in mind, they developed a method that determines the widths using the Euclidean distance between centers and second-order derivatives of a function. However, calculating the width of each neuron is computationally expensive. Instead of assigning each neuron a distinct width, it makes more sense to assign different widths to the neurons that represent different clusters for computational efficiency. Therefore, Yao et al. [
23] further proposed a method to optimize widths by dividing a global optimization problem into several subspace optimization problems that can be solved concurrently and then coordinated to converge to a global optimum. Similarly, Zhang et al. [
24] introduced a two-stage fuzzy clustering method to split the input space into multiple overlapping regions that are then used to construct a local Gaussian-radial-basis function network. Another method that should be mentioned is the variable projection [
25], which is used to reduce the number of parameters in the optimization problem.
However, the aforementioned methods all suffer from the curse of dimensionality. As the input dimension grows, the selection of optimal neurons itself can become cumbersome. To compound the problem, the number of optimal neurons can also rise exponentially when approximating high-dimensional and geometrically complex functions. Furthermore, the methods are designed for CPU-based, general-purpose computing machines but are not appropriate for tapping into the modern GPU-oriented machine-learning tools [
26,
27] whose computational efficiency drops significantly when handling branching statements and dynamical memory allocation. This gap motivates us to reevaluate the structure of GRBFNN. As stated previously, the localized property of Gaussian functions is beneficial for identifying the parsimonious structure of GRBFNN with low input dimensions, but it also leads to the blow-up of the number of neurons in high dimensional situations.
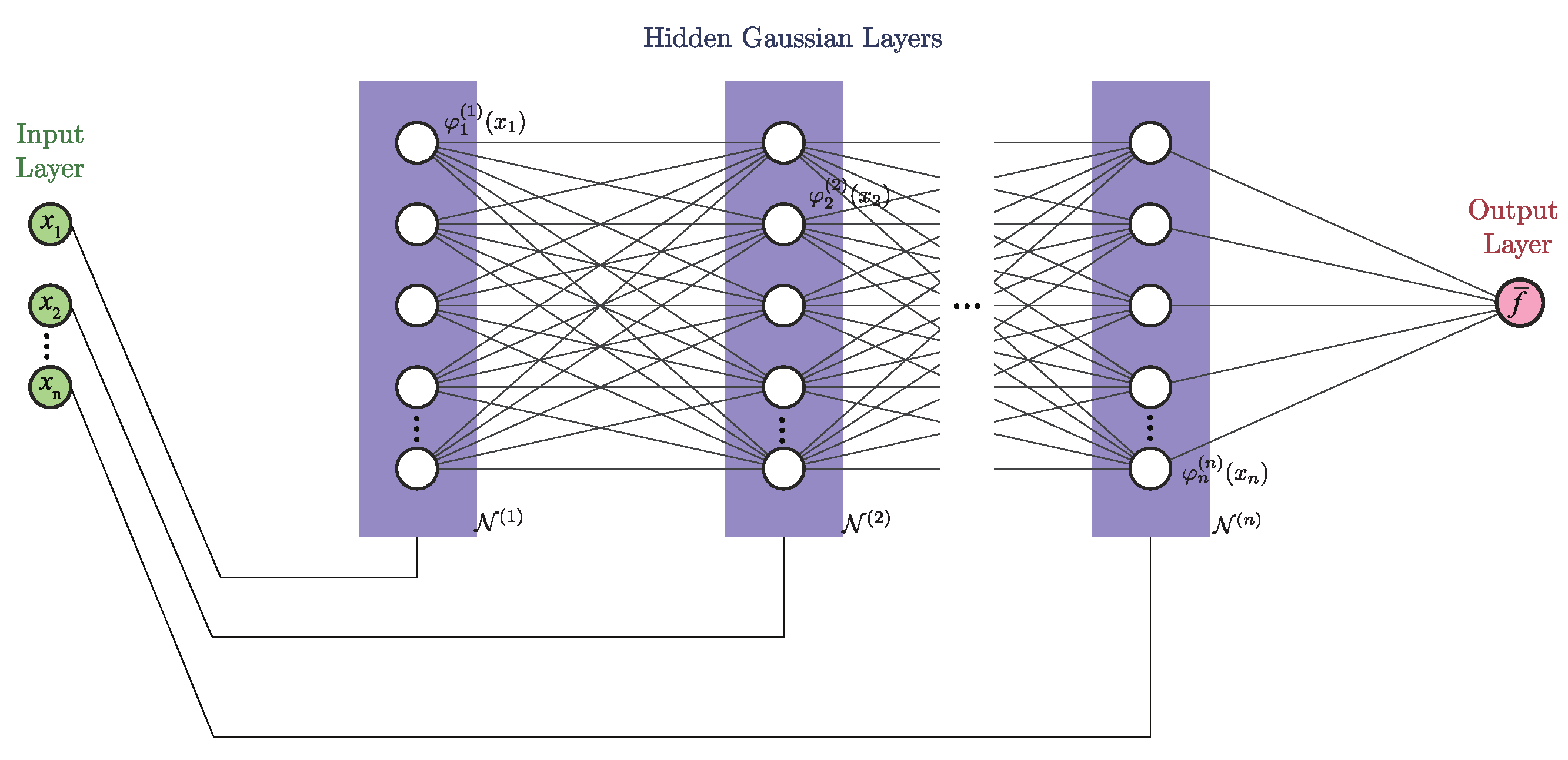
Given that the recent development of deep neural networks has shown promise in solving such problems,
the main goal of this paper is to develop a deep-neural-network representation of GRBFNN or at least a good approximation of it with significant improvement of computational efficiency such that the network can be used for solving very high dimensional problems. We approach this problem by utilizing the separable property of Gaussian radial basis functions. That is, every Gaussian-radial-basis function can be decomposed into the product of multiple uni-variate Gaussian functions. Based on this property, we construct a new neural network, namely separable Gaussian neural network (SGNN), whose number of layers is equal to the number of input dimensions, with the neurons of each layer formed by the corresponding uni-variate Gaussian functions. Through dividing the input into multiple columns by their dimensions and feeding them into the corresponding layers, the output equivalent to that of a GRBFNN is constructed from multiplications and summations of uni-variate Gaussian functions in the forward propagation. It should be noted that Poggio and Girosi [
15] have reported the separable property of Gaussian-radial-basis functions and proposed using it for neurobiology even in 1990.
SGNN offers several advantages.
The number of neurons of SGNN is and increases linearly with the dimension of the input, while the number of neurons of GRBFNN given by grows exponentially. This reduction in neurons also decreases the number of trainable variables from to , yielding a more compact network than GRBFNN.
The reduction in trainable variables further decreases the computational load during the training and testing of neural networks. As shown in
Section 3, this has led to 100 times speedup of training time for approximating tri-variate functions.
SGNN is much easier to tune than other MLPs. Since the number of layers in SGNN is equal to the number of dimension of the input data, the only tunable network-structural hyper-parameter is the layer width, i.e., the number of neurons in a layer. This can significantly alleviate the tuning workload as compared to other MLPs that must simultaneously tune the width and depth of layers.
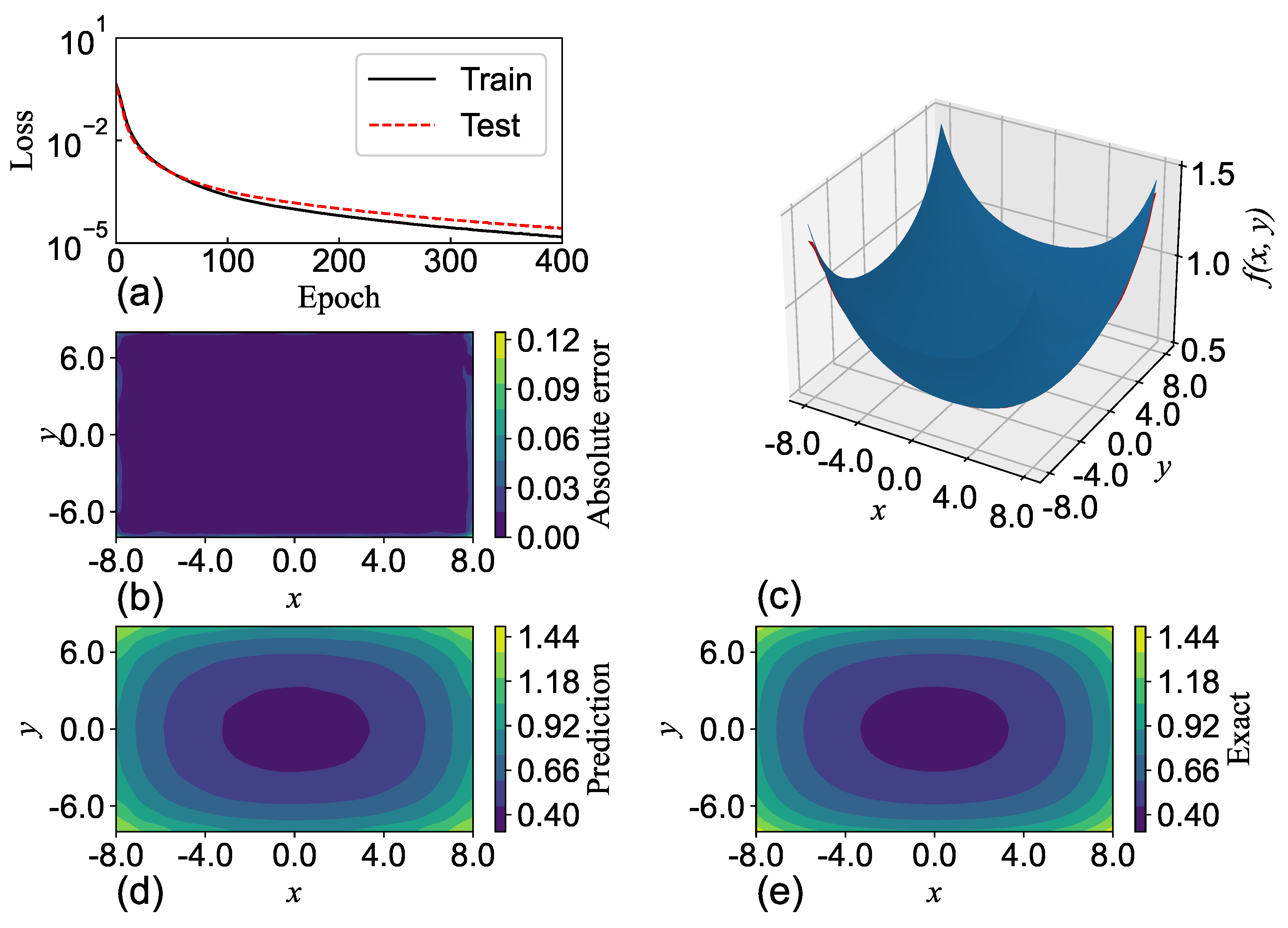
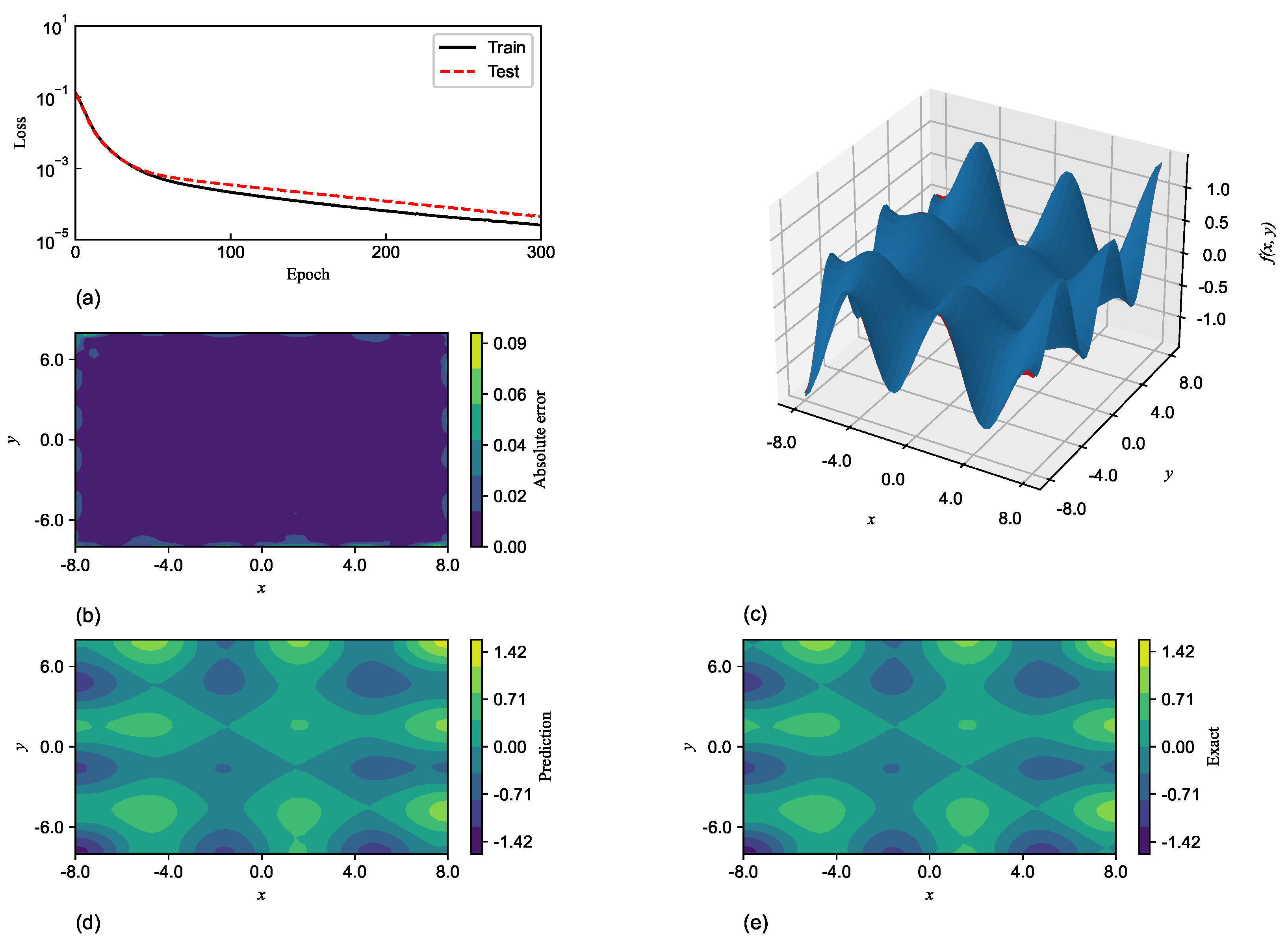
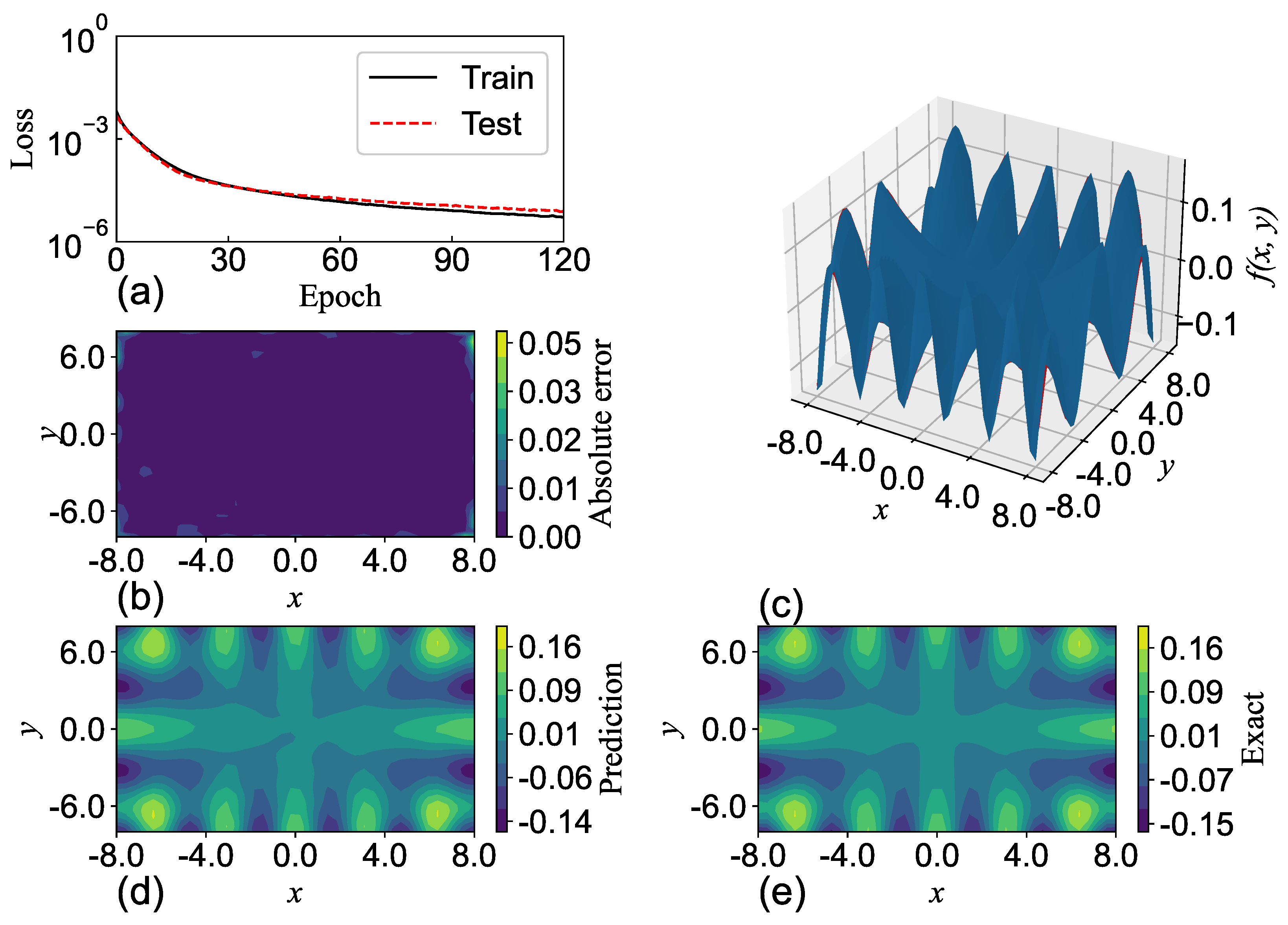
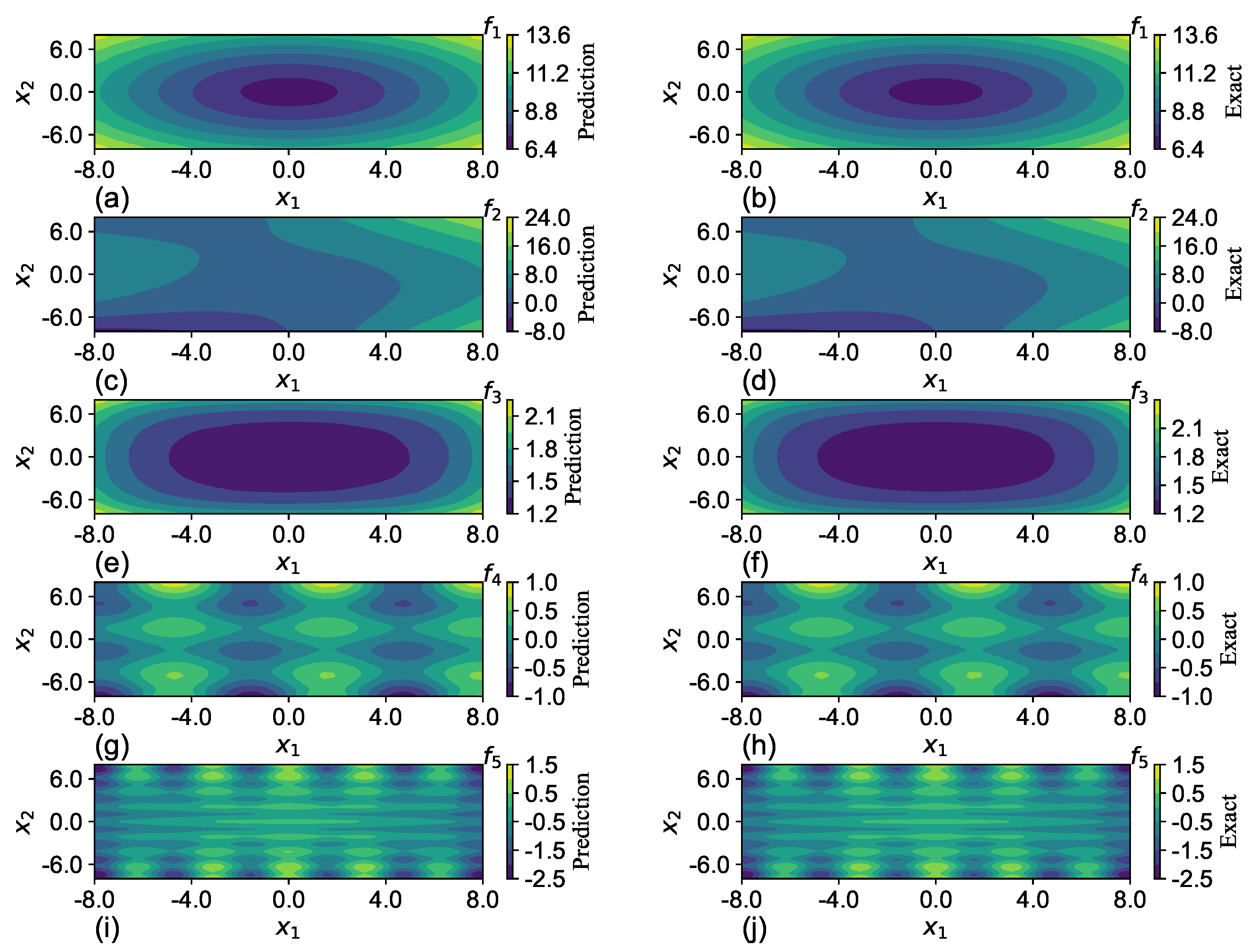
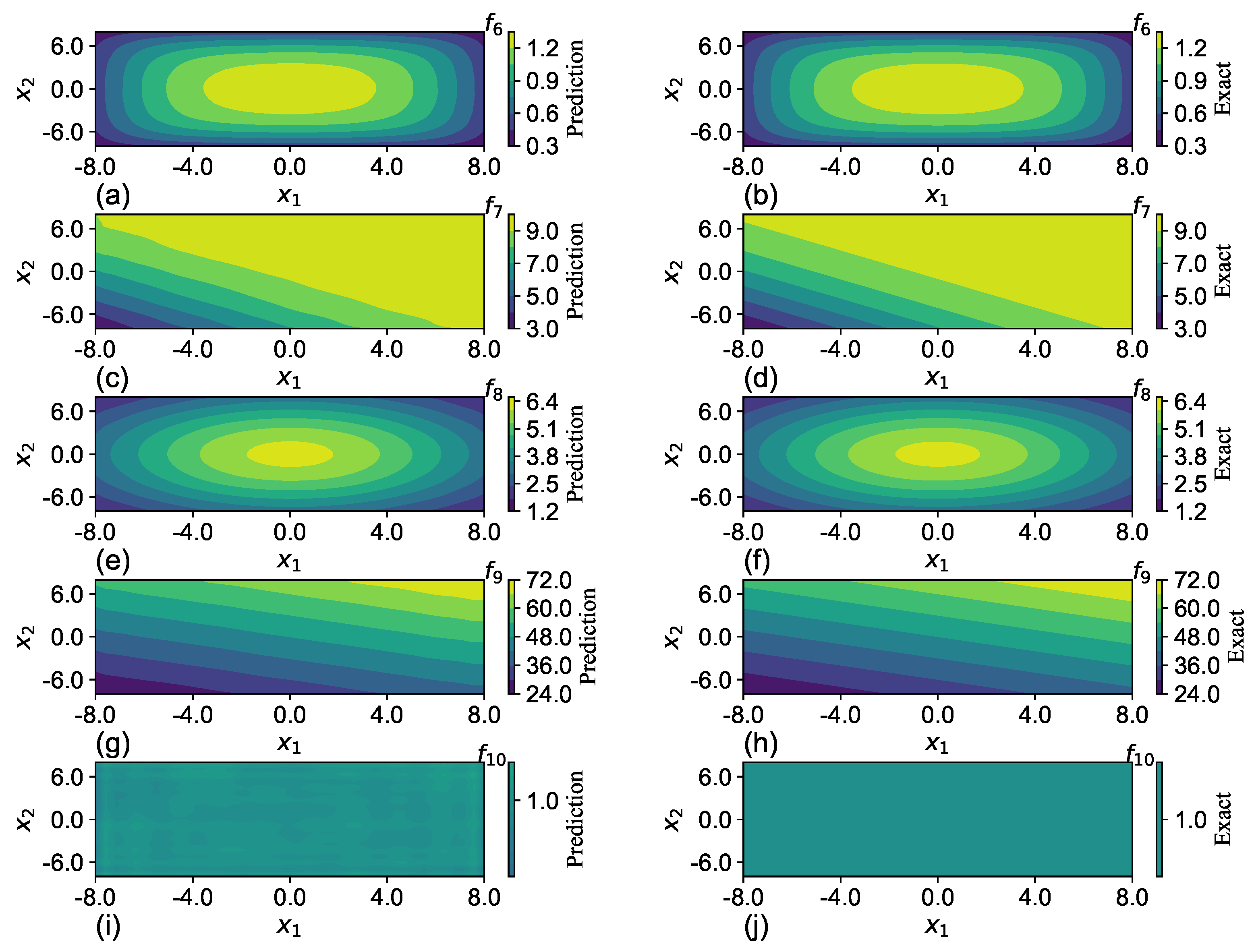
SGNN holds a similar level of accuracy as GRBFNN, making it particularly suitable for approximating multi-variate functions with complex geometry. In
Section 7, it is shown that SGNN can yield approximations for complex functions that are three orders of magnitude more accurate than those MLPs yield with ReLU and Sigmoid functions.
The rest of this paper is organized as follows. In
Section 2, we introduce the structure of SGNN and use it to approximate a multi-variate real-value function. In
Section 3, we compare SGNN and GRBFNN regarding the number of trainable variables and the computational complexity of forward and backward propagation. In
Section 4, we show that SGNN can preserve the dominant sub-eigenspace of the Hessian of GRBFNN in the gradient descent search. This property can help SGNN maintain a similar level of accuracy as GRBFNN while substantially improving the computational efficiency. In
Section 5, we show that the computational time of SGNN scales linearly with the increase in dimension and demonstrate its efficacy in function approximations through numerous examples. In
Section 6 and
Section 7, extensive comparisons between SGNN and GRBFNN and between SGNN and MLPs are performed. At last, the conclusion is summarized in
Section 8.
4. Subspace Gradient Descent
The aim of this section is to discuss the high performances of SGNNs over GRBFNNs in terms of computational efficiency and accuracy through the lens of gradient descent. As illustrated in
Section 3, SGNN has exponentially fewer trainable variables than the associated GRBFNN for high-dimensional input. In other words, GRBFNN may be over-parameterized. The recent work [
29,
30,
31] has shown that optimizing a loss function constructed by an over-parameterized neural network can lead to Hessian matrices that possess few dominant eigenvalues with many near-zero ones before and after training. This means the gradient descent can happen in a small subspace. Inspired by their work, we consider the infinitesimal variation of the loss function
J for GRBFNN a
where
represents a vector of all trainable weights, and
is the associated Hessian matrix. The centers and widths of Gaussian functions are assumed to be constant for simplicity. Since the Hessian matrix
is symmetric, we can represent it in the form
where
are the
k dominant eigenvalues padded by
non-dominant ones (assuming
), and
=
are the rest non-dominant eigenvalues.
Let
be the weights of SGNN. The variation of the mapping from
to
in Equation (
12) reads
where
:
. It should be noted that
is a super sparse matrix.
Substitution of Equation (
31) into Equation (
28)
with
Let
where
and
. Substituting Equation (
34) into Equation (
33) yields
Therefore, the dominant eigenvalues of the Hessian of GRBFNN are also included in the corresponding SGNN. This means that the gradient of SGNN can descend in the mapped dominant non-flat subspace of GRBFNN, which may contribute to the comparable accuracy and training efficiency of SGNN as opposed to GRBFNN, as discussed in
Section 3.
7. Comparison with Deep NNs
In this section, we compare the performance of SGNN with deep ReLU and Sigmoid NNs, which are two popular choices of activation functions. Through the approximation of four-dimensional candidate functions, SGNN shows much better trainability and approximability over deep ReLU and Sigmoid NNs.
Table 6 presents the training time per epoch, total epoch for training, and loss after training of three deep NNs by averaging the results of 30 runs. All NNs possess four hidden layers with 20 neurons per layer. The training-set size is fixed to 16,384, with a mini-batch size of 256. As opposed to SGNN and Sigmoid-NN, which have stable training times per epoch across all candidate functions, the time of ReLU-NN fluctuates. This might be led by the difference in calculating the derivatives of a ReLU unit with an input less or greater than zero. SGNN has a longer training time per epoch because of the computation of a Gaussian function and derivative of
and
. One may argue that this comparison is unfair because SGNN has extra trainable variables. However, SGNN has fewer trainable weights (see
Table 7) because no weights connect the input and first layer, and the output layer is not trainable.
Table 6.
Performance comparison of SGNN and deep neural networks with ReLU and Sigmoid activation functions. Data are generated by averaging the results of 30 runs. All NNs have four hidden layers, with 20 neurons per layer.
Table 6.
Performance comparison of SGNN and deep neural networks with ReLU and Sigmoid activation functions. Data are generated by averaging the results of 30 runs. All NNs have four hidden layers, with 20 neurons per layer.
| | SGNN | ReLU-NN | Sigmoid-NN |
|---|
| | Seconds/Epoch | Epoch | Loss | Seconds/Epoch | Epoch | Loss | Seconds/Epoch | Epoch | Loss |
| 0.099 | 218 | | 0.054 | 150 | | 0.063 | 39 | |
| 0.098 | 262 | | 0.054 | 119 | | 0.054 | 166 | |
| 0.106 | 193 | | 0.312 | 167 | | 0.054 | 169 | |
| 0.100 | 196 | | 0.234 | 161 | | 0.056 | 101 | |
| 0.097 | 392 | | 0.173 | 94 | | 0.066 | 29 | |
| 0.101 | 147 | | 0.293 | 115 | | 0.053 | 187 | |
| 0.099 | 246 | | 0.241 | 139 | | 0.054 | 145 | |
| 0.100 | 173 | | 0.344 | 109 | | 0.054 | 243 | |
| 0.099 | 245 | | 0.439 | 126 | | 0.054 | 374 | |
| 0.101 | 158 | | 0.135 | 143 | | 0.054 | 173 | |
Table 7.
Comparison of SGNN and ReLU-based NN in approximation of Results are generated by averaging the data of 30 runs.
Table 7.
Comparison of SGNN and ReLU-based NN in approximation of Results are generated by averaging the data of 30 runs.
| | Layers | Neuron/Layer | Parameters | Seconds/Epoch | Epoch | Loss | Min Loss |
|---|
| SGNN | 4 | 20 | 1360 | 0.097 | 392 | | |
| 4 | 40 | 5120 | 0.130 | 149 | | |
| ReLU-NN | 4 | 20 | 1381 | 0.056 | 96 | 0.497 | 0.477 |
| 4 | 40 | 5161 | 0.067 | 99 | 0.458 | 0.409 |
| 7 | 40 | 10,081 | 0.082 | 135 | 0.336 | 0.273 |
| 10 | 40 | 15,001 | 0.176 | 112 | 0.324 | 0.258 |
| 10 | 50 | 23,251 | 0.156 | 100 | 0.309 | 0.253 |
| 10 | 60 | 33,301 | 0.250 | 96 | 0.288 | 0.232 |
| 10 | 70 | 45,151 | 0.120 | 97 | 0.278 | 0.215 |
| 10 | 80 | 58,801 | 0.261 | 89 | 0.291 | 0.205 |
Although SGNN has appreciably larger training epochs, this also leads to more accurate predictions. The loss values of SGNN after training are uniformly smaller than those of ReLU-NN and Sigmoid-NN, except for . In fact, for , , , and , the accuracy of SGNN is even two orders of magnitude better than the other two models.
Despite the efficient training speed of Sigmoid-NN, the network is more difficult to train with random weight initialization for and . In fact, the approximation of by Sigmoid-NN is nowhere close to the group truth after training. When functions become more complex, SGNN outperforms ReLU-NN and Sigmoid-NN in minimizing loss through stochastic gradient descent. This could be attributed to the locality of Gaussian functions that increase the active neurons, reducing the flat subspace whose gradients diminish. Sigmoid-NN aborts with significantly fewer epoch numbers. This could be led by the small derivatives of Sigmoid functions when input stays within the saturation region, which makes it more difficult to train the network.
Next, we further compare the trainability of SGNN with ReLU-DNN. We train the two networks with different configurations to approximate the function
, which has a more complex geometry and is more difficult to approximate. The configuration of the NNs and the training performance are listed in
Table 7.
Because the layer of SGNN is fixed by the number of function variables, its only tunable network hyper-parameter is the number of neurons per layer. Doubling the neurons/layer of SGNN from 20 to 40 decreases the loss by two orders of magnitude. Although the training time per epoch increases by 30%, the number of epochs reduces by 60%. Consequently, the total training time is cut by almost 50%, from 38.2 to 19.4 s.
However, the accuracy of ReLU-DNN slightly increases with the increase in width and depth of the model. Close to 50% loss reduction is achieved by adding seven more layers and 50 neurons per layer. However, the error is still three orders of magnitude higher than the error by a four-layer SGNN with one-tenth of trainable variables, with half the training time per epoch. According to the universal approximation theorem, although one can keep expanding the network structure to improve the accuracy, it is against the observation in the last row. This is because the convergence of gradient descent can be a practical obstacle when the network becomes over-parametrized. In this situation, the network may impose a very high requirement on the initial weights to yield optimal solutions.
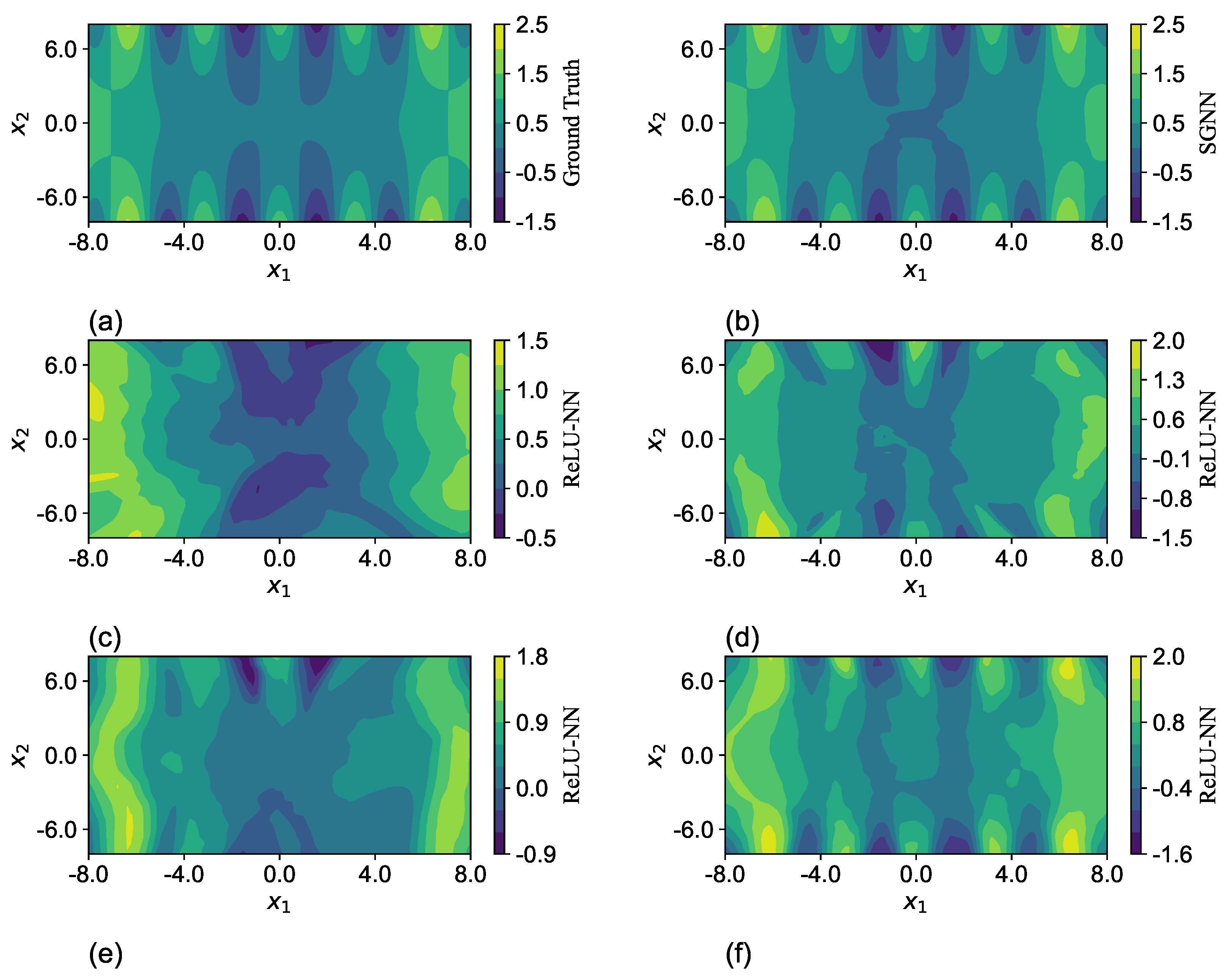
To visualize the differences in the expressiveness between SGNN and ReLU-NN, the predictions of one run in
Table 7 are selected and plotted through a cross-sectional cut in the
plane with the other two variables
and
fixed at zero, as shown in
Figure 7. The network configurations are listed in
Table 8. The predictions of SGNN in
Figure 7b match well the ground truth in
Figure 7a. Despite the minor differences in colors near the origin, their maximum magnitude is less than 0.1. The ReLU-NN with the same structure has a much worse approximation. Although the network gradually captures the main geometric features of
by significantly augmenting its structure to 10 layers and 70 neurons per layer, the difference of magnitude can still be as large as 0.5, as shown in
Figure 7f.