Abstract
Designing advanced health monitoring systems is still an active research topic. Wearable and remote monitoring devices enable monitoring of physiological and clinical parameters (heart rate, respiration rate, temperature, etc.) and analysis using cloud-centric machine-learning applications and decision-support systems to predict critical clinical states. This paper moves from a totally cloud-centric concept to a more distributed one, by transferring sensor data processing and analysis tasks to the edges of the network. The resulting solution enables the analysis and interpretation of sensor-data traces within the wearable device to provide actionable alerts without any dependence on cloud services. In this paper, we use a supervised-learning approach to detect heartbeats and classify arrhythmias. The system uses a window-based feature definition that is suitable for execution within an asymmetric multicore embedded processor that provides a dedicated core for hardware assisted pattern matching. We evaluate the performance of the system in comparison with various existing approaches, in terms of achieved accuracy in the detection of abnormal events. The results show that the proposed embedded system achieves a high detection rate that in some cases matches the accuracy of the state-of-the-art algorithms executed in standard processors.
1. Introduction
The design of advanced health-monitoring systems has always been a topic of active research. During the past decades, numerous portable devices have been introduced for the early detection and diagnosis of heart failure, since it is a common, costly, disabling, and deadly syndrome. Advanced heart-monitoring devices are capable of providing reliable, accurate heart monitoring and are able to detect sporadic events during periods of time when things would otherwise be unclear. Functioning as a black box, these devices have the potential to enhance the decision-making process, by notifying doctors when patients need readmission and in this way constituting an effective alternative capable of saving patients’ lives.
Various portable devices have been introduced by the industry (e.g., Wahoo Fitness (https://www.wahoofitness.com), Polar H10 Heart Rate Sensor (https://www.polar.com/en/products/accessories/H10_heart_rate_sensor), and Fitbit PurePulse (https://www.fitbit.com/purepulse)) to monitor and record variations in the heart rate, rhythm, and other patterns of its operation, by detecting electrical impulses generated by the polarization and depolarization of cardiac tissue. Such devices rely on nearby smartphones that act as a gateway for transmitting the recorded sensor signals to cloud services, where appropriate signal processing, analysis, and classification algorithms are applied. The analyzed electrocardiogram (ECG) can be used in decision-support systems to assist physicians and cardiologists to evaluate irregular heart rhythm, potentially diagnose cardiac abnormalities, and predict critical clinical states [1].
Existing portable heart monitoring devices are very inaccurate as a result of electromagnetic noise produced by the environment (e.g., mobile phones, electrical wires, appliances) or from the noise produced by the muscular movement of the human while wearing the device [2,3]. Moreover, depending on the accuracy of the ECG device, even a short recording may lead to a large volume of data, thus affecting the battery performance of the portable devices as well as that of the accompanying smartphone while storing and transmitting the recording wirelessly [4,5], thus limiting their diagnostic use to 24 h monitoring or even less [6]. Finally, the concept of transmitting the sensor data to cloud services for processing and analysis creates a number of security issues that need to be addresses as they are directly related with the privacy of the users [7,8,9]. Existing portable solutions are totally cloud-centric: all personal data collected are stored on the cloud and in most cases users no longer own the data they produce. This approach severely limits the ability of the user to maintain control of personal data. Now, more than ever, there is a need for privacy-preserving applications where users are always in control of their sensitive data [8,10].
The major challenge in the evolving landscape of wearable devices is to increase the accuracy of the biosignals collected [2], broaden the capabilities for interpretation of data [11], whilst at the same time improving the long-term operativity [5] and guaranteeing the privacy of the users [12]. With the advent of wearable-sensing technologies [13] and smart textiles [14,15], the miniaturization of embedded systems and energy generators [16], and the introduction of new communication protocols [17] and access technologies [18,19,20], the development of wearable devices that can provide accurate and precise measurements is becoming a realistic possibility.
The goal of this paper is to develop new, light-weight algorithms for the analysis and interpretation of ECG sensor data that can be executed in the embedded processor provided by a wearable device. The motivation here is to transfer data processing and analysis tasks to the edges of the network so that we can increase the longevity of the wearable devices by reducing energy requirements through reduced transmissions of large sensor data [21,22]. In this way, the wearable device becomes capable of analyzing and interpreting sensor-data traces to provide actionable alerts without any dependence on cloud services. Therefore, we evolve the current paradigm for developing wearable solutions from a totally cloud-centric one to a more distributed one. As a result, our approach also reinforces the privacy of confidential data by storing the user’s sensitive data within the private memory of the edge devices.
In this work, we present an end-to-end system that is suitable for execution within a resource-constrained wearable device. The system processes the ECG sensor data, automatically detects the heartbeats, and conducts an arrhythmia classification using a combination of supervised-learning approaches. The new algorithm presented uses a window-based feature definition to achieve high detection rates, which in some cases match the accuracy of the state-of-the-art algorithms.
We implement the system on a prototyping board that supports an asymmetric multicore embedded processor with one generic processing core and a dedicated core for hardware assisted pattern matching. The hardware assisted core enables the machine-learning algorithms to operate with much lower energy requirements. Examples of such asymmetric embedded processors are the Intel® Curie™ module [23] and the NXP MPC8572E module [24]. Given the real-world implementation, we evaluate the performance of the system of heartbeat classification in comparison with various existing approaches, in terms of achieved accuracy in the detection of abnormal events. We measure the correctness of the automatic classification according to precision and recall using as comparative measure the average between them, using a comprehensive ECG database following AAMI recommended practice [25]. The results indicate that our approach is viable as regards both moving data processing and analysis into the wearable device, in terms of the accuracy achieved as well as regarding energy efficiency.
The rest of the paper is organized as follows. In Section 2, we briefly present the physiological aspects of the operation of the heart and the arrhythmia classification problem, and we provide a limited number of related previous systems on automated heartbeat detection and arrhythmia classification. In Section 3, we present the high-level architecture of the proposed system, along with the description of the main building blocks. We provide the details of the implementation of the heartbeat detection and arrhythmia classification using machine learning techniques in Section 4. We show the results of the proposed system in comparison to previously published systems in Section 5. Finally, we conclude in Section 6, where we also list some future research directions.
2. Preliminaries and Previous Work
Electrocardiography monitoring is the dominant method of understanding variations in heart rate, heart rhythm, and other patterns of the heart’s electrical impulses. ECG records the electrical activity of the heart, by detecting electrical impulses generated by the polarization and depolarization of cardiac tissue. The impulses are then translated into a waveform, which eventually enables physicians or cardiologists to evaluate irregular heart rhythm, thus diagnosing cardiac abnormalities.
Arrhythmia literally means without rhythm, but this word is used to denote any abnormal rhythm. In terms of heart activity, the term arrhythmia refers to any change from the normal sequence of electrical impulses. In this sense, arrhythmias refer to a collection of conditions that are characterized by heart rhythms that do not follow a normal pattern. The electrical impulses may happen too fast (tachycardia), too slowly (bradycardia), too early (premature contraction), or irregularly (flutter or fibrillation), causing the heart to beat too fast, too slowly, too early, or irregularly [26]. The ECG provides the most accurate means of identifying a cardiac arrhythmia. The arrhythmias can be classified into two major categories. The first category consists of arrhythmias formed by a single irregular heartbeat, herein called morphological arrhythmia. The other category consists of arrhythmias formed by a set of irregular heartbeats, herein called rhythmic arrhythmias [1].
An ECG recording is analyzed based on the three waves: P, , and T, which occur in a sequential way. Figure 1 provides a graphical representation of the complex. The P wave has small magnitude and represents the impulse of the two regions of the heart’s atria that transfers the blood from the veins. It is characterized by depolarization of the atrial muscles. The complex that has high magnitude and spikes because of the rise in the conduction velocity, measures the depolarization of the ventricles. Lastly, the repolarization and revitalization of the ventricles takes place during the T wave. The assessment of cardiac function is linked with the detection of significant fiducial points of a heartbeat: the P wave, the complex, and the T wave. Moreover, the recognition of the onset and offset points of the P wave, the onset and offsets point of complex, and the onset and offset point of the T wave of a complex is diagnostically relevant. Furthermore, the wave recognition points serve also as references for all amplitude measurements, for example, the distances between fiducial points of a heartbeat such as the P wave, the interval, the width, the interval, the amplitude of P, the amplitude of , level, and the amplitude of T.
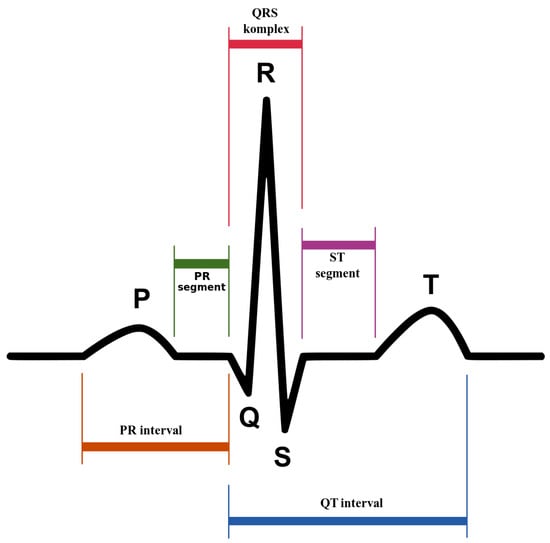
Figure 1.
complex: the pattern of electrical activity of the heart during one cardiac cycle as recorded by electrocardiography. Created by Anthony Atkielski, source: https://www.wikilectures.eu/w/Electrocardiogram.
The design of algorithms for the analysis of heart arrhythmias based on ECG recordings has been studied for many years [1]. All such algorithms rely on the analysis of the complex. The most prominent in terms of amplitude is the wave and is therefore the easiest to detect. Hence, the heartbeat detection problem is equivalent to the detection. The -detection problem, on its standard formulation, takes as input an ECG sample and computes whether it resides in a wave. It is necessary at this point to clarify that the detection is complicated mainly because of the different kinds of noise present in the ECG signal and not only because of the pysiological variability of the complexes. The aforementioned noise sources include muscle noise, wave variations caused by slight translocation of electrodes, power-line interference, baseline wander, and T waves with high-frequency characteristics similar to complexes. Therefore, detection methods typically apply appropriate digital signal processing algorithms to raw signals to reduce noise and amplify the signal. One of the earliest and most established algorithms introduced by Pan and Tompkins [27] identifies the complexes based upon digital analysis of the slope, the amplitude, and the width of the ECG data. The algorithm periodically adjusts the thresholds and the parameters used to detect changes in the morphology and heart rate. For this reason, this approach is also known as the adaptive-thresholds approach. Another method is the dual-slope detection, which calculates the slopes on both sides of the complex. The algorithm presented by Wang et al. [28] uses low computational complexity techniques to calculate the steepness, the shape, and the height of the signal to locate each complex. Yet another approach relies on the use of the wavelet transform [29,30]. The wavelet transform decomposes a time-variant signal into components appearing at various scales (or resolutions). The components in each scale represent the signal temporal features, on the basis of their frequency contents. In contrast to the previous techniques, which require prior knowledge about the frequency spectrum, the multistage multiscale mathematical morphology filtering used by Zhang et al. [31] allows for the detection of the various components of the complex by suppressing the impulsive noise and uses the multi-frame differential modulus accumulation to remove the baseline drift and to enhance the signal.
There exist many algorithms for computerized detection of abnormal heart activity which are based on the interpretation of ECG recordings. However, Shah et al. [32] report that they are very inaccurate. This explains why it is necessary for the diagnosis to be performed by medical specialists and physicians. Very recently, Saini et al. [33] investigated the use of machine-learning techniques to reduce the errors in the computerized electrocardiogram interpretation of the cardiac rhythm. An application of the k-Nearest Neighbor (KNN) algorithm as a classifier for the detection of the -complex in ECG produced very high detection rates. Pławiak [34] developed an evolutionary neural system which makes the efficient classification of cardiac disorders possible. The best results were observed with the use of an evolutionary neural system, which was based on the SVM classifier and obtained a recognition sensitivity at a level of . The same author also developed revolutionarily effective ensembles of classifiers for the automatic recognition of myocardium dysfunctions based on ECG signals [35]. The best genetic ensemble of classifiers optimized by sets obtained a recognition sensitivity of 17 myocardium dysfunctions at a level of , which is the best result achieved to date against the background of the current scientific literature. Yıldırım et al. [36] proposed another deep-learning approach for the detection of cardiac arrhythmia, which is based on a one-dimensional convolutional neural-network model. The proposed method is simple, efficient, and fast (it allows for real-time classification), thus making it very practical. More recently, Rajpurkar et al. [37] applied a 34-layer convolutional neural network, which maps a sequence of ECG samples to a sequence of rhythm classes. They make a comparison between the performance of the deep-learning network and that of certified cardiologists in detecting a wide range of heart arrhythmias from electrocardiograms recorded with a wearable monitor. Interestingly, their system outperforms the average cardiologist performance in both recall (sensitivity) and precision (positive predictive value).
The design of portable health-monitoring devices has closely followed the electronics miniaturization evolution over the years [38]. Wearable monitoring devices that are able to provide accurate and precise measurements are gaining support from physicians and healthcare units. Existing solutions lack computational power to locally process the ECG recordings and to detect abnormal behavior. With the use of such devices, the recorded signal is transmitted to a nearby gateway device, which in turn relays the recording over to the cloud, where advanced analysis algorithms are executed [4,39]. Given the nature of the heart activity, the large majority of the trace periods correspond to normal heart activity. Therefore, in most cases, large amounts of data transmitted over the network infrastructure and processed at the cloud level are simply dropped, leading to a significant waste, not only of the battery resources of the wearable device, but also of network and cloud resources [40,41]. It is evident that minimal possible latency, network bandwidth preservation, and efficient data-storage resource utilization are elements of paramount importance for any IoT-related application [42,43].
The design of efficient health-monitoring systems that are capable of locally processing the ECG recordings has been a topic of active research over the last few years. Most researchers have tried to merge wireless sensor networks with smart gateways [44,45], while others have proposed the use of smartphones as gateways for developing personal health monitoring systems [4,46]. More recently, researchers have developed specialized diagnosis techniques for specific types of health-monitoring applications [47] and platforms that use dedicated IoT communication protocols [48]. However, all the aforementioned solutions somehow fail to harness the full extent of capabilities that fog computing offers towards providing a holistic solution that will be applicable to a larger number of cases, involving multiple actors.
3. System Design
We organize the components of the end-to-end system based on the fog-computing approach in three layers given their functionality and their physical proximity to the user. In particular, we have (1) the wearable device that is always connected to the user, (2) the edge device that is located within the house premises of the user, and (3) the cloud services. Figure 2 provides a graphical representation of the three layers and the corresponding components as well as their interaction using different communication technologies such as WIFI, bluetooth low-energy (BLE) wireless connection, and LORA.
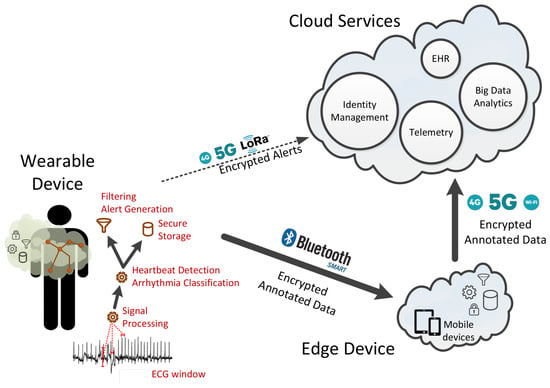
Figure 2.
A Fog-computing based organization of the system components and their interaction utilizing different networking technologies.
The wearable device is comprised of several hardware components such as the ECG sensors, the asymmetric multicore processor, the memory unit, the network interface, the battery and possible power nano-generator. In this work, the software is responsible for (1) collecting the ECG data from the analog-to-digital converter (ADC) based on a given sampling rate, (2) processing of the signal received from the ADC to reduce the noise and amplify the signal, and (3) detecting the heartbeats and classification of arrhythmias using hardware-assisted machine learning techniques. Other aspects of the software firmware such as the encryption, storage, and retrieval of ECG recorded sessions and ECG metadata, as well as the communication with the edge devices and the cloud platform, are not within the scope of this paper.
The edge-device software is installed on the patient’s device (either a smartphone/tablet or a fixed device; for instance, a computer in the patient’s home) and is connected to both the wearable device and the cloud services. The edge device communicates with the wearable device over a well defined API via a secure bluetooth low-energy wireless connection. It has internal storage space for storing the data collected from the ECG sensor and is capable of processing them, allowing for additional (and, typically, more elaborate) computation to that performed in the wearable device. It can also generate alerts, sent to the user either in small packages or in batch mode. As with the wearable device, data stored within the edge device are cryptographically encrypted, and message exchanges across the different layers are always performed in a secure and encrypted manner.
Finally, the cloud services offer all the necessary features to facilitate authentication, authorization, access control, asynchronous notifications, data integration with electronic health records, and other services relying on privacy-preserving data integration.
In this work, we focus on the software executed on the wearable device, and specifically on the processing and analysis of the data collected from the ECG sensors that are conducted either within the wearable or with the edge device. The processing and analysis of the ECG data is based on three modules:
- Signal-processing module —The first module is responsible for processing the signals collected from each ECG sensor lead to reduce the noise and amplify the signal (see Section 2). The goal is to have a well extracted ECG morphology, with as little information loss as possible. The module produces ECG records composed of a number n of channels, each channel corresponding to a recording received from a specific ECG sensor lead, therefore composing an n-dimensional vector. The elements of the channels represent the amplitude of the signal at any given time, these are called samples. This module is executed within the wearable device.More specifically, all the signals initially pass through a bandpass filter to reduce the influence of muscle noise, 60 Hz interference, baseline wander, and T-wave interference. The desirable passband to maximize the energy is approximately 5–15 Hz [27]. It is implemented with a cascade of low-pass and high-pass filters to achieve a 3 dB passband from about 5–12 Hz, reasonably close to the desirable passband. Subsequently, a derivative filter processes the output to provide the complex slope information. After differentiation, the signal is squared point by point, based on the formula: . This transformation makes all data points non-negative and performs a nonlinear amplification of the output of the derivative, emphasizing the higher frequencies, in our case the ECG frequencies and the QRS complex in particular. The result of the preprocessing stage for each step are depicted in Figure 3.
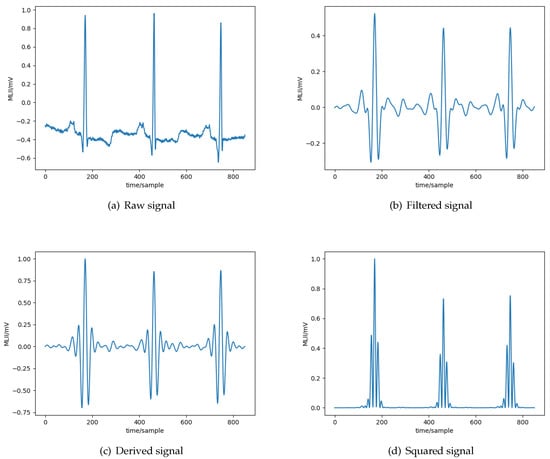 Figure 3. Different phases of the processing of the ECG signal.
Figure 3. Different phases of the processing of the ECG signal. - Heartbeat-detection module—The second stage of the arrhythmia-detection system consists in extracting the heartbeats from the ECG samples produced by the previous module. The ECG samples retrieved are segmented in contiguous regions of fixed duration (see Section 4 for more details on the length of the regions). Given this transformation, each segment is examined to identify if it contains a complex. Given a complex, the R peak location is then detected from such regions using the formula: . In words, the location of the R peak is the one corresponding to the maximum absolute value in the region detected as a complex. This is because, given that a time segment has been detected as a wave, it is very likely that, in such a small portion of the signal, the R peak is located at the point of maximum amplitude. The output of the module is a sequence of locations of R peaks.The identification of the complexes is implemented based on the KNN algorithm, as it can be executed at hardware level in one of the recently introduced asymmetric multicore embedded processors that provide a dedicated core for hardware assisted pattern matching (e.g., the NXP MPC8572E module [24] and the Intel® Curie™ module [23]). The KNN implementation supported by the hardware is based on a simple majority voting to identify the label that will be associated to a given value. Note that the use of the KNN algorithm in this module is different from the one used by Saini et al. [33], as the latter does not use a window but examines each ECG sample individually. It is also different from the one used by Akrivopoulos et al. [49], in which the KNN algorithm is used to characterize whether the complex is normal or abnormal. The design aspects and the technical details of the heartbeat detection module are presented in more details in Section 4.
- Arrhythmia classification module—The third module conducts the final stage of arrhythmia classification. The module examines the R peaks detected by the previous module, identifies the signal around an R peak location, and extracts the corresponding region from the raw signal. In this way, the sequence of R peaks is transformed into a tridimensional stream of the form: where b is the total number of detected beats, d is the number of previous beats to consider so as to make a prediction, and s is the raw signal that makes up the specific beat. Essentially, we generate a sliding window over the sequence of beats, of length equal to t beats.Given that the ECG data are sequential and that determining whether a heartbeat is arrhythmic or not depends not only on its morphology but also on those of the previous beats, we designed a particular kind of recurrent neural network (RNN) for the arrhythmia classification purpose, based on long–short term memory (LSTM) units [50]. LSTM-based architectures are very powerful for applications on time-series data, we therefore use them for our problem. The transformed sequence is passed to the neural-network classifier, which has been designed and trained in order to identify normal and arrhythmic beats. The detected arrhythmic beats are then classified in more fine-grained classes, defined by a well-known standard for cardiac algorithm evaluation [25]. The methodology for training and validating the RNN is presented in more details in Section 4.
4. Implementation Details
In this section, we present in detail the machine-learning techniques used for the heartbeat-detection and the arrhythmia-classification modules. The preliminary analysis of the dataset for the fine tuning of the modules and the training of the classifiers was conducted on a workstation using an Intel i5 processor with 8 GB of RAM, under a generic linux kernel v4.4.0-130. The initial models were developed using Python version 3.5.2 in combination with scikit-learn (https://scikit-learn.org/stable/) library v0.18.1 and keras https://keras.io/) library v1.2.2. For the evaluation of the performance using a hardware-assisted core, an Arduino/Genuino 101 development board https://store.arduino.cc/genuino-101) was used using the Intel® Curie™ ODK Z tree firmware (https://github.com/01org/CODK-Z).
4.1. ECG Data
The database considered in this study is a collection of annotated ECG recordings obtained by the Arrhythmia Laboratory of Boston’s Beth Israel Hospital, known as the MIT-BIH Arrhythmia Database [51]. We now provide some details on how this dataset was created as reported in Reference [51]. First, the creators of the dataset selected 48 half-hour excerpts of two channels (24 h of signal in total) of ECG recordings from 47 subjects: out of which 22 are women aged 23–89 years and the remaining 25 are men aged 32–89 years. The electrodes were placed on the chest as is standard practice for ambulatory ECG recording. The recordings were digitized at 360 samples per second per channel with 11-bit resolution over a 10 mV range. The samples include two leads—the modified limb lead-II and one of the modified leads , , , or with a resolution of 5 l V/bit. On the basis of these recordings, a total of 25 traces were selected such that they include arrhythmic cardiac activity. Out of these, four also include paced beats, meaning that they were produced by a pacemaker. Finally, following a random selection process, an additional 23 samples were chosen. Each of these samples corresponds to a 2D vector of 650 K entries. The vector is composed of two channels, therefore it is two dimensional.
After the creation of the recordings, a simple slope-sensitive detector was used to automatically annotate the recordings. Then, two cardiologists were consulted independently to correct the resulting annotations, as well as provide corrections and additions regarding the classification of the cardia rhythm and the signal quality. The two new sets of annotations were compared to identify discrepancies between the opinions of the two physiologists and generate a unifying version. In Reference [51], it is reported that from 48 recordings, within six of them, for a total of 33 heartbeats, the two physiologists could reach an agreement on the classification. Moreover, it is reported that a total of seven episodes of loss of signal or noise that was so severe in both channels simultaneously, that complexes could not be detected; these episodes were all quite short and have a total duration of about 10 s. In all of the remaining data, every complex was annotated, numbering about in total.
According to the Association for the Advancement of Medical Instrumentation (AAMI) standard [25] for testing and reporting performance results of cardiac rhythm and segment measurement algorithms, the evaluation of an arrhythmia detector algorithm should be made on the classification performance on five major categories of heartbeats:
- N includes beats originating in the sinus node (normal and bundle branch block beat types);
- S includes supraventricular ectopic beats (SVEBs);
- V includes ventricular ectopic beats (VEBs);
- F includes beats that result from fusing normal and VEBs;
- Q includes any heartbeat not in the N, S, V, or F categories, essentially the undefined heartbeats as well as beats resulting from the use of a pacemaker.
Note that the last category includes beats that are a result of a pacemaker operating under the skin of the patient. These beats are regulated and it is not meaningful to use them for arrhythmia classification. As stated above, the MIT-BIH database includes four patients who are using a pacemaker. For this reason, these four patients were excluded from our study. In this way, the resulting Q category includes only 33 beats. Because of the lack of data, we excluded all these beats from our study. Please also note that this is a common practice in other state-of-the-art studies; see, for example, the work of Park et al. [52]. Therefore, we are left with four beat categories.
Each of these four categories contains more fine-grained types of arrhythmic beats. Table 1 lists the complete definitions for each of the four categories.

Table 1.
Beat classification categories and total number of annotations included in the Arrhythmia Laboratory of Boston’s Beth Israel Hospital (MIT-BIH) dataset for each category.
Please note that the ECG records for patients 102, 104, 107, 217 have been excluded from this study because these recordings do not contain any arrhythmic beats.
4.2. Heartbeat-Detection Module
The KNN classifier used by the heartbeat-detection module is based on a feature matrix, comprised of m segments of ECG signals retrieved from n sensor leads (i.e., channels). Each segment s used to train the classifier is labeled based on the annotations provided by the physicians:
Because of to the unique characteristics of each individual person, the ECG signals produced by each heart are unique. For this reason, the training step is performed with an intra-patient perspective: a separated classification instance is used for each patient. Each KNN classifier is trained using an ECG segment of t seconds and a five-fold cross-validation is employed to avoid overfitting on the training set. In the five-fold cross-validation, the original training set is randomly partitioned into five equally sized subsamples. Of the five subsamples, a single subsample is retained as the validation data for testing the model, and the remaining four subsamples are used as training data. The cross-validation process is then repeated five times, with each of the five subsamples being used exactly once as the validation data. The five results are then averaged to produce a single estimation.
To fine-tune the performance of the module, we evaluated the performance using standard statistical indices to determine the precision and recall derived from three parameters: correctly detected peaks (), total detected peaks (), and actual number of peaks ().
Note that the annotations of the R peak locations in the database were not placed precisely on the maximum of the complex by the annotators. More precisely, in some cases, the R peak is not properly annotated, although it is guaranteed to be inside the complex. For this reason, we consider a peak detection correct if it resides inside a range of s around the actual peak annotation. We chose such an amount of sample such as to be comparable to the average width (for details, refer to the book of Walraven [53], Chapter 2, Section 41).
Clearly, the duration t of the training set is an important parameter for the overall performance of the module. Keeping t at minimum levels is essential, as a small training set is preferable for faster computation. For this reason, we conducted a series of experiments to identify the minimum amount of signal necessary for the training step without compromising the accuracy of the results. Figure 4a depicts the precision, the recall, and the F1 score achieved for different training-set lengths. Given the outcome of the experiment, a training set of s (corresponding to 3% of the ECG signal) suffices to produce very high accuracy—higher than 90%. On average, this corresponds to about 56 beats.
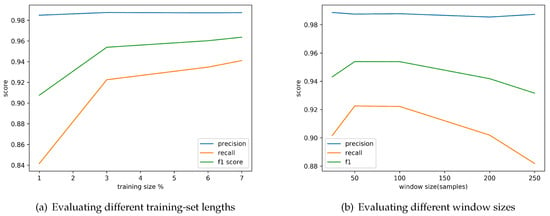
Figure 4.
Fine tuning of the performance of the heartbeat detection module by evaluating the achieved precision, recall and F1 score when using different training set lengths (t) expressed as a percentage of the total size of the ECG recording and different window sizes.
Given that the ECG signal is split into contiguous and fixed size regions, another important parameter, whose value we need to choose appropriately, is the window size. Thus, we conducted a second set of experiments to identify the precision and recall obtained for different window sizes. Figure 4b depicts the results of this experiment. Given the outcome of the fine-tuning, a window size of 50 samples maximizes the F1 Score.
To better understand the cases where the heartbeat-detection module fails to properly detect an R peak, we identified a set of examples from the dataset for which the module fails. As an example, we present here the situation for the ECG recording with label “101.” Figure 5a shows the detected peak from a specific segment of its ECG recording and Figure 5b indicates the k nearest regions (here ) to the segment in which the detected peak resides (here we use the standard Euclidean distance). To understand why the module fails in this case, it is important to look at the labels assigned to the neighbors in the training set. Recall that the KNN implementation is based on a simple majority voting. In this case, the neighbor region contains a peak, so the module decides that the region contains a peak. The detected peak is set to the maximum of the absolute value of the detected region.
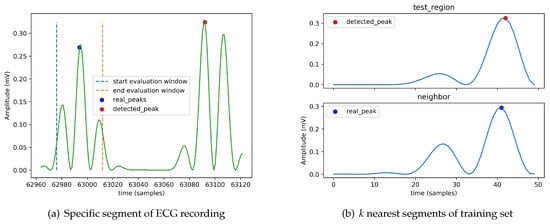
Figure 5.
Erroneous detection of an R peak on a specific segment of ECG recording labeled “101” of the dataset when k = 1 and using simple majority voting. The signal plotted is the result of the filtering and the squaring phases.
4.3. Arrhythmia-Classification Module
For the arrhythmia-classification module, we designed a set of classifiers using a LSTM neural-network architecture. We trained the classifiers following an intra-patient approach, resulting in a unique classifier model suitable for all patients. For this purpose, we partitioned the MIT-BIT dataset vertically such as to include a fraction of heartbeats from each patient; in particular, we used the first 50% of each ECG record for training the classifier. We tuned the model hyperparameters using the subsequent 10% of the dataset. The remaining part of the dataset was used for evaluating the overall performance. In this module we did not employ a cross-validation process.
The relative proportions of the classes of the available training examples influences the performance of an LSTM classifier. If a few classes dominate the training-data examples, then the classifier training process is heavily influenced by these classes [54]. The operation of the heart is characterized by large periods of normal activity and short periods of arrhythmic activity. Therefore, the ECG data collected will contain a large volume of normal activity and a small volume (if any at all) of abnormal activity. Many techniques are known to overcome this issue; for example by adopting a penalization model, or by undersampling/oversampling data. We decided to balance the MIT-BIH dataset by augmenting the number of samples in the least populated classes. We increased the number of samples in the under-represented class c as follows:
- r = random vector s.t. and
where is the set of beats for training, is the generic element of the random vector r, and the operator + stands for vector addition. The main idea used is that the shape of a heartbeat is the most relevant feature to discriminate between arrhythmic beats. Adding the same value to each sample in the beat would in fact preserve the shape, providing more training data to the network, as if they were new data.
Note that an approach that simply replicates the instances of the minority classes without any further processing would not suffice as it would lead the network to overfit on such data, memorizing the specific amplitudes without being capable of generalizing the unseen data. For this reason, the approach presented above uses randomization. Although this approach does not eliminate the problem of overfitting completely (additional real data would be required for that), it reduces the effect. Moreover, to reduce the number of instances belonging to the over-represented class (i.e., the normal class (N)), we use a random-sampling technique, which consists of randomly selecting a subset of heartbeats in the normal class. We reduce the cardinality of the normal class by a reduction factor, more precisely: , where denotes the cardinality of the set of instances initially belonging to the normal class, is the cardinality of the reduced set, and r is the reduction factor.
We measure the performance of the classifiers using the following measures: per-class precision, per-class recall, per-class F1 Score, average precision, average recall, and average F1 Score, which are defined as follows:
where c is the class index, and , , , and stand for true positive, false positive, true negative, and false negative outcomes, respectively, for class c. In accordance with the AAMI recommendations, we calculated two sets of performance measurements. It is worth noting that the AAMI-recommended calculations do not reward or penalize a classifier for the classification of ventricular fusion (F) or unknown beats (Q), such as VEBs.
The validation stage is devoted to the search of the best combination of hyperparameters, that is, the one that results in a model with the best performance on the validation set. We define as the best combination the one with the best average F1 score, as it is the measure that takes into account both the precision and the recall of the result for each class. The hyperparameters that have been fine-tuned are the window size, the number of channels, the timesteps, the number of LSTM layers, the number of fully connected layers, the number of neurons for each layer, the batch Size, the dropout probability, and the hidden-units activation function. We trained the model using the Adam optimization algorithm, an extension to stochastic gradient descent, using the categorical cross entropy as the loss function, defined as: , where M is the number of classes, is a binary indicator (0 or 1) if class label c is the correct classification for observation o, and is the predicted probability observation that o is of class c.
The value of the number of epochs parameter varied on the various executions. We employed an early stopping criterion: the model stops training when the value of the loss function does not decrease for ten epochs in the validation step. When this happens, we restore the weights of the network corresponding to the lowest loss value.
Because of the elevated number of candidate combinations, it was not feasible to perform an exhaustive grid search. For this reason, we tuned each parameter in isolation, maintaining the other ones fixed. When we chose the value for a given parameter, we kept it for all the subsequent runs.
First, we examined the effect of the window size. We define a heartbeat as a fixed number of samples around an R peak location. There is not an appropriate value for the number of samples agreed on in the literature, so we examine different values. Figure 6a depicts the achieved performance for different window sizes. The best value in terms of average F1 Score on the validation dataset is 200 samples.
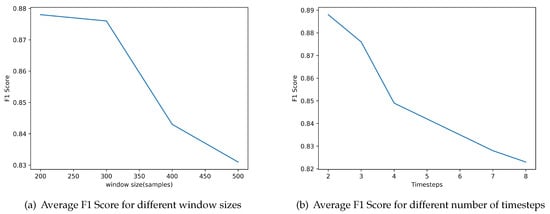
Figure 6.
Evaluation of the classifier parameters regarding Window size and number of Timesteps based on the performance achieved using the average F1 Score.
The second parameter that we examined was the number of timesteps considered in the feature vector, that is, the number of previous beats included for making the prediction for the current beat. Figure 6b depicts the evaluation of the performance of the classifier for different number of timesteps included. The results indicate that the best value is 2.
The next parameter we examined was the number of LSTM layers that we connect in a sequential manner in the classifier model. We also tested whether the inclusion of standard fully connected layers after the LSTMs improves the results. We also examined the use of dropout regularization layers between the fully connected layers. The dropout probabilities considered are zero (no dropout layers), 0.1, 0.3, 0.5, and 0.8. Note that the batch-size parameter influences the correctness of the results; it is the number of instances after which the update of parameters is performed. The results of these evaluations are depicted in Figure 7. The optimal number of LSTMs and fully connected layers are 4 and 3, respectively. Furthermore, the best F1 score is obtained if we include dropout layers with dropout probability 0.1. Finally, optimal batch Size is 128. The recurrent neural network optimization lasted 2347 s for a total of 15 epochs.
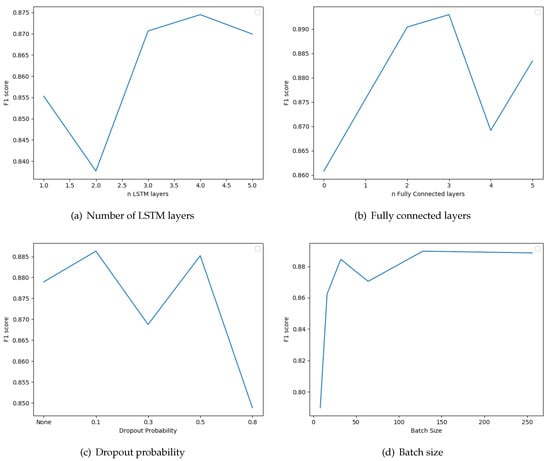
Figure 7.
Evaluation of the classifier parameters regarding number of ong-short term memory (LSTM) layers, number of fully connected layers, dropout probablity, and batch size, based on the performance achieved using the average F1 Score.
A visualization of the resulting Recurrent Neural Network is included in Figure 8, as generated by the TensorBoard (https://www.tensorflow.org/guide/summaries_and_tensorboard), a graphical tool for visualizing and debugging Neural Networks.
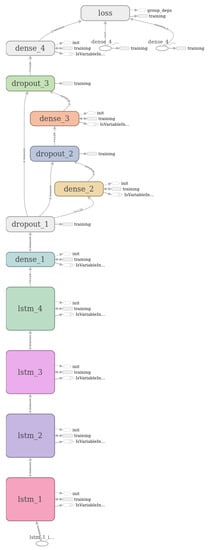
Figure 8.
Graphical visualization of the resulting classification model used by the Arrhythmia Classification Module generated by the TensorBoard graphical tool.
5. Results and Discussion
To evaluate the effectiveness of our system in detecting, analyzing, and interpreting ECG traces, we conducted a series of experiments using the MIT-BIH dataset. First, we evaluated the performance of the heartbeat-detection module as a standalone module and subsequently the performance of the entire system, that is, the heartbeat-detection followed by the arrhythmia-classification module. We evaluated the performance by comparing our approach with selected state-of-the-art solutions from the literature.
5.1. Heartbeat Detection
To better understand the performance of the heartbeat-detection module (HDM), we conducted a comparative evaluation with (1) the algorithm of Pan and Tompkins [27] (PT), as it is one of the earliest and the most established algorithm, (2) the algorithm of Saini Singh, and Khosla [33] (SSK), as it is also based on the KNN algorithm, and (3) on a naive algorithm implementation using the first-order difference technique (FOD), which does not require any dependencies on any algorithmic library, and thus can be supported by low-power embedded processors available in wearable devices. For each of the four algorithms, we measure the performance in terms of precision, recall, and F1 score, as defined in Section 4.2, using the ECG records of the MIT-BIH database. Note that our module, the HDM, uses 3% of the data of each recording as a training set, while the SSK module follows an inter-patient training approach and uses the complete record of patient 100 for training. In contrast, the PT and FOD modules do not require a training phase. Therefore, to make the comparison fair, all four algorithms were evaluated using the exact same input. For this reason, we used the 97% of the recordings of each patient as test set, excluding patient 100. In Table 2, we present the results of the comparative evaluation.

Table 2.
Performance of the heartbeat-detection module (HDM), the Pan and Tompkins algorithm (PM), Saini, Singh and Khosla algorithm (SSK), and the algorithm based on the first order difference technique (FOD) in terms of precision, recall, and F1 score.
The results indicate that HDM achieves a high level of precision, even higher than the PT algorithm, which is one of the most established algorithms in the bibliography, implying that almost all beats detected are correct. The results also indicate that HDM fails to detect only a very small number of beats, whereas the PT algorithm performs slightly better. For this reason, the resulting F1 score for HDM is slightly lower than that of PT. The results indicate that the PT module achieves a higher F1 score compared to our module. However, it is important to recall that our module is executed within the dedicated hardware core where the energy consumption is much lower in contrast to the PT algorithm that is executed within the generic core thus requiring much more power.
Although the overall performance of the module proposed in this work is slightly lower than the one achieved by the state-of-the-art model, the important element to consider is that it can be executed within the dedicated core for hardware assisted pattern matching provided by the asymmetric multicore embedded processors (e.g., the NXP MPC8572E module [24] and the Intel® Curie™ module [23]). Given that the signal-processing module can also be implemented using hardware elements, the main cores of the embedded processors can be used for other computation and communication services required by the wearable device. Moreover, the power consumption of the HDM algorithm executed within the dedicated hardware core is extremely low, in contrast to the consumption achieved when executed in the main cores. Considering the evaluation conducted by Akrivopoulos et al. [49] using the Intel® Quark™ SE C1000 asymmetric multicore embedded processor operating at 32 MHz, the parallel broadcast mode of accessing the neurons of the pattern-matching engine enables the execution of the KNN algorithm in real-time—it requires about 3.2 ms to classify a sample, requiring a power of 15.24 mW. However, when HDM is executed in the x86 main core, the power consumption increases to 210.24 mW.
At this point, recall that the training of HDM is conducted following an intra-patient approach. In this way, the KNN classifier is trained for the morphology of each individual patient. Therefore, it is natural to expect that the results are improved. However, this implies that the KNN has to be trained for each specific patient and no global training data can be used. In future work, we plan to build classifiers that are able to be applied to new patients without using labeled data from them.
5.2. Arrhythmia Classification
After the heartbeat is detected by the previous module, the sequence is transformed into a tridimensional stream and passed to the RNN classifier. Figure 9a depicts the confusion matrix of the classifier achieved for each classification category considered and Figure 9b provides the normalized version of the matrix.
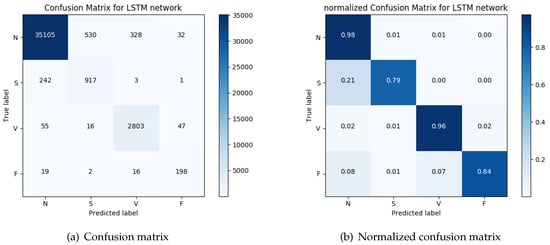
Figure 9.
The actual beats included in the MIT-BIH database for each category and the classification of the beats produced by the arrhythmia-classification module.
The overall performance of the system we presented here (ACM) indicates a very high accuracy, reaching 96% for VEBs and 79% for SVEBs. To better understand the overall performance, we compare the results with those achieved by the classification system based on morphology and interval features (MIF) presented by Chazal et al. [55] and the classifier using a mixture of experts (MOE) approach presented by Hu et al. [56]. The results of the evaluation of the ACM module are compared to the results of the evaluation of MOE and MIF as reported in Reference [55,56]. The comparison of the results are presented in Table 3.

Table 3.
Performance of the arrhythmia-classification module (ACM), the classification system based on morphology and interval features (MIF), and the classifier using a mixture of experts (MOE) in terms of average precision, average recall, and average F1 score.
As in the case of the HDM, the training of this module is conducted following an intra-patient approach. Here as well, this training approach improves the results. Again, this requires the RNN to be trained for each specific patient.
These results demonstrate that the particular approach to signal processing, heartbeat-detection, and feature generation, based only on the detected beats is an improvement on previously reported results for automated heartbeat-classification systems, especially when considering systems of low capabilities. In particular, the ACM module uses two features whereas the MIF [55] uses 15 features and MOE [56] uses eight features. To further evaluate the performance of the ACM module within an embedded processor, we used an Arduino/Genuino 101 development board (https://store.arduino.cc/genuino-101) to assess the performance of the feature generation when executed on the x86 core of the Intel® Curie™ asymmetric embedded processor. The evaluation indicates that the embedded processor is capable of processing the raw signal and generating the two features in real time. The measured power consumption is 28.84 mW and the total execution time is 6.7 ms.
6. Conclusions and Future Work
We presented a system for automatic heartbeat detection and arrhythmia classification based on wearable devices. The solution takes advantage of recent technological developments in microcontrollers and in particular asymmetric embedded processors that support hardware-assisted execution of machine-learning methods. In this way, the computational resources available at the edges of the network are used to locally process the collected signal and perform signal analysis, heartbeat detection, and arrhythmia classification. The performance evaluation indicates that the achieved accuracy is high and is, in fact, enough to conduct a first level analysis of the signal in order to identify normal signals that do not need to be stored or transmitted to the cloud services. Because only the final result is transmitted, the solution conserves bandwidth, limits the need for storage, and decreases latency. These findings are a positive indication that the fog-computing approach can indeed help address the problems resulting from an excess volume of data that are transferred from IoT devices to cloud services.
We evaluated the capability of the end-to-end method to identify the heartbeats and their classification into four groups: normal beats, VEBs, SVEBs, and fusion of normal and VEBs, on the MIT-BIH arrhythmia database. The resulting system achieves a precision of and recall of on the detection of heartbeats, and a precision of and recall of on the arrythmia classification. These results are an indication that the computational power available at the edges of the network can be used to achieve results comparable to state-of-the-art techniques while at the same time significantly improving the overall network utilization. Moreover, it is important to keep in mind that the solutions proposed here take advantage of the hardware-assisted execution of machine-learning methods in order to minimize the power consumption. Given the dependence of the wearable device on a battery, clearly energy efficiency is also an important requirement. Although there exist other methods that achieve much better results, such as in References [34,35], such solutions do not utilize the special-purpose hardware core. Instead, they are executed within the generic core of the embedded processor leading to higher power consumption. In this sense, our findings indicate that there is a compromise between energy consumption and accuracy in the domain of embedded processors.
Future investigations in this area will involve the improvement of the precision and accuracy of the detection and classification phases of the system. To this end, the use of evolutionary computation may help optimize the parameters of the classifiers. It is also important to look into the intra-patient training methodology and attempt to minimize the size of the training set. Another avenue for future work is the implementation of other types of classifiers within an asymmetric multicore embedded processor in order to further improve the accuracy of the entire system while at the same time keeping the energy consumption low. Additionally, despite the current bibliography mainly focusing on detection (and hence, heart-rate calculation), there are also other approaches that can be used within a constrained resources device. Finally, it is also important to evaluate the performance of this technique using different data sets.
Author Contributions
The conceptualization, methodology, experiment design, writing and editing were conducted by A.A. and I.C. The software, experiment execution, analysis and data curation by A.S. and F.T.
Funding
Partially supported by ERC Advanced Grant 788893 AMDROMA “Algorithmic and Mechanism Design Research in Online Markets”.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Luz, E.J.; Schwartz, W.R.; Cámara-Chávez, G.; Menotti, D. ECG-based heartbeat classification for arrhythmia detection: A survey. Comput. Methods Programs Biomed. 2016, 127, 144–164. [Google Scholar] [CrossRef] [PubMed]
- El-Amrawy, F.; Nounou, M.I. Are currently available wearable devices for activity tracking and heart rate monitoring accurate, precise, and medically beneficial? Healthc. Inform. Res. 2015, 21, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Mariappan, P.M.; Raghavan, D.R.; Aleem, S.H.A.; Zobaa, A.F. Effects of electromagnetic interference on the functional usage of medical equipment by 2G/3G/4G cellular phones: A review. J. Adv. Res. 2016, 7, 727–738. [Google Scholar] [CrossRef]
- Chatzigiannakis, I.; Valchinov, E.S.; Antoniou, A.; Kalogeras, A.; Alexakos, C.; Konstantinopoulos, P. Advanced observation and telemetry heart system utilizing wearable ECG device and a Cloud platform. In Proceedings of the 2015 IEEE Symposium on Computers and Communication (ISCC), Larnaca, Cyprus, 6–9 July 2015; pp. 25–30. [Google Scholar]
- Shcherbina, A.; Mattsson, C.M.; Waggott, D.; Salisbury, H.; Christle, J.W.; Hastie, T.; Wheeler, M.T.; Ashley, E.A. Accuracy in wrist-worn, sensor-based measurements of heart rate and energy expenditure in a diverse cohort. J. Pers. Med. 2017, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Amaxilatis, D.; Chatzigiannakis, I.; Mavrommati, I.; Vasileiou, E.; Vitaletti, A. Delivering elder-care environments utilizing TV-channel based mechanisms. JAISE 2017, 9, 783–798. [Google Scholar] [CrossRef]
- Chatzigiannakis, I.; Pyrgelis, A.; Spirakis, P.G.; Stamatiou, Y.C. Elliptic curve based zero knowledge proofs and their applicability on resource constrained devices. In Proceedings of the 2011 IEEE 8th International Conference on Mobile Adhoc and Sensor Systems (MASS), Valencia, Spain, 17–22 October 2011; pp. 715–720. [Google Scholar]
- Angeletti, F.; Chatzigiannakis, I.; Vitaletti, A. Towards an Architecture to Guarantee Both Data Privacy and Utility in the First Phases of Digital Clinical Trials. Sensors 2018, 18, 4175. [Google Scholar] [CrossRef] [PubMed]
- Akribopoulos, O.; Chatzigiannakis, I.; Koninis, C.; Theodoridis, E. A web services-oriented architecture for integrating small programmable objects in the web of things. In Proceedings of the 2010 IEEE Developments in E-systems Engineering (DESE), London, UK, 6–8 September 2010; pp. 70–75. [Google Scholar]
- Angeletti, F.; Chatzigiannakis, I.; Vitaletti, A. The role of blockchain and IoT in recruiting participants for digital clinical trials. In Proceedings of the 25th International Conference on Software, Telecommunications and Computer Networks (SoftCOM 2017), Split, Croatia, 21–23 September 2017; pp. 1–5. [Google Scholar] [CrossRef]
- Erdmier, C.; Hatcher, J.; Lee, M. Wearable device implications in the healthcare industry. J. Med Eng. Technol. 2016, 40, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Motti, V.G.; Caine, K. Users’ privacy concerns about wearables. In Proceedings of the International Conference on Financial Cryptography and Data Security, San Juan, PR, USA, 26–30 January 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 231–244. [Google Scholar]
- Wang, T.; Yang, H.; Qi, D.; Liu, Z.; Cai, P.; Zhang, H.; Chen, X. Mechano-Based Transductive Sensing for Wearable Healthcare. Small 2018, 14, 1702933. [Google Scholar] [CrossRef] [PubMed]
- Schneegass, S.; Amft, O. (Eds.) Smart Textiles—Fundamentals, Design, and Interaction; Human-Computer Interaction Series; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar] [CrossRef]
- Qi, D.; Liu, Z.; Leow, W.R.; Chen, X. Elastic substrates for stretchable devices. MRS Bull. 2017, 42, 103–107. [Google Scholar] [CrossRef]
- Kang, Y.J.; Wang, B.; Dai, S.; Liu, G.; Pu, Y.; Hu, C. Folded Elastic Strip-Based Triboelectric Nanogenerator for Harvesting Human Motion Energy for Multiple Applications. ACS Appl. Mater. Interfaces 2015, 7, 20469–20476. [Google Scholar] [CrossRef]
- Amaxilatis, D.; Chatzigiannakis, I. Design and Analysis of Adaptive Hierarchical Low-Power Long-Range Networks. J. Sens. Actuator Netw. 2018, 7, 51. [Google Scholar] [CrossRef]
- Centenaro, M.; Vangelista, L.; Zanella, A.; Zorzi, M. Long-Range Communications in Unlicensed Bands: the Rising Stars in the IoT and Smart City Scenarios. IEEE Wirel. Commun. 2015, 23. [Google Scholar] [CrossRef]
- Raza, U.; Kulkarni, P.; Sooriyabandara, M. Low power wide area networks: An overview. IEEE Commun. Surv. Tutor. 2017, 19, 855–873. [Google Scholar] [CrossRef]
- Sanchez-Iborra, R.; Sanchez-Gomez, J.; Ballesta-Viñas, J.; Cano, M.D.; Skarmeta, A.F. Performance Evaluation of LoRa Considering Scenario Conditions. Sensors 2018, 18, 772. [Google Scholar] [CrossRef] [PubMed]
- Akrivopoulos, O.; Chatzigiannakis, I.; Tselios, C.; Antoniou, A. On the deployment of healthcare applications over fog computing infrastructure. In Proceedings of the 2017 IEEE 41st Annual Computer Software and Applications Conference (COMPSAC), Turin, Italy, 4–8 July 2017; Volume 2, pp. 288–293. [Google Scholar]
- Akrivopoulos, O.; Amaxilatis, D.; Mavrommati, I.; Chatzigiannakis, I. Utilising Fog Computing for Developing a Person-Centric Heart Monitoring System. In Proceedings of the 14th International Conference on Intelligent Environments, Rome, Italy, 25–28 June 2018; pp. 9–16. [Google Scholar] [CrossRef]
- Intel® Curie™ Module. 2017. Available online: https://www.intel.com/content/dam/support/us/en/documents/boardsandkits/curie/intel-curie-module-datasheet.pdf (accessed on 1 March 2017).
- MPC8572E PowerQUICC III Integrated Processor. 2016. Available online: http://www.nxp.com/docs/en/data-sheet/MPC8572EEC.pdf (accessed on 1 March 2016).
- Association for the Advancement of Medical Instrumentation. ANSI/AAMI EC57:2012—Testing and Reporting Performance Results of Cardiac Rhythm and ST Segment Measurement Algorithms; American National Standard 2013; American National Standards Institute: New York, NY, USA, 2013. [Google Scholar]
- Sandøe, E.; Sigurd, B. Arrhythmia: A Guide to Clinical Electrocardiology; Publishing Partners Verlags GmbH: Bingen, Germany, 1991. [Google Scholar]
- Pan, J.; Tompkins, W.J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, 32, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Deepu, C.J.; Lian, Y. A computationally efficient QRS detection algorithm for wearable ECG sensors. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology, Boston, MA, USA, 30 August–3 September 2011; pp. 5641–5644. [Google Scholar]
- Sutha, P.; Jayanthi, V. Fetal electrocardiogram extraction and analysis using adaptive noise cancellation and wavelet transformation techniques. J. Med Syst. 2018, 42, 21. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Kumar, B.V.; Coimbra, M.T. Combining general multi-class and specific two-class classifiers for improved customized ECG heartbeat classification. In Proceedings of the 21st International Conference on Pattern Recognition (ICPR2012), Tsukuba, Japan, 11–15 November 2012; pp. 2428–2431. [Google Scholar]
- Zhang, F.; Lian, Y. QRS detection based on multiscale mathematical morphology for wearable ECG devices in body area networks. IEEE Trans. Biomed. Circuits Syst. 2009, 3, 220–228. [Google Scholar] [CrossRef]
- Shah, A.P.; Rubin, S.A. Errors in the computerized electrocardiogram interpretation of cardiac rhythm. J. Electrocardiol. 2007, 40, 385–390. [Google Scholar] [CrossRef]
- Saini, I.; Singh, D.; Khosla, A. QRS detection using K-Nearest Neighbor algorithm (KNN) and evaluation on standard ECG databases. J. Adv. Res. 2013, 4, 331–344. [Google Scholar] [CrossRef]
- Pławiak, P. Novel methodology of cardiac health recognition based on ECG signals and evolutionary-neural system. Expert Syst. Appl. 2018, 92, 334–349. [Google Scholar] [CrossRef]
- Pławiak, P. Novel genetic ensembles of classifiers applied to myocardium dysfunction recognition based on ECG signals. Swarm Evol. Comput. 2018, 39, 192–208. [Google Scholar] [CrossRef]
- Yıldırım, Ö.; Pławiak, P.; Tan, R.S.; Acharya, U.R. Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput. Biol. Med. 2018, 102, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Rajpurkar, P.; Hannun, A.Y.; Haghpanahi, M.; Bourn, C.; Ng, A.Y. Cardiologist-Level Arrhythmia Detection with Convolutional Neural Networks. arXiv, 2017; arXiv:1707.01836. [Google Scholar]
- Nemati, E.; Deen, M.J.; Mondal, T. A wireless wearable ECG sensor for long-term applications. IEEE Commun. Mag. 2012, 50, 36–43. [Google Scholar] [CrossRef]
- Miao, F.; Cheng, Y.; He, Y.; He, Q.; Li, Y. A Wearable Context-Aware ECG Monitoring System Integrated with Built-in Kinematic Sensors of the Smartphone. Sensors 2015, 15, 11465–11484. [Google Scholar] [CrossRef] [PubMed]
- Chatzigiannakis, I.; Kinalis, A.; Nikoletseas, S. An adaptive power conservation scheme for heterogeneous wireless sensor networks with node redeployment. In Proceedings of the Seventeenth Annual ACM Symposium on Parallelism in Algorithms and Architectures, Las Vegas, NE, USA, 18–20 July 2005; pp. 96–105. [Google Scholar]
- Chatzigiannakis, I.; Kinalis, A.; Nikoletseas, S. Power conservation schemes for energy efficient data propagation in heterogeneous wireless sensor networks. In Proceedings of the 38th Annual Simulation Symposium, San Diego, CA, USA, 4–6 April 2005; pp. 60–71. [Google Scholar]
- Cisco Systems Inc. Fog Computing and the Internet of Things: Extend the Cloud to Where the Things Are. 2015. Available online: http://www.cisco.com/c/dam/enus/solutions/trends/iot/docs/computing-overview.pdf (accessed on 1 December 2015).
- Theodoridis, E.; Mylonas, G.; Chatzigiannakis, I. Developing an IoT Smart City framework. In Proceedings of the IISA 2013, Piraeus, Greece, 10–12 July 2013; pp. 180–185. [Google Scholar] [CrossRef]
- Chen, Y.; Shen, W.; Huo, H.; Xu, Y. A Smart Gateway for Health Care System Using Wireless Sensor Network. In Proceedings of the SENSORCOMM 2010, Venice, Italy, 18–25 July 2010; pp. 545–550. [Google Scholar] [CrossRef]
- Mohapatra, S.; Rekha, K.S. Sensor-cloud: A hybrid framework for remote patient monitoring. Int. J. Comput. Appl. 2012, 55, 7–11. [Google Scholar] [CrossRef]
- Yang, S.; Gerla, M. Personal gateway in mobile health monitoring. In Proceedings of the 2011 IEEE International Conference on Pervasive Computing and Communications Workshops (PERCOM Workshops), Seattle, WA, USA, 21–25 March 2011; pp. 636–641. [Google Scholar] [CrossRef]
- Gia, T.N.; Jiang, M.; Rahmani, A.M.; Westerlund, T.; Liljeberg, P.; Tenhunen, H. Fog Computing in Healthcare Internet of Things: A Case Study on ECG Feature Extraction. In Proceedings of the 2015 IEEE CIT/IUCC/DASC/PICOM, Liverpool, UK, 26–28 October 2015; pp. 356–363. [Google Scholar] [CrossRef]
- Yi, D.; Binwen, F.; Xiaoming, K.; Qianqian, M. Design and implementation of mobile health monitoring system based on MQTT protocol. In Proceedings of the 2016 IEEE Advanced Information Management, Communicates, Electronic and Automation Control Conference (IMCEC), Xi’an, China, 3–5 Ocotber 2016; pp. 1679–1682. [Google Scholar] [CrossRef]
- Akrivopoulos, O.; Amaxilatis, D.; Antoniou, A.; Chatzigiannakis, I. Design and Evaluation of a Person-Centric Heart Monitoring System over Fog Computing Infrastructure. In Proceedings of the First International Workshop on Human-Centered Sensing, Networking, and Systems, HumanSys@SenSys 2017, Delft, The Netherlands, 5 November 2017; pp. 25–30. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef]
- Park, K.; Cho, B.; Lee, D.; Song, S.; Lee, J.; Chee, Y.; Kim, I.; Kim, S. Hierarchical support vector machine based heartbeat classification using higher order statistics and hermite basis function. In Proceedings of the 2008 IEEE Computers in Cardiology, Bologna, Italy, 14–17 September 2008; pp. 229–232. [Google Scholar]
- Walraven, G. Basic Arrhythmias, 8th ed.; Pearson: London, UK, 2016. [Google Scholar]
- Ripley, B.D. Pattern Recognition and Neural Networks; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- De Chazal, P.; O’Dwyer, M.; Reilly, R.B. Automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 2004, 51, 1196–1206. [Google Scholar] [CrossRef]
- Hu, Y.H.; Palreddy, S.; Tompkins, W.J. A patient-adaptable ECG beat classifier using a mixture of experts approach. IEEE Trans. Biomed. Eng. 1997, 44, 891–900. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).