Atomic Force Microscopy Imaging of Elastin Nanofibers Self-Assembly
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparing Elastin Samples
2.1.1. Preparation of Stock Suspensions
2.1.2. Elastin Sample Preparation
2.2. Atomic Force Microscopy
2.2.1. AFM Imaging
2.2.2. Image Processing
2.2.3. Tip Geometry
3. Results
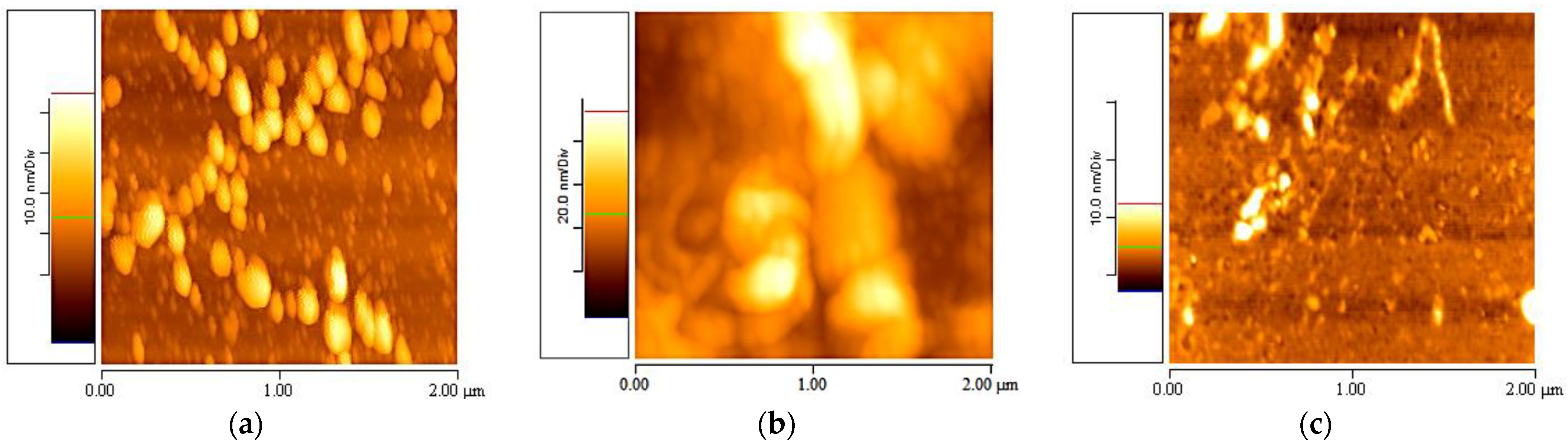
3.1. Elastin in Different Solvents
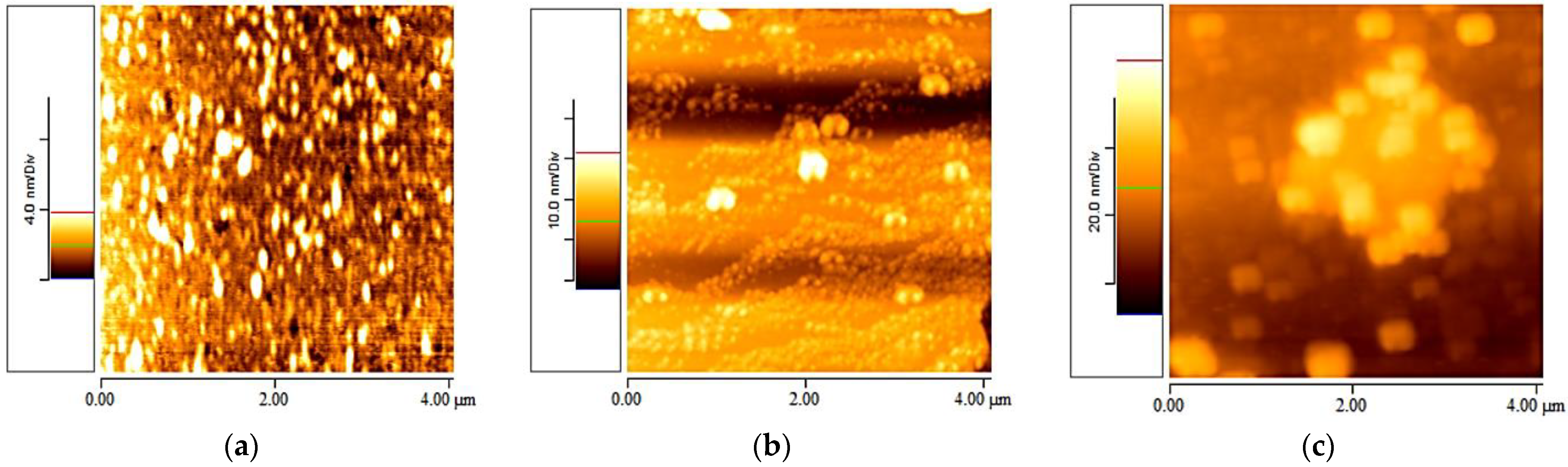
3.2. Elastin Observed at Different Concentrations
3.3. Elastin Observed at Different Temperatures
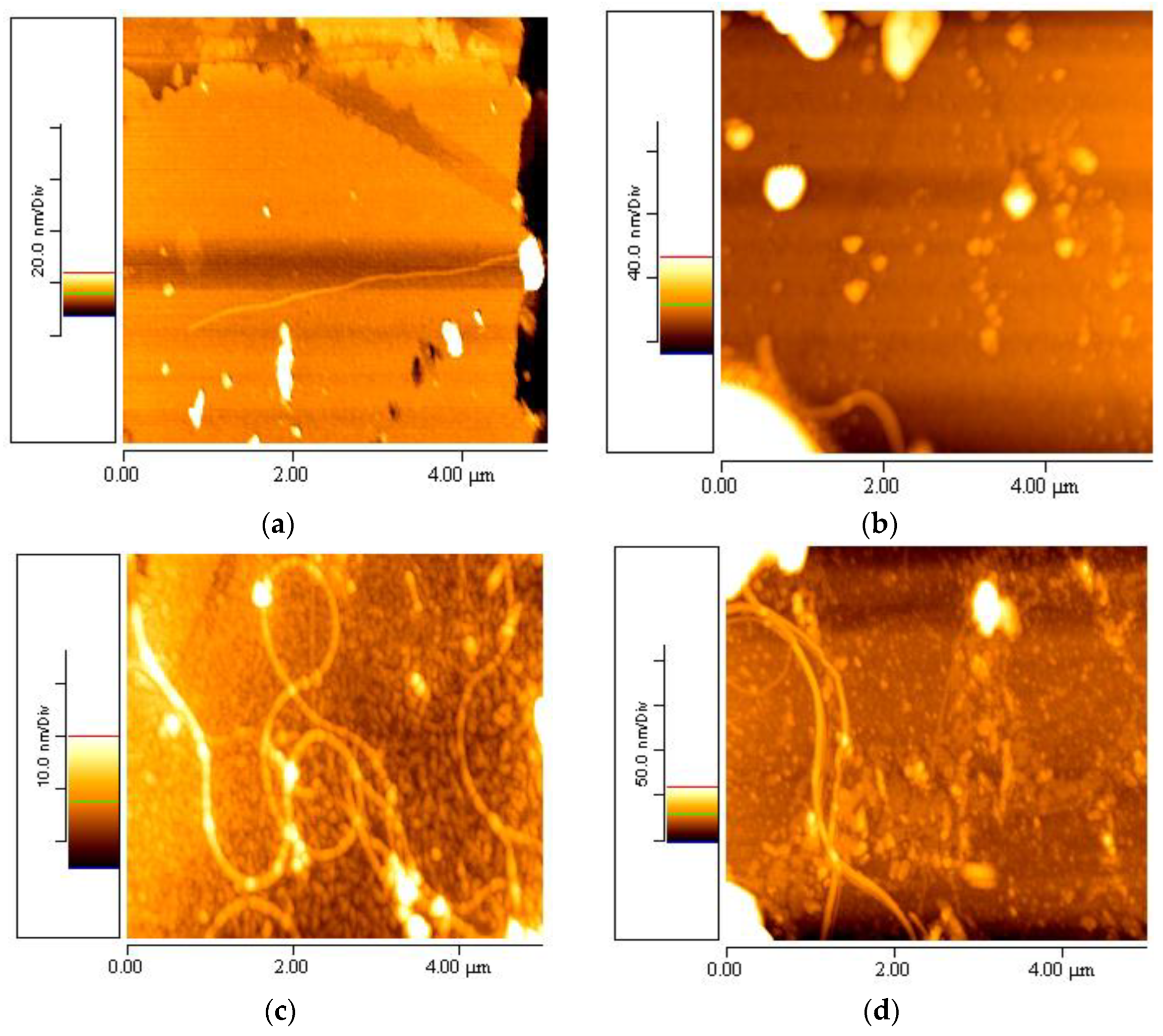
3.4. Elastin Observed at Different Periods of Time
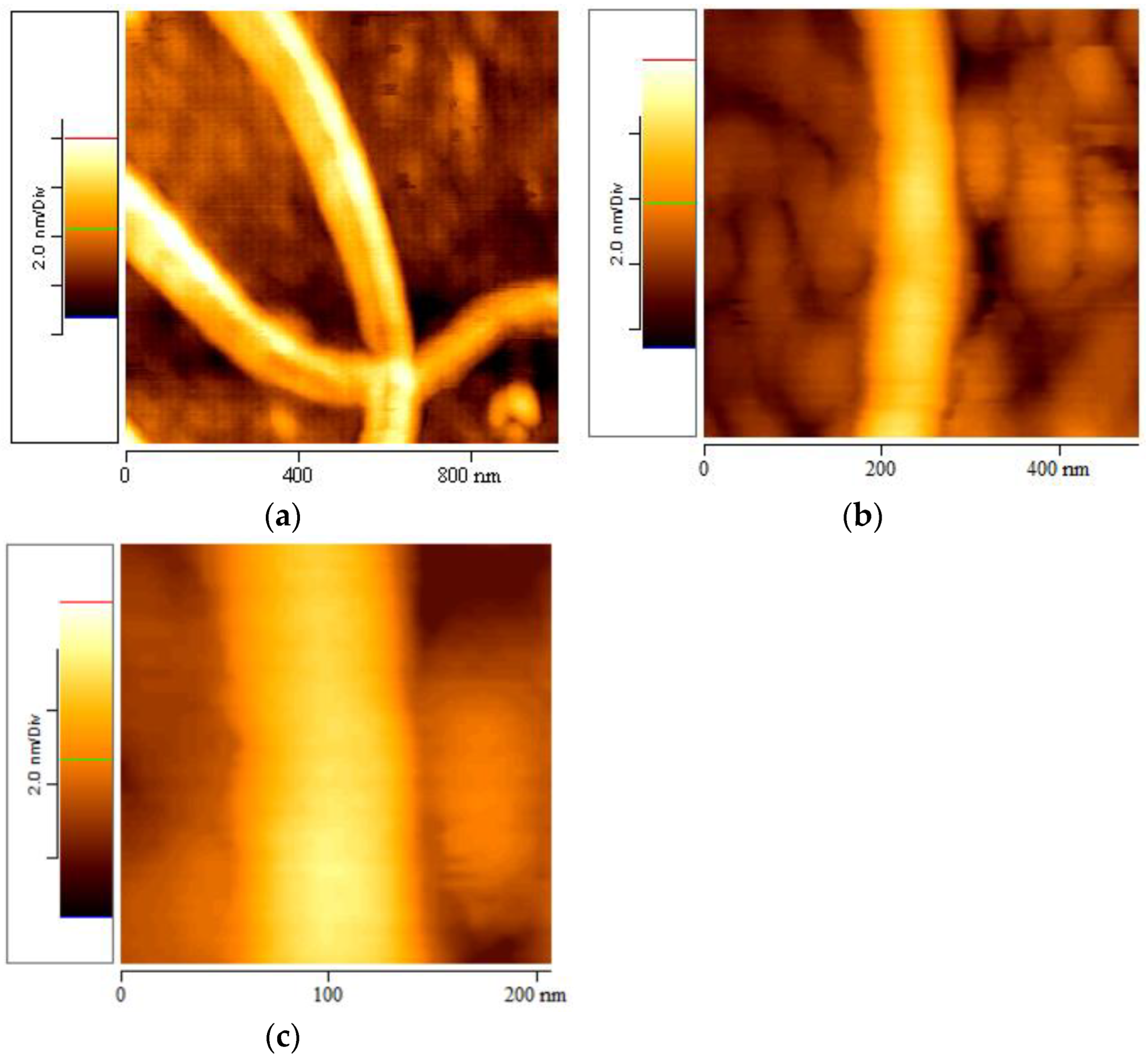
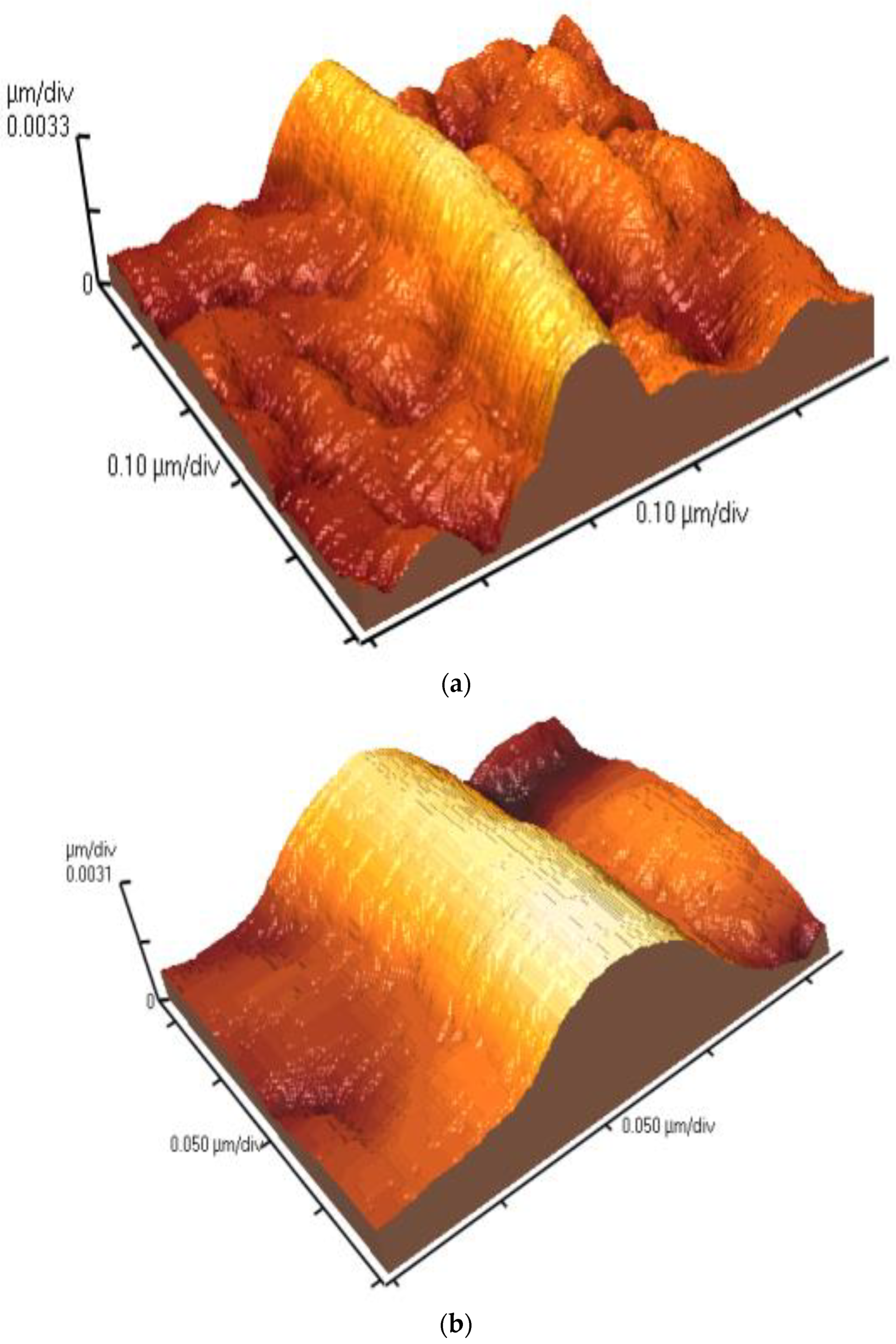
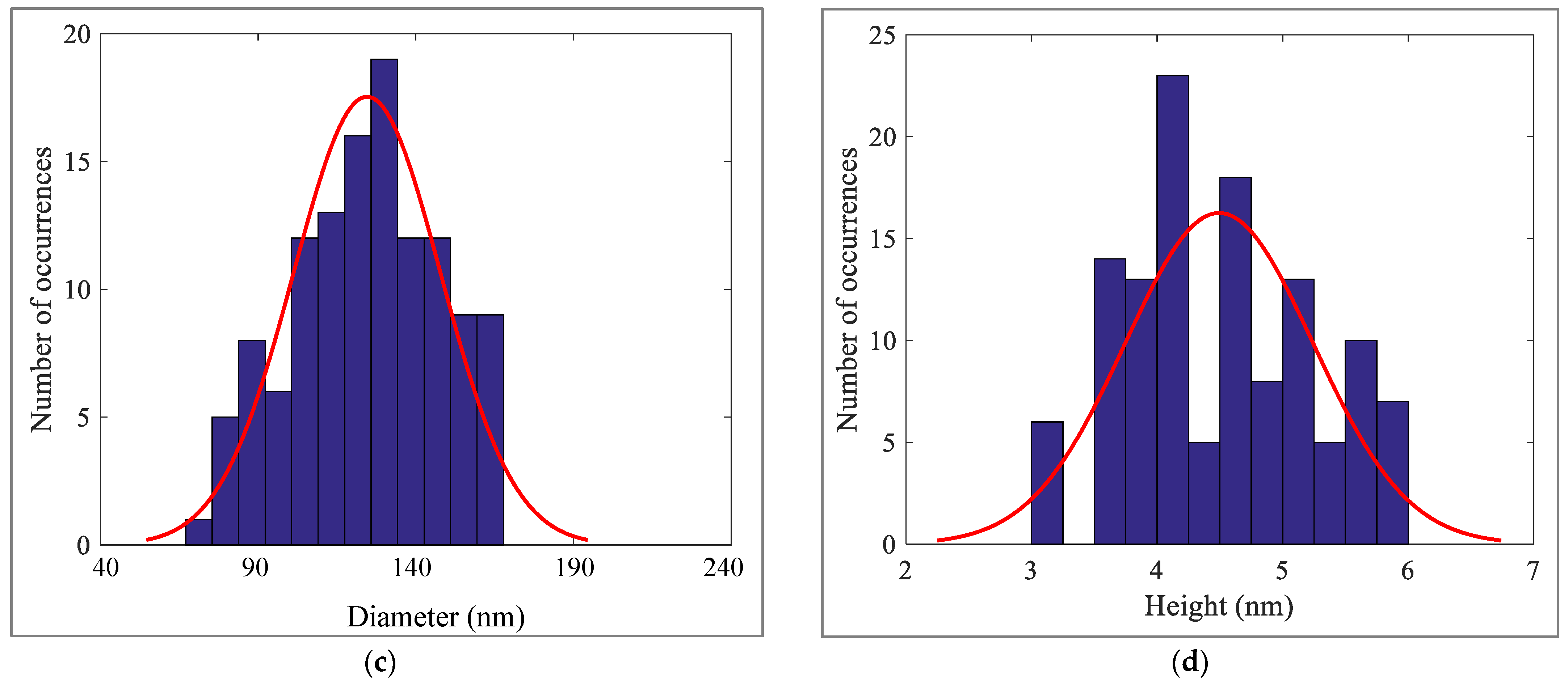
3.5. Geometrical Characteristics of Elastin Nanofibrils
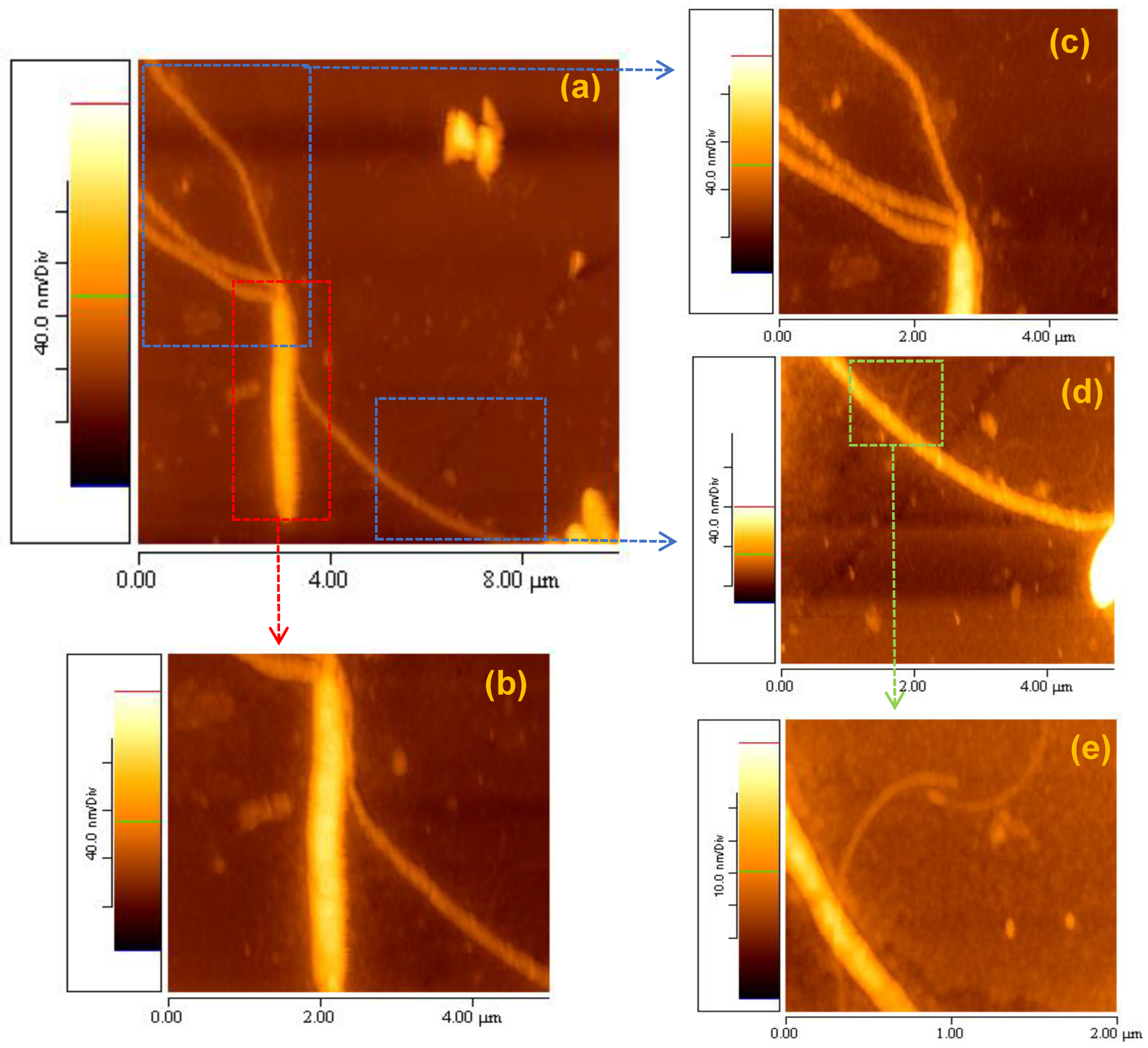
3.6. Self-Assembly Imaging of Elastin
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodriguez-Cabello, J.C.; Martin, L.; Girotti, A.; Garca-Arévalo, C.; Arias, F.J.; Alonso, M. Emerging applications of multifunctional elastin-like recombinamers. Nanomedicine 2011, 6, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Nettles, D.L.; Chilkoti, A.; Setton, L.A. Applications of elastin-like polypeptides in tissue engineering. Adv. Drug Deliv. Rev. 2010, 62, 1479–1485. [Google Scholar] [CrossRef] [PubMed]
- Pepe, A.; Bochicchio, B. An Elastin-Derived Self-Assembling Polypeptide. J. Soft Matter 2013, 2013, 732157. [Google Scholar] [CrossRef]
- Kowalczyk, T.; Hnatuszko-Konka, K.; Gerszberg, A.; Kononowicz, A.K. Elastin-like polypeptides as a promising family of genetically-engineered protein based polymers. World J. Microbiol. Biotechnol. 2014, 30, 2141–2152. [Google Scholar] [CrossRef] [PubMed]
- Asai, D.; Xu, D.; Liu, W.; Garcia Quiroz, F.; Callahan, D.J.; Zalutsky, M.R.; Craig, S.L.; Chilkoti, A. Protein polymer hydrogels by in situ, rapid and reversible self-gelation. Biomaterials 2012, 33, 5451–5458. [Google Scholar] [CrossRef]
- Wu, X.; Sallach, R.; Haller, C.A.; Caves, J.A.; Nagapudi, K.; Conticello, V.P.; Levenston, M.E.; Chaikof, E.L. Alterations in physical crosslinking modulate mechanical properties of two-phase protein polymer networks. Biomacromolecules 2005, 6, 3037–3044. [Google Scholar] [CrossRef]
- Lim, D.W.; Nettles, D.L.; Setton, L.A.; Chilkoti, A. Rapid crosslinking of elastin-like polypeptides with (hydroxymethyl)phosphines in aqueous solution. Biomacromolecules 2007, 8, 1463–1470. [Google Scholar] [CrossRef]
- Bochicchio, B.; Armenante, M.R.; Crudele, M.A.; Pepe, A. Molecular Determinants for the Self-Assembly of Elastin Peptides. Conf. Pap. Sci. 2014, 2014, 214235. [Google Scholar] [CrossRef]
- Wagenseil, J.E.; Mecham, R.P. New insights into elastic fiber assembly. Birth Defects Res. (Part C)—Embryo Today Rev. 2007, 81, 229–240. [Google Scholar] [CrossRef]
- Almine, J.F.; Bax, D.V.; Mithieux, S.M. Elastin-based materials. Chem. Soc. Rev. 2010, 39, 3371–3379. [Google Scholar] [CrossRef]
- Rodriguez-Cabello, J.C.; Prieto, S.; Reguera, J.; Arias, F.J.; Ribeiro, A. Biofunctional design of elastin-like polymers for advanced applications in nanobiotechnology. J. Biomater. Sci. Polym. Ed. 2007, 18, 269–286. [Google Scholar] [CrossRef]
- Maeda, I.; Fukumoto, Y.; Nose, T.; Shimohigashi, Y.; Nezu, T.; Terada, Y.; Kodama, H.; Kaibara, K.; Okamoto, K. Structural requirements essential for elastin coacervation: Favorable spatial arrangements of valine ridges on the three-dimensional structure of elastin-derived polypeptide (VPGVG)n. J. Pept. Sci. 2011, 17, 735–743. [Google Scholar] [CrossRef]
- Beckmann, A.; Xiao, S.; Müller, J.P.; Mercadante, D.; Nüchter, T.; Kröger, N.; Langhojer, F.; Petrich, W.; Holstein, T.W.; Benoit, M.; et al. A fast recoiling silk-like elastomer facilitates nanosecond nematocyst discharge. BMC Biol. 2015, 13, 3. [Google Scholar] [CrossRef]
- Stylianou, A.; Kontomaris, S.V.; Grant, C.; Alexandratou, E. Atomic Force Microscopy on Biological Materials Related to Pathological Conditions. Scanning 2019, 2019, 8452851. [Google Scholar] [CrossRef]
- Kontomaris, S.V. The Hertz Model in AFM Nanoindentation Experiments: Applications in Biological Samples and Biomaterials. Micro Nanosyst. 2018, 10, 11–22. [Google Scholar] [CrossRef]
- Stylianou, A.; Yova, D.; Alexandratou, E. Investigation of the influence of UV irradiation on collagen thin films by AFM imaging. Mater. Sci. Eng. C 2014, 45, 455–468. [Google Scholar] [CrossRef]
- Wenger, M.P.E.; Bozec, L.; Horton, M.A.; Mesquidaz, P. Mechanical properties of collagen fibrils. Biophys. J. 2007, 93, 1255–1263. [Google Scholar] [CrossRef]
- Lekka, M. Atomic force microscopy: A tip for diagnosing cancer. Nat. Nanotechnol. 2012, 7, 691–692. [Google Scholar] [CrossRef]
- Ramalho, R.; Rankovic, S.; Zhou, J.; Aiken, C.; Rousso, I. Analysis of the mechanical properties of wild type and hyperstable mutants of the HIV-1 capsid. Retrovirology 2016, 13, 1–7. [Google Scholar] [CrossRef]
- Ganser, B.K.; Li, S.; Klishko, V.Y.; Finch, J.T.; Sundquist, W.I. Assembly and analysis of conical models for the HIV-1 core. Science 1999, 283, 80–83. [Google Scholar] [CrossRef]
- Sundquist, W.I.; Kräusslich, H.G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012, 2, 7. [Google Scholar] [CrossRef] [PubMed]
- Lekka, M. Discrimination Between Normal and Cancerous Cells Using AFM. BioNanoScience 2016, 6, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Kontomaris, S.V.; Stylianou, A.; Chliveros, G.; Malamou, A. Determining Spatial Variability of Elastic Properties for Biological Samples Using AFM. Micromachines 2023, 14, 182. [Google Scholar] [CrossRef] [PubMed]
- Hasirci, V.; Vrana, E.; Zorlutuna, P.; Ndreu, A.; Yilgor, P.; Basmanav, F.B.; Aydin, E. Nanobiomaterials: A review of the existing science and technology, and new approaches. J. Biomater. Sci. Polym. Ed. 2006, 17, 1241–1268. [Google Scholar] [CrossRef]
- Markeiwicz, P.; Coh, M.C. Identifying location on a substrate for the repeated positioning of AFM samples. Ultramicroscopy 1997, 68, 215–221. [Google Scholar] [CrossRef]
- Stylianou, A.; Yova, D. Surface Nanoscale Imaging of Collagen Thin by Atomic Force Microscopy. Mater. Sci. Eng. C 2013, 33, 2947–2957. [Google Scholar] [CrossRef]
- Stylianou, A.; Kontomaris, S.V.; Yova, D. Assessing Collagen Nanoscale Thin Films Heterogeneity by AFM Multi-mode Imaging and Nanoindetation for NanoBioMedical Applications. Micro Nanosyst. 2014, 6, 95–102. [Google Scholar] [CrossRef]
- Kontomaris, S.V.; Yova, D.; Sambani, K.; Stylianou, A. XIV Mediterranean Conference on Medical and Biological Engineering and Computing, AFM Investigation of the Influence of Red Light Irradiation on Collagen 2016, IFMBE Proceedings; Springer: Berlin, Germanym, 2016; Volume 57, pp. 269–274. [Google Scholar] [CrossRef]
- Horcas, I.; Fernández, R.; Gómez-Rodríguez, J.M.; Colchero, J.; Gómez-Herrero, J.; Baro, A.M. Review of Scientific Instruments, WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007, 78, 013705. [Google Scholar] [CrossRef]
- Bellotti, R.; Picotto, G.B.; Ribotta, L. AFM Measurements and Tip Characterization of Nanoparticles with Different Shapes. Nanomanuf. Metrol. 2022, 5, 127–138. [Google Scholar] [CrossRef]
- Canet-Ferrer, J.; Coronado, E.; Forment-Aliaga, A.; Pinilla-Cienfuegos, E. Correction of the tip convolution effects in the imaging of nanostructures studied through scanning force microscopy. Nanotechnology 2014, 25, 395703. [Google Scholar] [CrossRef]
- Rodriguez-Cabello, J.C.; Gonzalez De Torre, I.; González-Pérez, M.; González-Pérez, F.; Montequi, I. Fibrous Scaffolds From Elastin-Based Materials. Front. Bioeng. Biotechnol. 2021, 9, 652384. [Google Scholar] [CrossRef]
- Waterhouse, A.; Wise, S.G.; Ng, M.K.C.; Weiss, A.S. Elastin as a nonthrombogenic biomaterial. Tissue Eng. Part B Rev. 2011, 17, 93–99. [Google Scholar] [CrossRef]
- Srokowski, E.M.; Woodhouse, K.A. Surface and adsorption characteristics of three elastin-like polypeptide coatings with varying sequence lengths. J. Mater. Sci. Mater. Med. 2013, 24, 71–84. [Google Scholar] [CrossRef]
- Dandurand, J.; Samouillan, V.; Lacabanne, C.; Pepe, A.; Bochicchio, B. Water structure and elastin-like peptide aggregation. J. Therm. Anal. Calorim. 2014, 120, 419–426. [Google Scholar] [CrossRef]
- Koga, T.; Nakamoto, K.; Odawara, K.; Matsuoka, T.; Higashi, N. Fabrication of Thermo-Responsive Molecular Layers from Self-Assembling Elastin-Like Oligopeptides Containing Cell-Binding Domain for Tissue Engineering. Polymers 2015, 7, 134–146. [Google Scholar] [CrossRef]
- Pepe, A.; Delaunay, F.; Bracalello, A.; Bochicchio, B. The Inhibitory Effect of Resveratrol on Elastin Amyloidogenesis. Conf. Pap. Sci. 2014, 2014, 410545. [Google Scholar] [CrossRef]
- del Mercato, L.L.; Maruccio, G.; Pompa, P.P.; Bochicchio, B.; Tamburro, A.M.; Cingolani, R.; Rinaldi, R. Amyloid-like Fibrils in Elastin-Related Polypeptides: Structural Characterization and Elastic Properties. Biomacromolecules 2008, 9, 796–803. [Google Scholar] [CrossRef]
- Bochicchio, B.; Pepe, A.; Delaunay, F.; Lorusso, M.; Baud, S.; Dauchez, M. Amyloidogenesis of proteolytic fragments of human elastin. RSC Adv. 2013, 3, 13273–13285. [Google Scholar] [CrossRef]
- Cao, M.; Shen, Y.; Wang, Y.; Wang, X.; Li, D. Self-Assembly of Short Elastin-like Amphiphilic Peptides: Effects of Temperature, Molecular Hydrophobicity and Charge Distribution. Molecules 2019, 24, 202. [Google Scholar] [CrossRef]
- Tarakanova, A.; Yeo, G.C.; Baldock, C.; Weiss, A.S.; Buehler, M.J. Tropoelastin is a Flexible Molecule that Retains its Canonical Shape. Macromol. Biosci. 2019, 19, 1800250. [Google Scholar] [CrossRef]
- Ozsvar, J.; Yang, C.; Cain, S.A.; Baldock, C.; Tarakanova, A.; Weiss, A.S. Tropoelastin and Elastin Assembly. Front. Bioeng. Biotechnol. 2021, 9, 643110. [Google Scholar] [CrossRef] [PubMed]
- Bracalello, A.; Secchi, V.; Mastrantonio, R.; Pepe, A.; Persichini, T.; Iucci, G.; Bochicchio, B.; Battocchio, C. Fibrillar Self-Assembly of a Chimeric Elastin-Resilin Inspired Engineered Polypeptide. Nanomaterials 2019, 9, 1613. [Google Scholar] [CrossRef] [PubMed]
- Alvisi, N.; Gutiérrez-Mejía, F.A.; Lokker, M.; Lin, Y.-T.; de Jong, A.M.; van Delft, F.; de Vries, R. Self-Assembly of Elastin-like Polypeptide Brushes on Silica Surfaces and Nanoparticles. Biomacromolecules 2021, 22, 1966–1979. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.F.; Sousa, M.G.D.C.; Rodrigues, G.R.; de Oliveira, K.B.S.; Pereira, A.M.; da Costa, A.; Machado, R.; Franco, O.L.; Dias, S.C. Elastin-like Polypeptides in Development of Nanomaterials for Application in the Medical Field. Front. Nanotechnol. 2022, 4, 874790. [Google Scholar] [CrossRef]
- Juanes-Gusano, D.; Santos, M.; Reboto, V.; Alonso, M.; Rodríguez-Cabello, J.C. Self- assembling systems comprising intrinsically disordered protein polymers like elastin-like recombinamers. J. Pept. Sci. 2022, 28, e3362. [Google Scholar] [CrossRef]
- Coenen, A.M.J.; Bernaerts, K.V.; Harings, J.A.W.; Jockenhoevel, S.; Ghazanfari, S. Elastic materials for tissue engineering applications: Natural, synthetic, and hybrid polymers. Acta Biomater. 2018, 79, 60–82. [Google Scholar] [CrossRef]
- MacEwan, S.R.; Chilkoti, A. Applications of elastin-like polypeptides in drug delivery. J. Control. Release 2014, 190, 314–330. [Google Scholar] [CrossRef]
- Wang, R.; de Kort, B.J.; Smits, A.I.P.M.; Weiss, A.S. Elastin in Vascular Grafts. In Tissue-Engineered Vascular Grafts; Reference Series in Biomedical Engineering; Walpoth, B., Bergmeister, H., Bowlin, G., Kong, D., Rotmans, J., Zilla, P., Eds.; Springer: Cham, Switzerland, 2019. [Google Scholar] [CrossRef]
- Shifren, A.; Mecham, R.P. The stumbling block in lung repair of emphysema: Elastic fiber assembly. Proc. Am. Thorac. Soc. 2006, 3, 428–433. [Google Scholar] [CrossRef]
- Sarangthem, V.; Singh, T.D.; Dinda, A.K. Emerging Role of Elastin-Like Polypeptides in Regenerative Medicine. Adv. Wound Care 2021, 10, 257–269. [Google Scholar] [CrossRef]





| Diameter (nm) | Height (nm) | |
|---|---|---|
| Fiber | ||
| Fibril | ||
| Nanofibril |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sambani, K.; Kontomaris, S.V.; Yova, D. Atomic Force Microscopy Imaging of Elastin Nanofibers Self-Assembly. Materials 2023, 16, 4313. https://doi.org/10.3390/ma16124313
Sambani K, Kontomaris SV, Yova D. Atomic Force Microscopy Imaging of Elastin Nanofibers Self-Assembly. Materials. 2023; 16(12):4313. https://doi.org/10.3390/ma16124313
Chicago/Turabian StyleSambani, Kyriaki, Stylianos Vasileios Kontomaris, and Dido Yova. 2023. "Atomic Force Microscopy Imaging of Elastin Nanofibers Self-Assembly" Materials 16, no. 12: 4313. https://doi.org/10.3390/ma16124313
APA StyleSambani, K., Kontomaris, S. V., & Yova, D. (2023). Atomic Force Microscopy Imaging of Elastin Nanofibers Self-Assembly. Materials, 16(12), 4313. https://doi.org/10.3390/ma16124313








