Silver Nanoparticles Induced Changes in DNA Methylation and Histone H3 Methylation in a Mouse Model of Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Treatment
2.2. ELISA
2.3. Analysis of miRNA and mRNA Expression by Real-Time PCR
2.4. Statistical Evaluation
3. Results
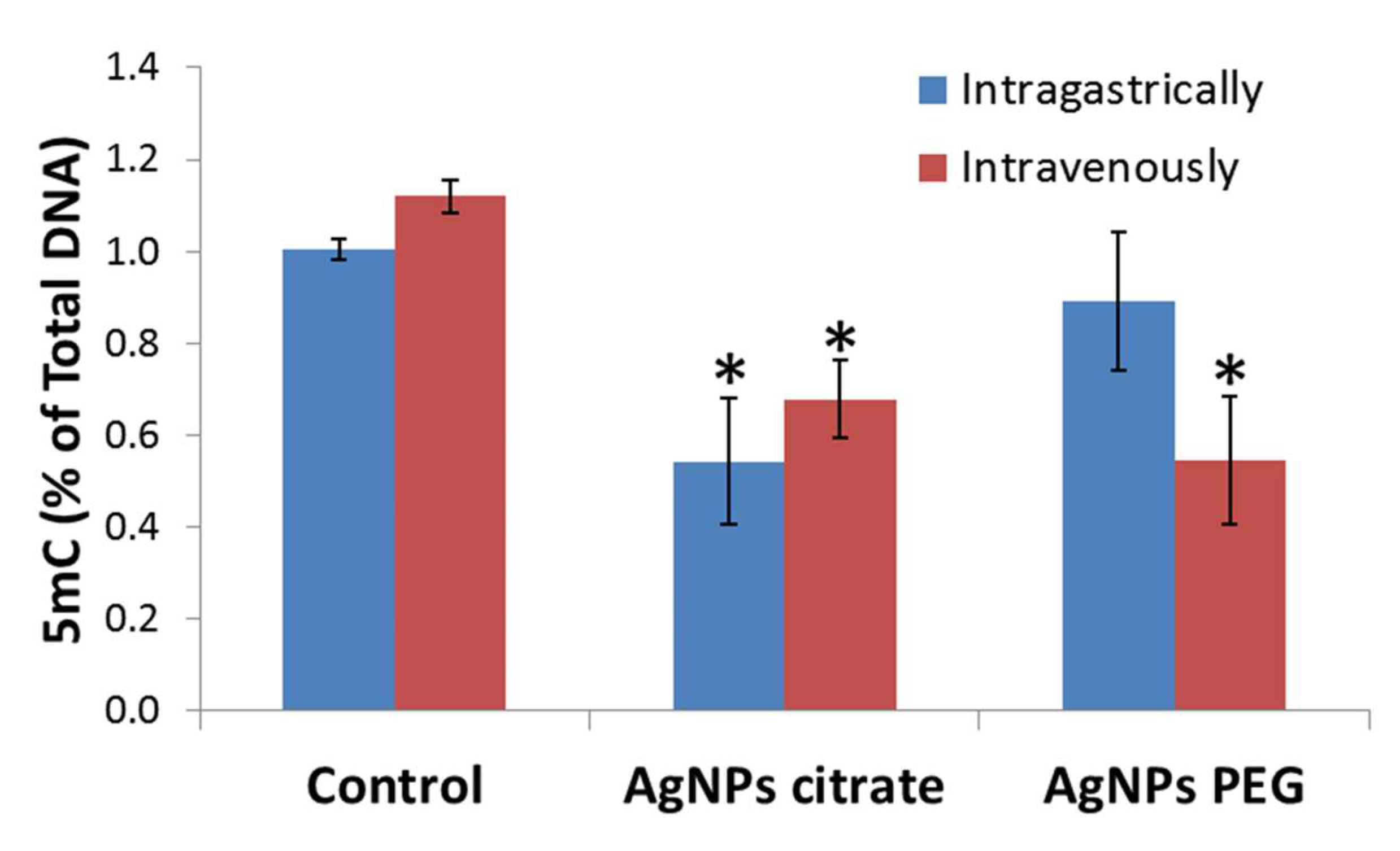
3.1. AgNPs Treatment Decreases the Tumor’s Global DNA Methylation
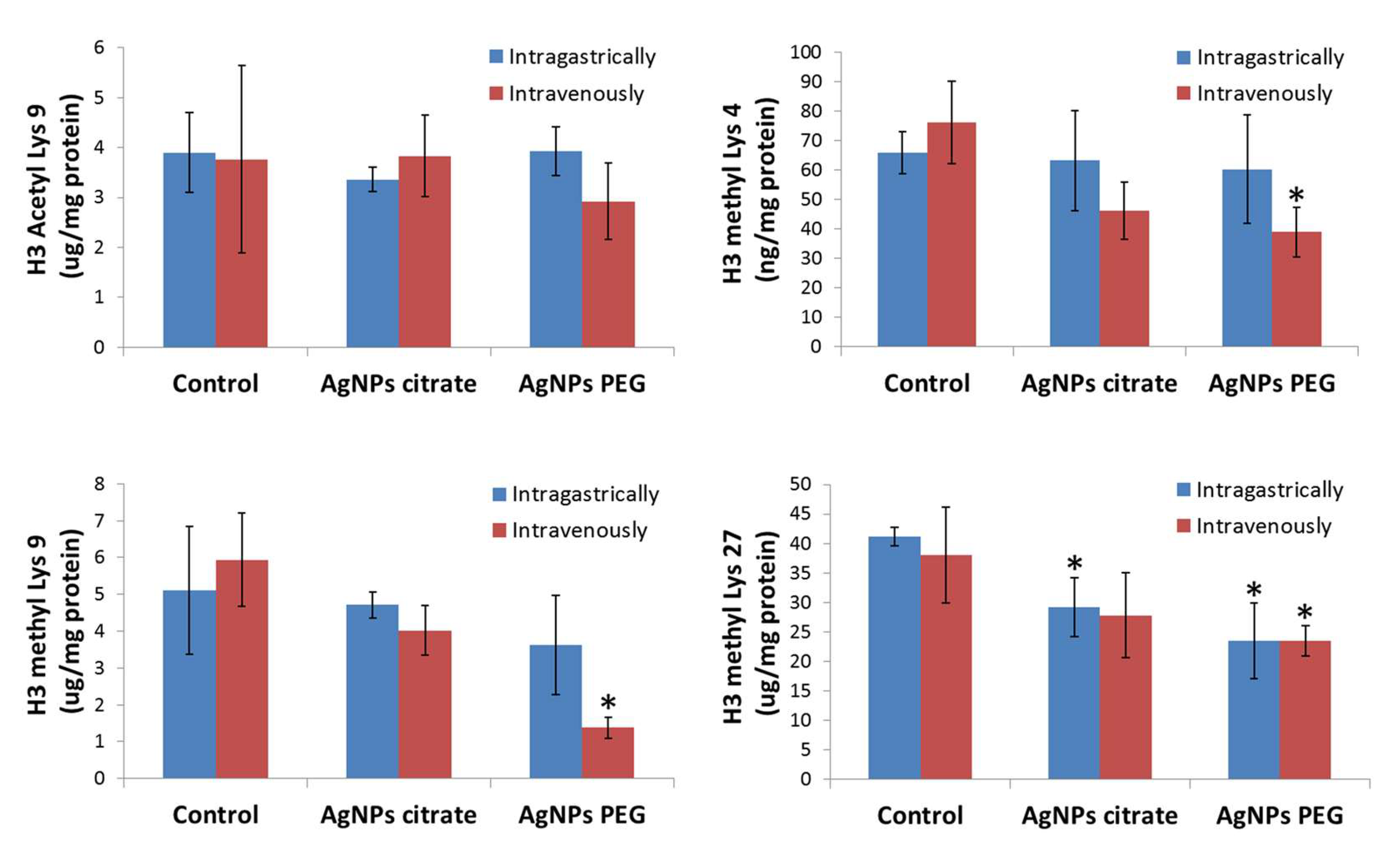
3.2. AgNPs Treatment Decreases Methylation but Not Acetylation of Histone H3 in Tumor Tissue
3.3. AgNPs Treatment Affects the Expression of Chromatin-Modifying Enzymes in Tumor Tissue
3.4. Effect of AgNPs Treatment on Expression of Oncogenes and Tumor Suppressor Genes in Tumor Tissue
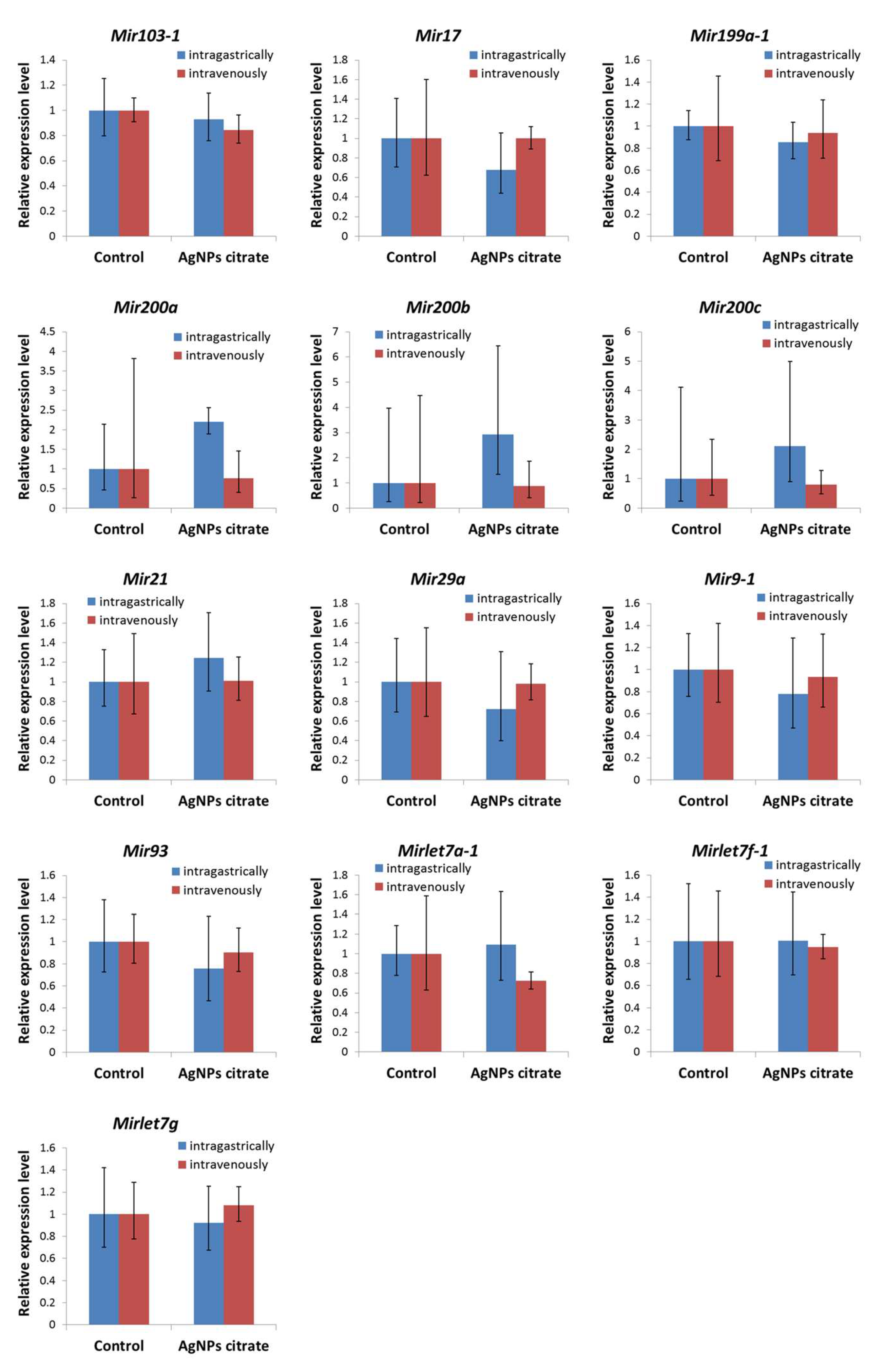
3.5. Expression of Metastasis-Related miRNAs in Tumor Tissue Was Not Affected by AgNPs Treatment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kruszewski, M.; Brzoska, K.; Brunborg, G.; Asare, N.; Dobrzynska, M.; Dusinska, M.; Fjellsbo, L.M.; Georgantzopoulou, A.; Gromadzka-Ostrowska, J.; Gutleb, A.C.; et al. Toxicity of Silver Nanomaterials in Higher Eukaryotes. Adv. Mol. Toxicol. 2011, 5, 179–218. [Google Scholar]
- Aueviriyavit, S.; Phummiratch, D.; Maniratanachote, R. Mechanistic study on the biological effects of silver and gold nanoparticles in Caco-2 cells—Induction of the Nrf2/HO-1 pathway by high concentrations of silver nanoparticles. Toxicol. Lett. 2014, 224, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Brzóska, K.; Grądzka, I.; Kruszewski, M. Impact of silver, gold, and iron oxide nanoparticles on cellular response to tumor necrosis factor. Toxicol. Appl. Pharmacol. 2018, 356, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Swanner, J.; Singh, R. Synthesis, Purification, Characterization, and Imaging of Cy3-Functionalized Fluorescent Silver Nanoparticles in 2D and 3D Tumor Models. Methods Mol. Biol. 2018, 1790, 209–218. [Google Scholar]
- Pucelik, B.; Sułek, A.; Borkowski, M.; Barzowska, A.; Kobielusz, M.; Dąbrowski, J.M. Synthesis and Characterization of Size- and Charge-Tunable Silver Nanoparticles for Selective Anticancer and Antibacterial Treatment. ACS Appl. Mater. Interfaces 2022, 14, 14981–14996. [Google Scholar] [CrossRef]
- Haase, A.; Dommershausen, N.; Schulz, M.; Landsiedel, R.; Reichardt, P.; Krause, B.C.; Tentschert, J.; Luch, A. Genotoxicity testing of different surface-functionalized SiO2, ZrO2 and silver nanomaterials in 3D human bronchial models. Arch. Toxicol. 2017, 91, 3991–4007. [Google Scholar] [CrossRef]
- Juarez-Moreno, K.; Chávez-García, D.; Hirata, G.; Vazquez-Duhalt, R. Monolayer (2D) or spheroids (3D) cell cultures for nanotoxicological studies? Comparison of cytotoxicity and cell internalization of nanoparticles. Toxicol. Vitr. 2022, 85, 105461. [Google Scholar] [CrossRef]
- Austin, C.A.; Umbreit, T.H.; Brown, K.M.; Barber, D.S.; Dair, B.J.; Francke-Carroll, S.; Feswick, A.; Saint-Louis, M.A.; Hikawa, H.; Siebein, K.N.; et al. Distribution of silver nanoparticles in pregnant mice and developing embryos. Nanotoxicology 2012, 6, 912–922. [Google Scholar] [CrossRef]
- Zhang, X.F.; Park, J.H.; Choi, Y.J.; Kang, M.H.; Gurunathan, S.; Kim, J.H. Silver nanoparticles cause complications in pregnant mice. Int. J. Nanomed. 2015, 10, 7057–7071. [Google Scholar]
- Chakraborty, B.; Pal, R.; Ali, M.; Singh, L.M.; Rahman, D.S.; Ghosh, S.K.; Sengupta, M. Immunomodulatory properties of silver nanoparticles contribute to anticancer strategy for murine fibrosarcoma. Cell. Mol. Immunol. 2016, 13, 191–205. [Google Scholar] [CrossRef]
- Manshian, B.B.; Jimenez, J.; Himmelreich, U.; Soenen, S.J. Presence of an Immune System Increases Anti-Tumor Effect of Ag Nanoparticle Treated Mice. Adv. Healthc. Mater. 2017, 6, 1601099. [Google Scholar] [CrossRef] [PubMed]
- Wani, I.A.; Ahmad, T.; Khosla, A. Recent advances in anticancer and antimicrobial activity of silver nanoparticles synthesized using phytochemicals and organic polymers. Nanotechnology 2021, 32, 462001. [Google Scholar] [CrossRef] [PubMed]
- Baran, A.; Keskin, C.; Baran, M.F.; Huseynova, I.; Khalilov, R.; Eftekhari, A.; Irtegun-Kandemir, S.; Kavak, D.E. Ecofriendly Synthesis of Silver Nanoparticles Using Ananas comosus Fruit Peels: Anticancer and Antimicrobial Activities. Bioinorg. Chem. Appl. 2021, 2021, 2058149. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.J.; Hosseinian, F.; Willmore, W.G. A Current Overview of the Biological and Cellular Effects of Nanosilver. Int. J. Mol. Sci. 2018, 19, 2030. [Google Scholar] [CrossRef] [PubMed]
- Flores-López, L.Z.; Espinoza-Gómez, H.; Somanathan, R. Silver nanoparticles: Electron transfer, reactive oxygen species, oxidative stress, beneficial and toxicological effects. Mini review. J. Appl. Toxicol. 2019, 39, 16–26. [Google Scholar] [CrossRef]
- Gedda, M.R.; Babele, P.K.; Zahra, K.; Madhukar, P. Epigenetic Aspects of Engineered Nanomaterials: Is the Collateral Damage Inevitable? Front. Bioeng. Biotechnol. 2019, 7, 228. [Google Scholar] [CrossRef]
- Moreira, L.; Costa, C.; Pires, J.; Teixeira, J.P.; Fraga, S. How can exposure to engineered nanomaterials influence our epigenetic code? A review of the mechanisms and molecular targets. Mutat. Res. Rev. Mutat. Res. 2021, 788, 108385. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Dubey, P.; Matai, I.; Kumar, S.U.; Sachdev, A.; Bhushan, B.; Gopinath, P. Perturbation of cellular mechanistic system by silver nanoparticle toxicity: Cytotoxic, genotoxic and epigenetic potentials. Adv. Colloid Interface Sci. 2015, 221, 4–21. [Google Scholar] [CrossRef]
- Chen, Y.; Sheng, F.; Wang, X.; Zhang, Z.; Qi, S.; Chen, L. Early Epigenetic Responses in the Genomic DNA Methylation Fingerprints in Cells in Response to Sublethal Exposure of Silver Nanoparticles. Front. Bioeng. Biotechnol. 2022, 10, 927036. [Google Scholar] [CrossRef]
- Brzóska, K.; Grądzka, I.; Kruszewski, M. Silver, Gold, and Iron Oxide Nanoparticles Alter miRNA Expression but Do Not Affect DNA Methylation in HepG2 Cells. Materials 2019, 12, 1038. [Google Scholar] [CrossRef] [PubMed]
- Brzóska, K.; Wojewódzka, M.; Szczygiel, M.; Drzał, A.; Sniegocka, M.; Michalczyk-Wetula, D.; Biela, E.; Elas, M.; Kucińska, M.; Piotrowska-Kempisty, H.; et al. Silver Nanoparticles Inhibit Metastasis of 4T1 Tumor in Mice after Intragastric but Not Intravenous Administration. Materials 2022, 15, 3837. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Del Castillo Falconi, V.M.; Torres-Arciga, K.; Matus-Ortega, G.; Díaz-Chávez, J.; Herrera, L.A. DNA Methyltransferases: From Evolution to Clinical Applications. Int. J. Mol. Sci. 2022, 23, 8994. [Google Scholar] [CrossRef]
- Zeng, X.; Zheng, W.; Sheng, Y.; Ma, H. UBE2B promotes ovarian cancer growth via promoting RAD18 mediated ZMYM2 monoubiquitination and stabilization. Bioengineered 2022, 13, 8000–8012. [Google Scholar] [CrossRef]
- Liu, Y.L.; Zheng, J.; Tang, L.J.; Han, W.; Wang, J.M.; Liu, D.W.; Tian, Q.B. The deubiquitinating enzyme activity of USP22 is necessary for regulating HeLa cell growth. Gene 2015, 572, 49–56. [Google Scholar] [CrossRef]
- Meng, H.; Cao, Y.; Qin, J.; Song, X.; Zhang, Q.; Shi, Y.; Cao, L. DNA methylation, its mediators and genome integrity. Int. J. Biol. Sci. 2015, 11, 604–617. [Google Scholar] [CrossRef]
- Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010, 28, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Maki, A.; Lin, Y.; Aoyama, M.; Sato, K.; Gao, J.Q.; Tsujino, H.; Nagano, K.; Higashisaka, K.; Tsutsumi, Y. Silver Nanoparticles Induce DNA Hypomethylation through Proteasome-Mediated Degradation of DNA Methyltransferase 1. Biol. Pharm. Bull. 2020, 43, 1924–1930. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Li, M.; Lin, X.; Li, Y.; Song, H.; Chen, H. AgNPs Aggravated Hepatic Steatosis, Inflammation, Oxidative Stress, and Epigenetic Changes in Mice with NAFLD Induced by HFD. Front. Bioeng. Biotechnol. 2022, 10, 912178. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xu, M.; Zhang, J.; Ma, J.; Gao, M.; Zhang, Z.; Xu, Y.; Liu, S. Genome-Wide DNA Methylation Variations upon Exposure to Engineered Nanomaterials and Their Implications in Nanosafety Assessment. Adv. Mater. 2017, 29, 1604580. [Google Scholar] [CrossRef] [PubMed]
- Blanco, J.; Lafuente, D.; Gómez, M.; García, T.; Domingo, J.L.; Sánchez, D.J. Polyvinyl pyrrolidone-coated silver nanoparticles in a human lung cancer cells: Time- and dose-dependent influence over p53 and caspase-3 protein expression and epigenetic effects. Arch. Toxicol. 2017, 91, 651–666. [Google Scholar] [CrossRef]
- Mytych, J.; Zebrowski, J.; Lewinska, A.; Wnuk, M. Prolonged effects of silver nanoparticles on p53/p21 pathway-mediated proliferation, DNA damage response, and methylation parameters in HT22 hippocampal neuronal cells. Mol. Neurobiol. 2017, 54, 1285–1300. [Google Scholar] [CrossRef]
- Qian, Y.; Zhang, J.; Hu, Q.; Xu, M.; Chen, Y.; Hu, G.; Zhao, M.; Liu, S. Silver nanoparticle-induced hemoglobin decrease involves alteration of histone 3 methylation status. Biomaterials 2015, 70, 12–22. [Google Scholar] [CrossRef]
- Wamucho, A.; Heffley, A.; Tsyusko, O.V. Epigenetic effects induced by silver nanoparticles in Caenorhabditis elegans after multigenerational exposure. Sci. Total Environ. 2020, 725, 138523. [Google Scholar] [CrossRef]
- González-Palomo, A.K.; Saldaña-Villanueva, K.; Cortés-García, J.D.; Fernández-Macias, J.C.; Méndez-Rodríguez, K.B.; Pérez Maldonado, I.N. Effect of silver nanoparticles (AgNPs) exposure on microRNA expression and global DNA methylation in endothelial cells EA.hy926. Environ. Toxicol. Pharmacol. 2021, 81, 103543. [Google Scholar] [CrossRef]
- Eom, H.J.; Chatterjee, N.; Lee, J.; Choi, J. Integrated mRNA and micro RNA profiling reveals epigenetic mechanism of differential sensitivity of Jurkat T cells to AgNPs and Ag ions. Toxicol. Lett. 2014, 229, 311–318. [Google Scholar] [CrossRef]
- Mahmood, M.; Li, Z.; Casciano, D.; Khodakovskaya, M.V.; Chen, T.; Karmakar, A.; Dervishi, E.; Xu, Y.; Mustafa, T.; Watanabe, F.; et al. Nanostructural materials increase mineralization in bone cells and affect gene expression through miRNA regulation. J. Cell. Mol. Med. 2011, 15, 2297–2306. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brzóska, K.; Sochanowicz, B.; Szczygieł, M.; Drzał, A.; Śniegocka, M.; Michalczyk-Wetula, D.; Elas, M.; Kapka-Skrzypczak, L.; Kruszewski, M. Silver Nanoparticles Induced Changes in DNA Methylation and Histone H3 Methylation in a Mouse Model of Breast Cancer. Materials 2023, 16, 4163. https://doi.org/10.3390/ma16114163
Brzóska K, Sochanowicz B, Szczygieł M, Drzał A, Śniegocka M, Michalczyk-Wetula D, Elas M, Kapka-Skrzypczak L, Kruszewski M. Silver Nanoparticles Induced Changes in DNA Methylation and Histone H3 Methylation in a Mouse Model of Breast Cancer. Materials. 2023; 16(11):4163. https://doi.org/10.3390/ma16114163
Chicago/Turabian StyleBrzóska, Kamil, Barbara Sochanowicz, Małgorzata Szczygieł, Agnieszka Drzał, Martyna Śniegocka, Dominika Michalczyk-Wetula, Martyna Elas, Lucyna Kapka-Skrzypczak, and Marcin Kruszewski. 2023. "Silver Nanoparticles Induced Changes in DNA Methylation and Histone H3 Methylation in a Mouse Model of Breast Cancer" Materials 16, no. 11: 4163. https://doi.org/10.3390/ma16114163
APA StyleBrzóska, K., Sochanowicz, B., Szczygieł, M., Drzał, A., Śniegocka, M., Michalczyk-Wetula, D., Elas, M., Kapka-Skrzypczak, L., & Kruszewski, M. (2023). Silver Nanoparticles Induced Changes in DNA Methylation and Histone H3 Methylation in a Mouse Model of Breast Cancer. Materials, 16(11), 4163. https://doi.org/10.3390/ma16114163







