Differences in the Structure and Antimicrobial Activity of Hydrazones Derived from Methyl 4-Phenylpicolinimidate
Abstract
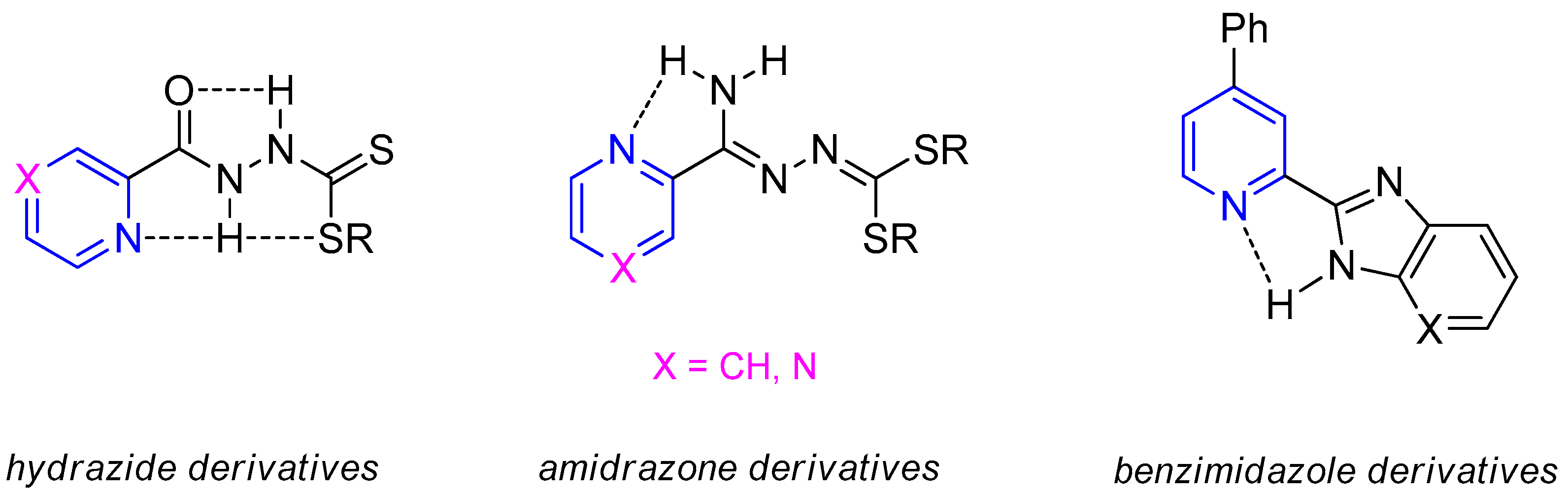
:1. Introduction
2. Materials and Methods
2.1. Chemistry
2.1.1. Methyl 4-Phenylpicolinimidate
2.1.2. 4-Phenylpicolinohydrazonamide (1)
2.1.3. 4-Phenylpicolinohydrazide (2)
2.1.4. General Procedure for the Synthesis of Hydrazones (3a,b–4a,b)
(E)-N′-((5-Nitrofuran-2-yl)methylene)-4-phenylpicolinohydrazonamide (3a)
(Z)-N′-((5-Nitrothiophen-2-yl)methylene)-4-phenylpicolinohydrazonamide (3b)
(E)-N′-((5-Nitrofuran-2-yl)methylene)-4-phenylpicolinohydrazide (4a)
(E)-N′-((5-Nitrothiophen-2-yl)methylene)-4-phenylpicolinohydrazide (4b)
2.2. ADMET
2.3. Biological Activities
2.3.1. In Vitro Antimicrobial Activity Assay
2.3.2. Tuberculostatic Activity Assay
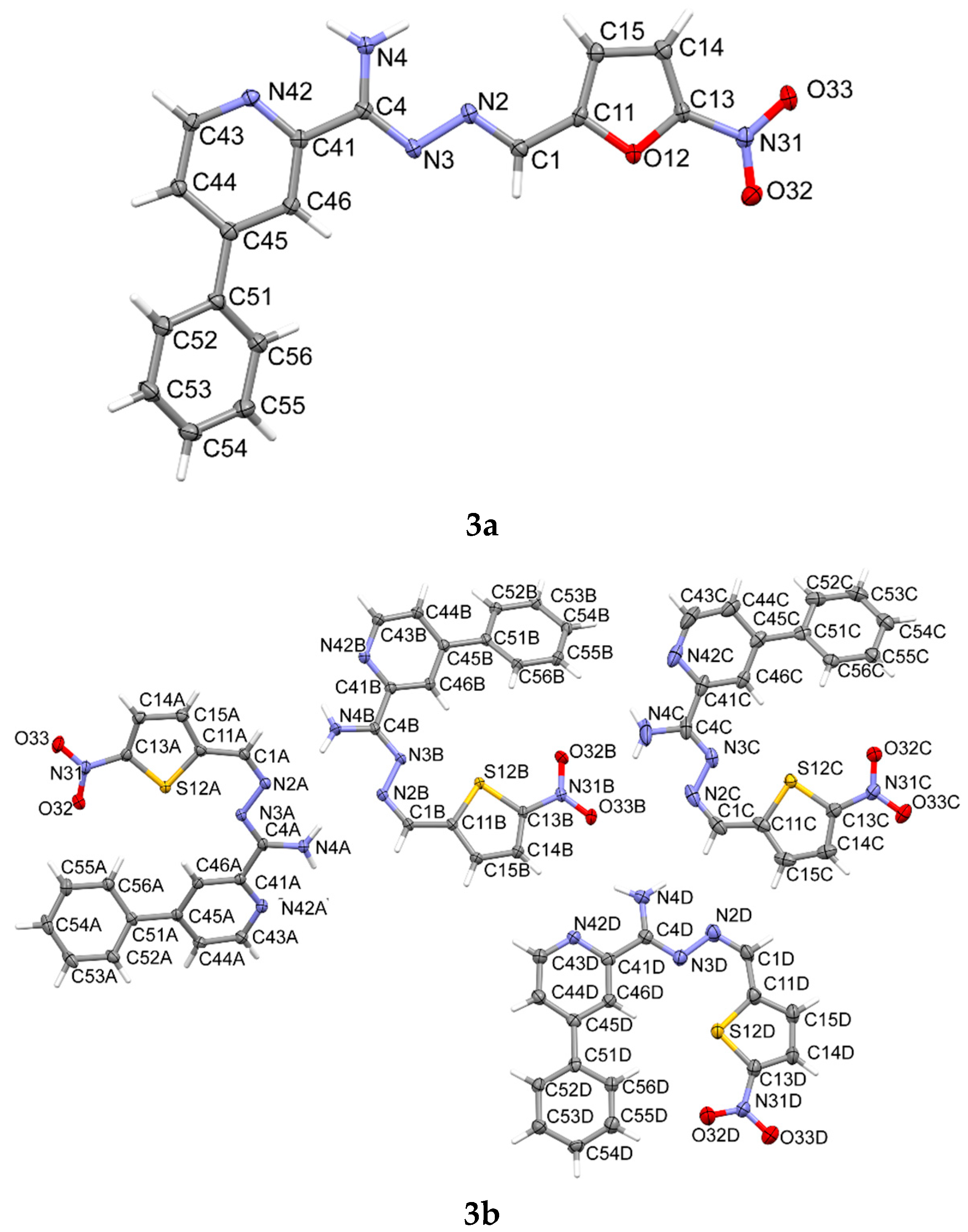
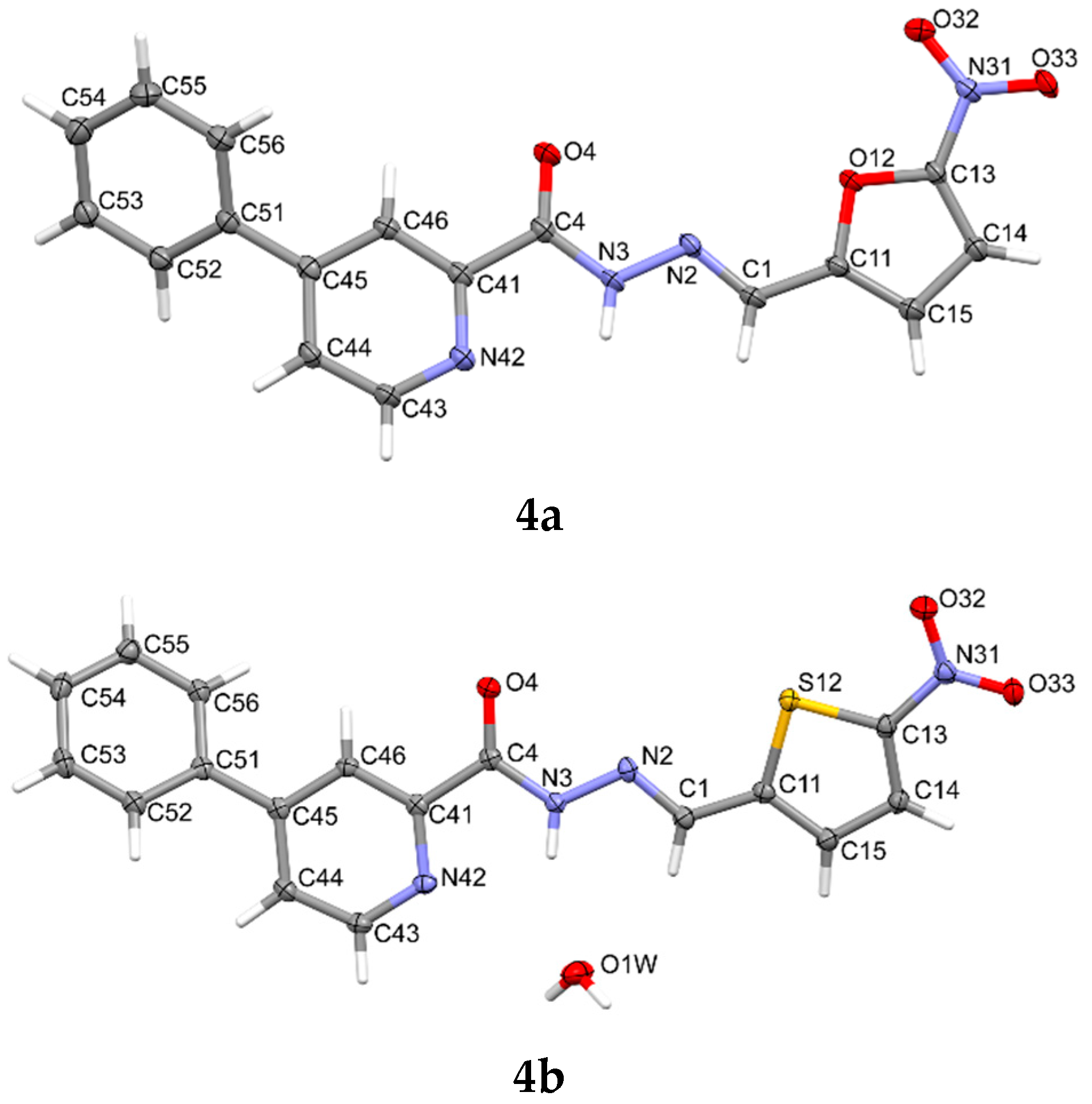
2.4. X-ray Study
2.5. DFT Calculations
3. Results and Discussion
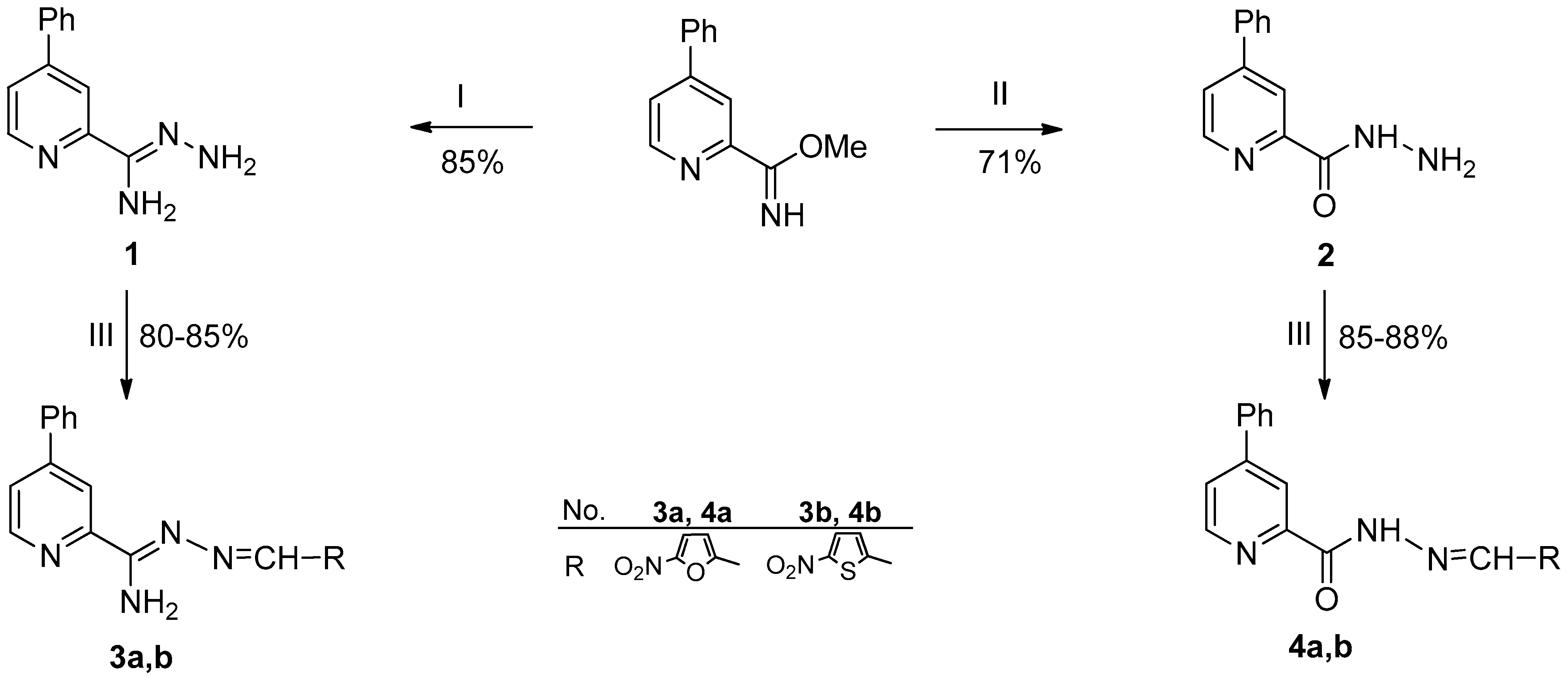
3.1. Synthesis
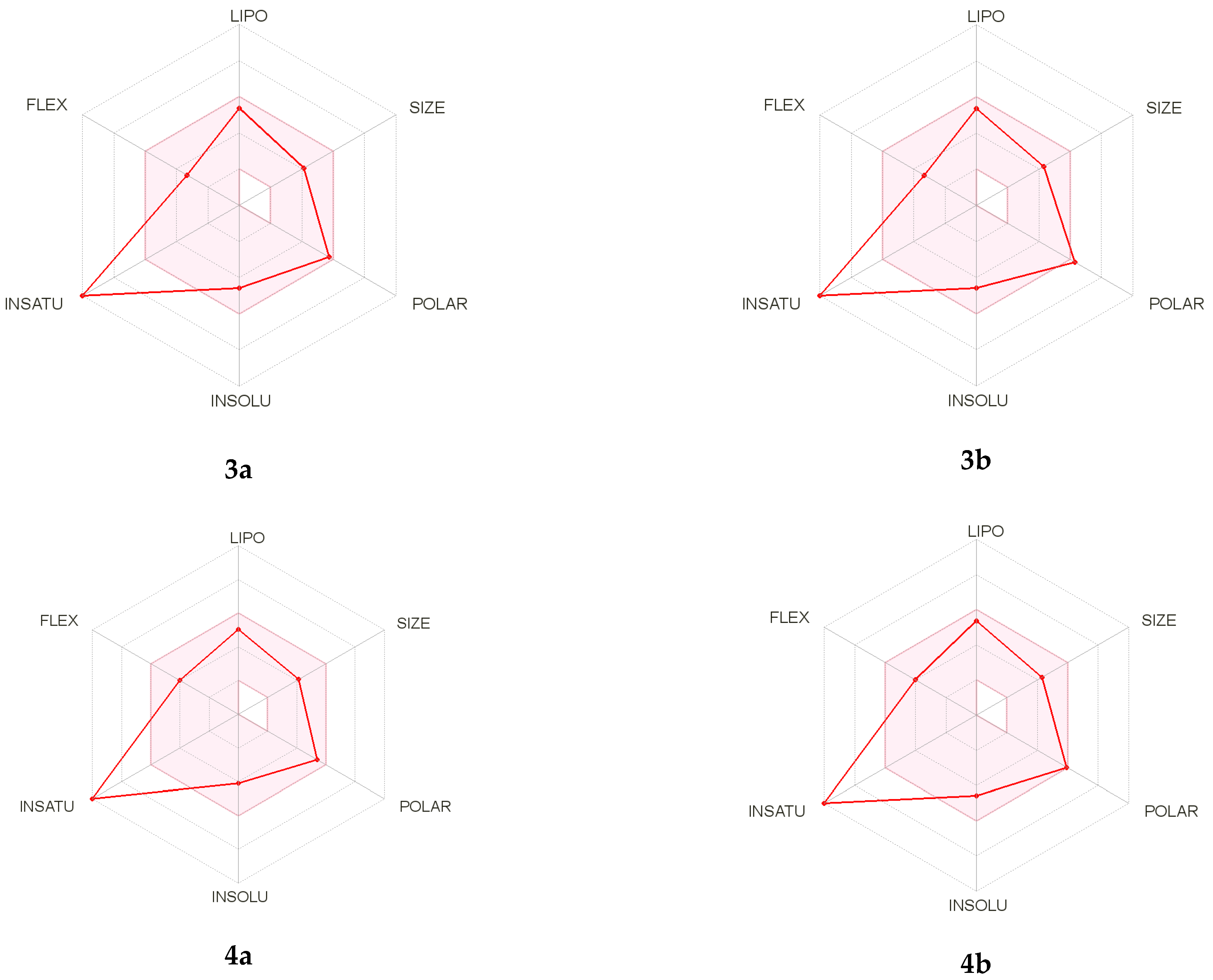
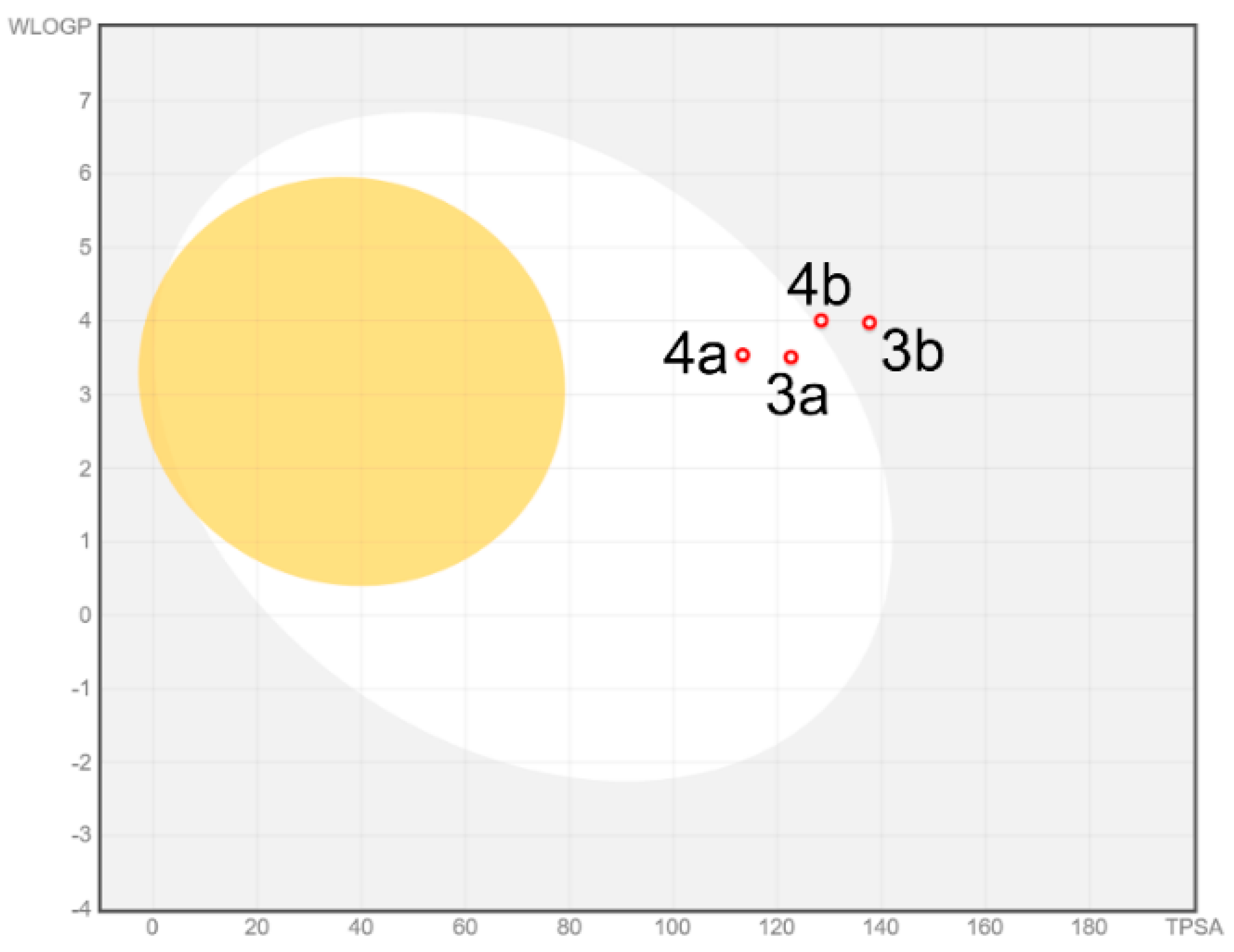
3.2. ADMET Analysis
3.3. Antimicrobial Activity
3.4. Tuberculostatic Activity
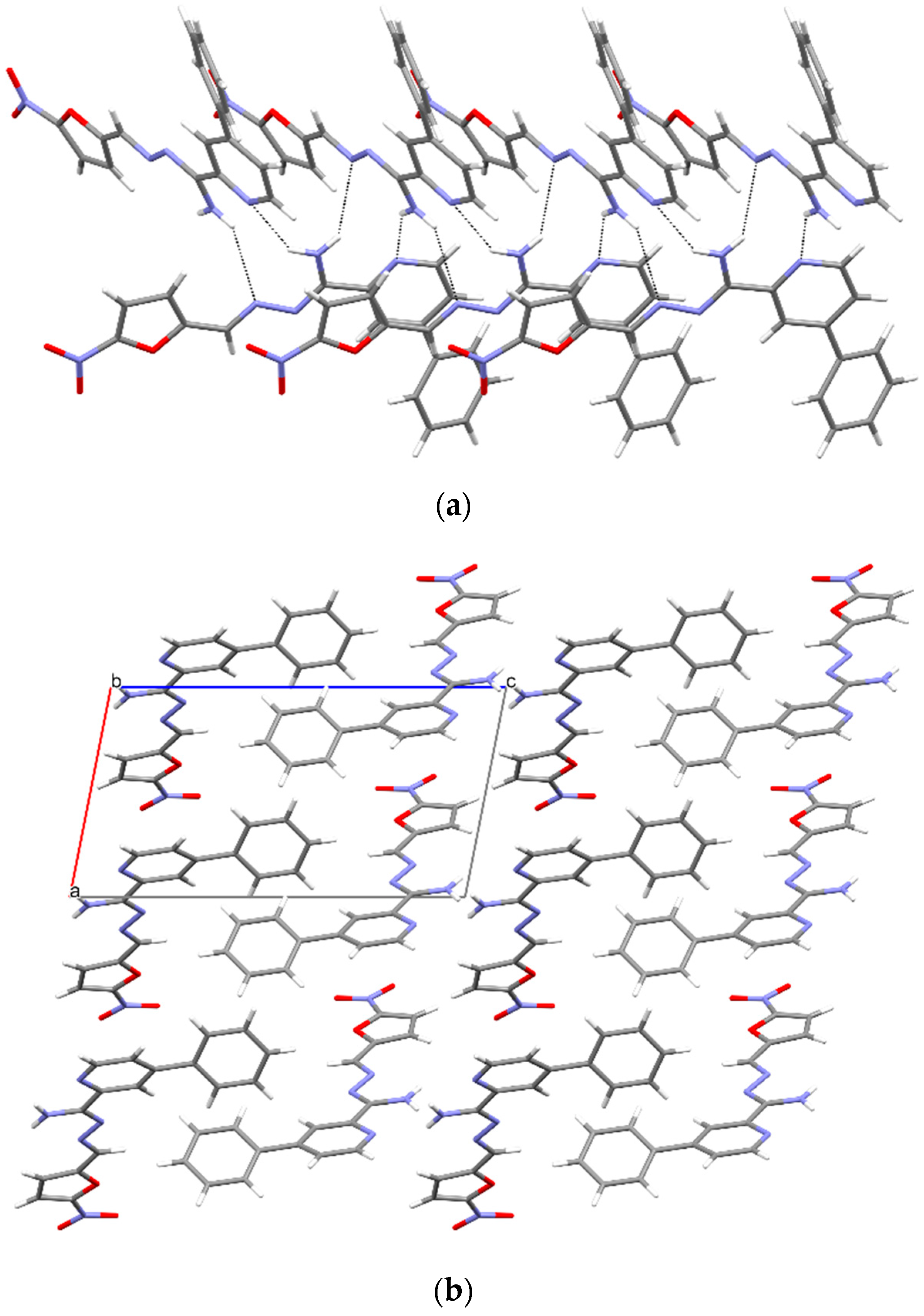
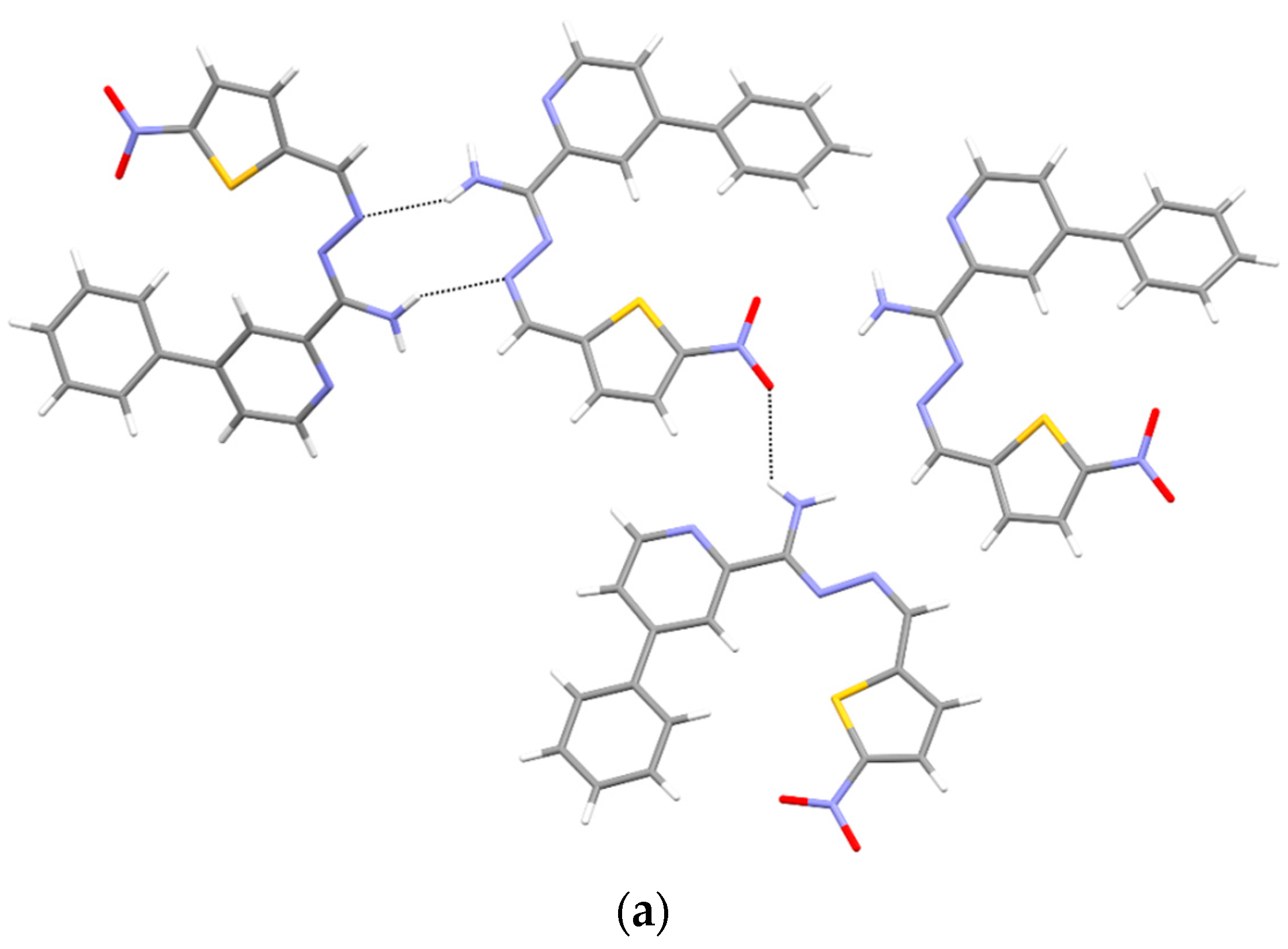
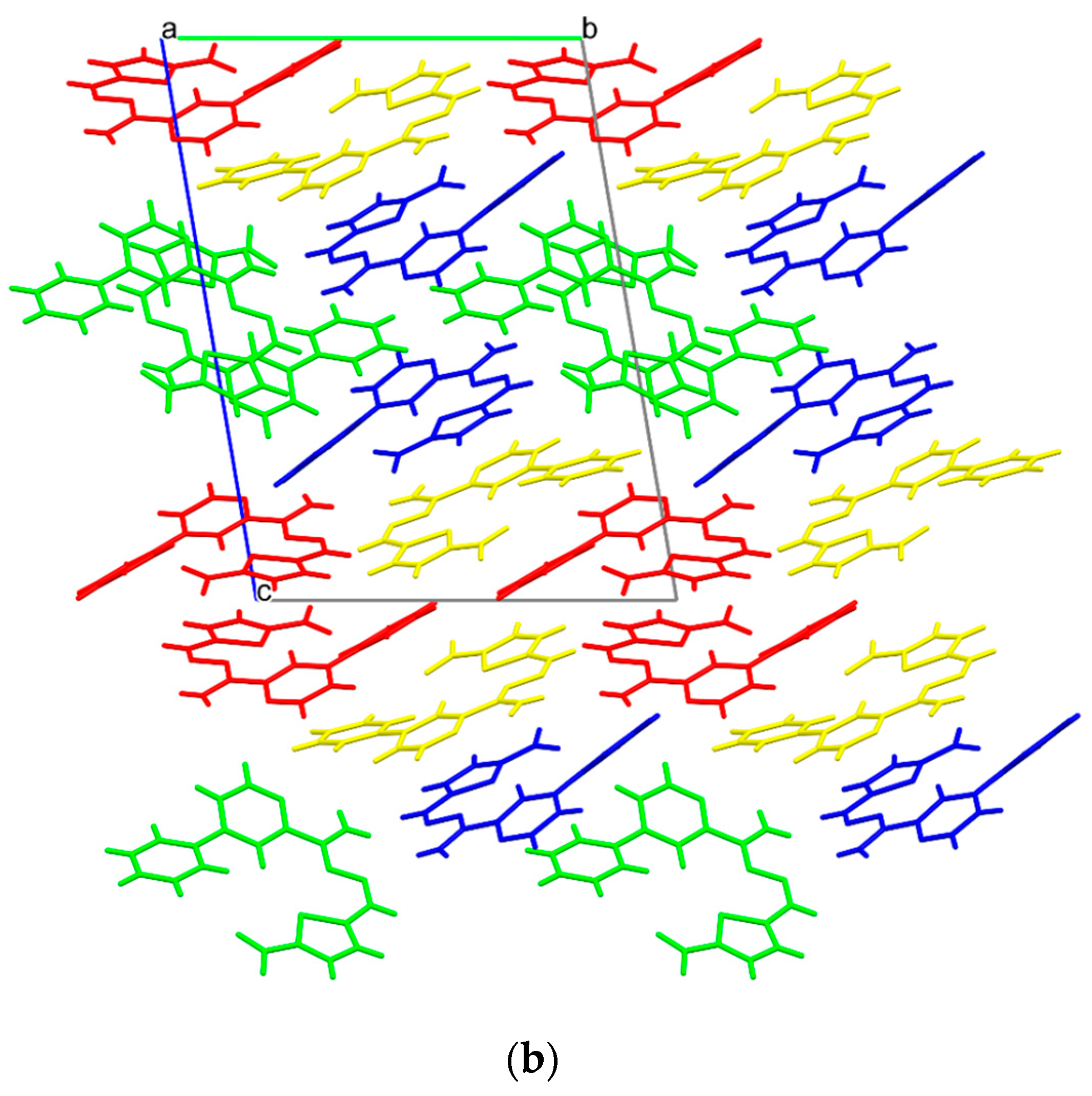
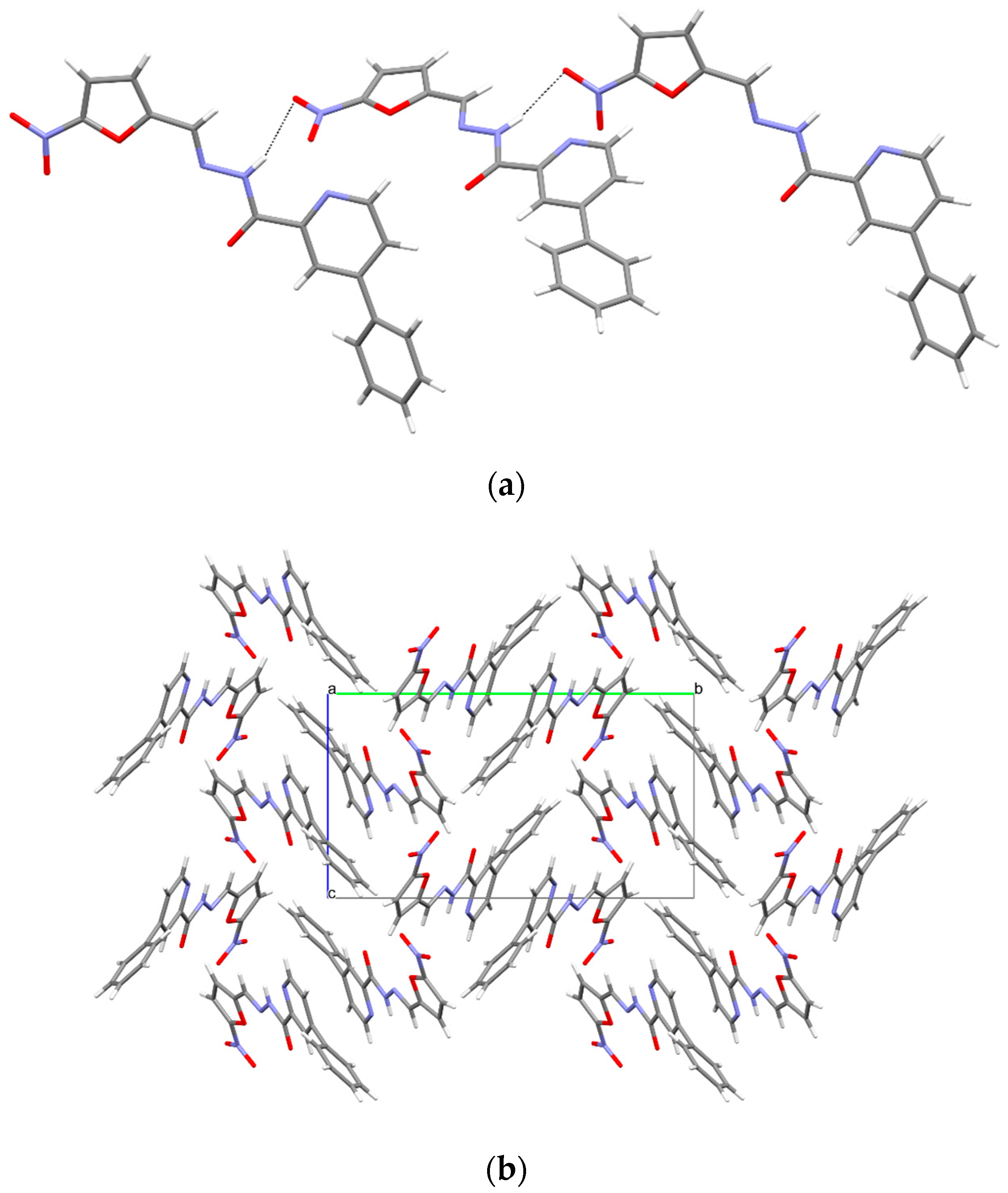
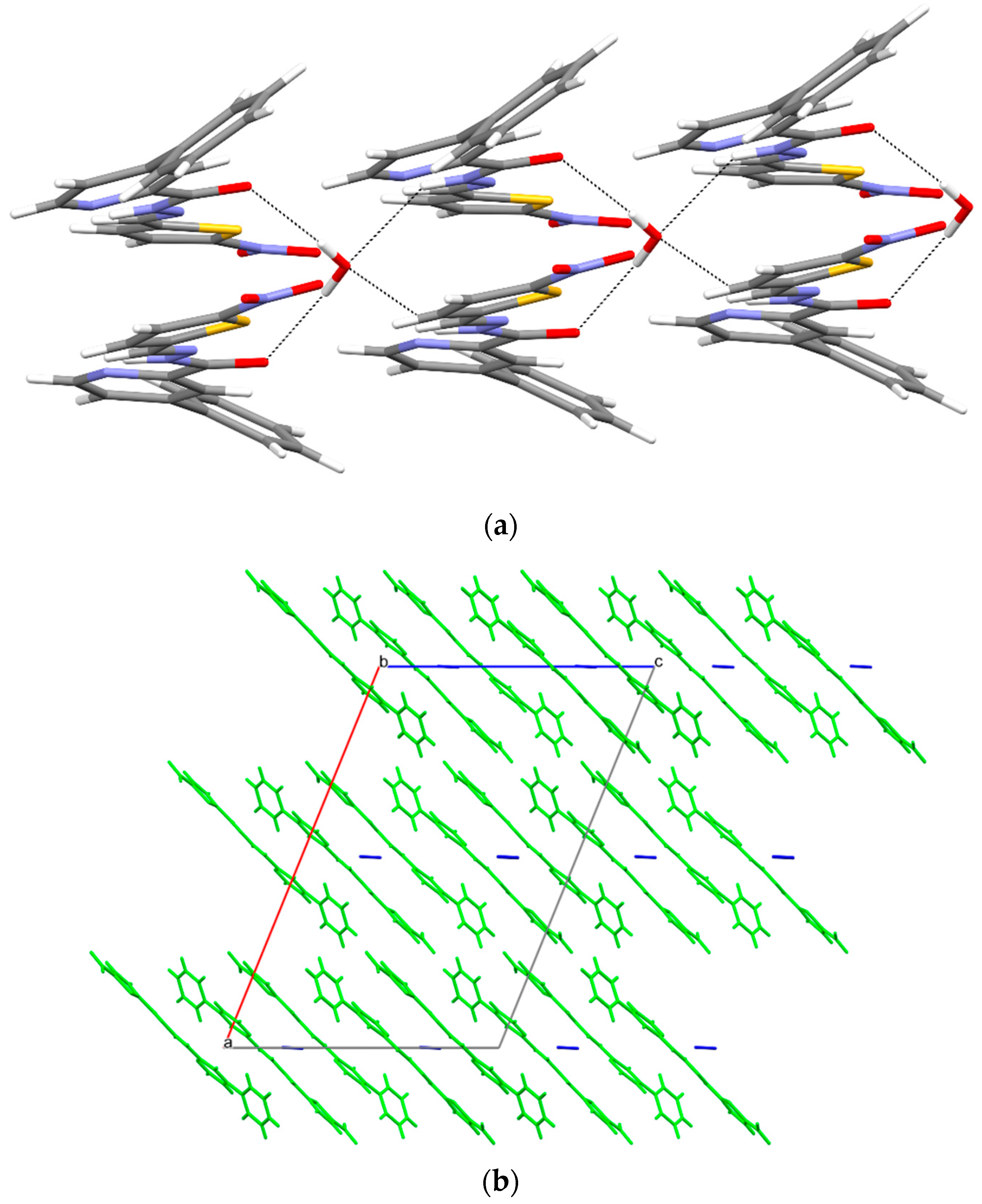
3.5. X-ray Study
3.6. Ab-Initio
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Islam, M.; Hameed, H.A.; Mugweru, J.; Chhotaray, C.; Wang, C.; Tan, Y.; Liu, J.; Li, X.; Tan, S.; Ojima, I.; et al. Drug resistance mechanisms and novel drug targets for tuberculosis therapy. J. Genet. Genom. 2017, 44, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Mai, T.Q.; Van Anh, N.T.; Hien, N.T.; Lan, N.H.; Giang, D.C.; Hang, P.T.T.; Marais, B.J.; Sintchenko, V. Drug resistance and Mycobacterium tuberculosis strain diversity in TB/HIV co-infected patients in Ho Chi Minh city, Vietnam. J. Glob. Antimicrob. Resist. 2017, 10, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Migliori, G.B.; Richardson, M.D.A.; Sotgiu, G.; Lange, C. Multidrug-Resistant and Extensively Drug-Resistant Tuberculosis in the West. Europe and United States: Epidemiology, Surveillance, and Control. Clin. Chest Med. 2009, 30, 637–665. [Google Scholar] [CrossRef] [PubMed]
- Lemos, A.C.M.; Matos, E.D. Multidrug-resistant tuberculosis. Braz. J. Infect. Dis. 2013, 17, 239–246. [Google Scholar] [CrossRef] [Green Version]
- Caminero, J.A.; Cayla, J.A.; García-García, J.-M.; García-Pérez, F.J.; Palacios, J.J.; Ruiz-Manzano, J. Diagnosis and Treatment of Drug-Resistant Tuberculosis. Arch. Bronconeumol. 2017, 53, 501–509. [Google Scholar] [CrossRef]
- Katzung, B.G.; Masters, S.B.; Trevor, A.J. Basic & Clinical Pharmacology, 6th ed.; McGraw-Hill Medical: New York, NY, USA, 2012. [Google Scholar]
- Grzemska, M. Tuberculosis drug therapy in adults. In Tuberculosis: A Comprehensive Clinical Reference; Schaaf, H., Zumla, A., Eds.; Saunders Elsevier: London, UK, 2009; pp. 638–648. [Google Scholar]
- Fatima, R.; Ashraf, M.; Ejaz, S.; Rasheed, M.A.; Altaf, I.; Afzal, M.; Batool, Z.; Saleem, U.; Anwar, K. In vitro toxic action potential of anti tuberculosis drugs and their combinations. Environ. Toxicol. Pharmacol. 2013, 36, 501–513. [Google Scholar] [CrossRef]
- Padmapriyadarsini, C.; Bhavani, P.; Tang, A.; Kumar, H.; Ponnuraja, C.; Narendran, G.; Hannah, E.; Ramesh, C.; Chandrasekar, C.; Wanke, C.; et al. Early changes in hepatic function among HIV–tuberculosis patients treated with nevirapine or efavirenz along with rifampin-based anti-tuberculosis therapy. Int. J. Infect. Dis. 2013, 17, e1154–e1159. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Global Tuberculosis Report 2021; World Health Organization: Geneva, Switzerland, 2021; Available online: https://www.who.int/publications/i/item/9789240037021 (accessed on 14 October 2021).
- Schaumburg, F.; Alabi, A.S.; Peters, G.; Becker, K. New epidemiology of Staphylococcus aureus infection in Africa. Clin. Microbiol. Infect. 2014, 20, 589–596. [Google Scholar] [CrossRef] [Green Version]
- Fernández, J.; Bert, F.; Nicolas-Chanoine, M.-H. The challenges of multi-drug-resistance in hepatology. J. Hepatol. 2016, 65, 1043–1054. [Google Scholar] [CrossRef] [Green Version]
- Kilbas, I.; Ciftci, I.H. Antimicrobial resistance of Enterococcus isolates in Turkey: A meta-analysis of current studies. J. Glob. Antimicrob. Resist. 2018, 12, 26–30. [Google Scholar] [CrossRef]
- Argudín, M.A.; Mendoza, M.C.; Martin, M.C.; Rodicio, M.R. Molecular basis of antimicrobial drug resistance in Staphylococcus aureus isolates recovered from young healthy carriers in Spain. Microb. Pathog. 2014, 74, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Gostev, V.; Kruglov, A.; Kalinogorskaya, O.; Dmitrenko, O.; Khokhlova, O.; Yamamoto, T.; Lobzin, Y.; Ryabchenko, I.; Sidorenko, S. Molecular epidemiology and antibiotic resistance of methicillin-resistant Staphylococcus aureus circulating in the Russian Federation. Infect. Genet. Evol. 2017, 53, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Vural, H.; Kara, M. Spectroscopic, optical, DNA, antimicrobial and density functional theory studies of 5-Bromo-2-(trifluoromethyl)pyridine. Optik 2017, 145, 479–488. [Google Scholar] [CrossRef]
- Jose, S.P.; Mohan, S. Vibrational spectra and normal co-ordinate analysis of 2-aminopyridine and 2-amino picoline. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2006, 64, 240–245. [Google Scholar] [CrossRef]
- Patel, N.B. Synthesis of 2-pyrazolines from pyridine based chalcone by conventional and microwave techniques: Their comparison and antimicrobial studies. J. Saudi Chem. Soc. 2016, 20, S451–S456. [Google Scholar] [CrossRef] [Green Version]
- Bhardwaj, V.; Noolvi, M.N.; Jalhan, S.; Patel, H.M. Synthesis, and antimicrobial evaluation of new pyridine imidazo [2,1b]-1,3,4-thiadiazole derivatives. J. Saudi Chem. Soc. 2016, 20, S406–S410. [Google Scholar] [CrossRef] [Green Version]
- Jose, G.; Kumara, T.H.S.; Sowmya, H.B.; Sriram, D.; Row, T.N.G.; Hosamani, A.A.; More, S.S.; Janardhan, B.; Harish, B.; Telkar, S.; et al. Synthesis, molecular docking, antimycobacterial and antimicrobial evaluation of new pyrrolo[3,2-c]pyridine Mannich bases. Eur. J. Med. Chem. 2017, 131, 275–288. [Google Scholar] [CrossRef]
- Chen, Q.; Zhu, X.-L.; Jiang, L.-L.; Liu, Z.-M.; Yang, G.-F. Synthesis, antifungal activity and CoMFA analysis of novel 1,2,4-triazolo[1,5-a]pyrimidine derivatives. Eur. J. Med. Chem. 2008, 43, 595–603. [Google Scholar] [CrossRef]
- Foks, H.; Orlewska, C.; Janowiec, M. Badania nad pochodnymi pirazyny XXVIII. Synteza i działanie tuberkulostatyczne 6-alkoksypirazyno-2-karbotioamidów. Acta. Pol. Pharm. 1992, 49, 47–50. [Google Scholar]
- Foks, H.; Mieczkowska, J.; Janowiec, M.; Zwolska, Z.; Andrzejczyk, Z. Synthesis and tuberculostatic activity of methyl 3-isonicotinoyldithiocarbazate and S’,S’-dimethyl dithiocarbonate isonictinoylhydrazone and their reactions with amines and hydrazines. Khim Geterosikl. Soed. 2002, 7, 918–925. [Google Scholar]
- Orlewska, C.; Foks, H.; Sowiński, P.; Martnowski, D.; Olczak, A.; Główka, M.L. Synthesis, structure and tuberculostatic activity of N’-(amino-pyridal-methylene)-hydrazinecarbodithioic acid methyl esters. Pol. J. Chem. 2001, 75, 1237–1245. [Google Scholar]
- Orlewska, C.; Foks, H.; Janowiec, M.; Zwolska-Kwiek, Z. Studies on pyrazine derivatives. XXIX. Synthesis of N1-thioamidosubstituted pyrazincarboxyamidrazones with expected tuberculostatic activity. Pharmazie 1995, 50, 565–566. [Google Scholar] [PubMed]
- Gobis, K.; Foks, H.; Serocki, M.; Augustynowicz-Kopeć, E.; Napiórkowska, A. Synthesis and evaluation of in vitro antimycobacterial activity of novel 1H-benzo[d]imidazole derivatives and analogues. Eur. J. Med. Chem. 2015, 89, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Gobis, K.; Foks, H.; Suchan, K.; Augustynowicz-Kopeć, E.; Napiórkowska, A.; Bojanowski, K. Novel 2-(2-phenalkyl)-1H-benzo[d]imidazoles as antitubercular agents. Synthesis, biological evaluation and structure–activity relationship. Bioorg. Med. Chem. 2015, 23, 2112–2120. [Google Scholar] [CrossRef]
- Gobis, K.; Foks, H.; Bojanowski, K.; Augustynowicz-Kopeć, E.; Napiórkowska, A. Synthesis of novel 3-cyclohexylpropanoic acid-derived nitrogen heterocyclic compounds and their evaluation for tuberculostatic activity. Bioorg. Med. Chem. 2012, 20, 137–144. [Google Scholar] [CrossRef]
- Olczak, A.; Szczesio, M.; Gołka, J.; Orlewska, C.; Gobis, K.; Foks, H.; Główka, M.L. Planarity of heteroaryldithiocarbazic acid derivatives showing tuberculostatic activity. II. Crystal structures of 3-[amino(pyrazin-2-yl)methylidene]-2-methylcarbazic acid esters. Cryst. Struct. Commun. 2010, 67, o37–o42. [Google Scholar] [CrossRef]
- Olczak, A.; Główka, M.L.; Gołka, J.; Szczesio, M.; Bojarska, J.; Kozłowska, K.; Foks, H.; Orlewska, C. Is planarity of pyridin-2-yl- and pyrazin-2-yl-formamide thiosemicarbazones related to their tuberculostatic activity? X-ray structures of two pyrazine-2-carboxamide-N′-carbonothioyl-hydrazones. J. Mol. Struct. 2007, 830, 171–175. [Google Scholar] [CrossRef]
- Vardanyan, R.; Hruby, V. Antimicrobial Drugs. Synthesis of Essential Drugs; Elsevier: Amsterdam, The Netherlands, 2006; pp. 499–523. [Google Scholar] [CrossRef]
- Kamal, A.; Shetti, R.V.; Azeeza, S.; Ahmed, S.K.; Swapna, P.; Reddy, A.M.; Khan, I.A.; Sharma, S.; Abdullah, S.T. Anti-tubercular agents. Part 5: Synthesis and biological evaluation of benzothiadiazine 1,1-dioxide based congeners. Eur. J. Med. Chem. 2010, 45, 4545–4553. [Google Scholar] [CrossRef]
- Silverman, R.B.; Holladay, M.W. The Organic Chemistry of Drug Design and Drug Action, 3rd ed.; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar] [CrossRef]
- Sharma, P.; Sharma, D.; Sharma, A.; Saini, N.; Goyal, R.; Ola, M.; Chawla, R.; Thakur, V. Hydrazone comprising compounds as promising anti-infective agents: Chemistry and structure-property relationship. Mater. Today Chem. 2020, 18, 100349. [Google Scholar] [CrossRef]
- Isoniazid. Tuberculosis 2008, 88, 112–116. [CrossRef]
- Brunner, H.; Störiko, R.; Rominger, F. Novel Chiral Oxazoline Ligands for Potential Charge-Transfer Effects in the Rh(I)-Catalysed Enantioselective Hydrosilylation. Eur. J. Inorg. Chem. 1998, 6, 771–781. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krause, M.; Foks, H.; Augustynowicz-Kopeć, E.; Napiórkowska, A.; Szczesio, M.; Gobis, K. Synthesis and Tuberculostatic Activity Evaluation of Novel Benzazoles with Alkyl, Cycloalkyl or Pyridine Moiety. Molecules 2018, 23, 985. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Cryst. 2015, C71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A 2008, A64, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Hübschle, C.B.; Sheldrick, G.M.; Dittrich, B. ShelXle: A Qt graphical user interface for SHELXL. J. Appl. Cryst. 2011, 44, 1281–1284. [Google Scholar] [CrossRef] [Green Version]
- Gobis, K.; Szczesio, M.; Olczak, A.; Pawlak, T.; Augustynowicz-Kopeć, E.; Krause, M.; Główka, M.L. Relationship between the Crystal Structure and Tuberculostatic Activity of Some 2-Amidinothiosemicarbazone Derivatives of Pyridine. Materials 2022, 15, 349. [Google Scholar] [CrossRef]
- Gordon, M.S.; Schmidt, M.W. Chapter 41. Advances In electronic structure theory: GAMESS a decade later. In Theory and Applications of Computational Chemistry, the First Forty Years; Dykstra, C.E., Frenking, G., Kim, K.S., Scuseria, G.E., Eds.; Elsevier: Amsterdam, The Netherlands, 2005; pp. 1167–1189. [Google Scholar]
- Becke, A.D. Density-Functional Thermochemistry. III. The Role of Exact Exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef] [Green Version]
- Vosko, S.H.; Wilk, L.; Nusair, M. Accurate spin-dependent electron liquid correlation energies for local spin density calculations: A critical analysis. Can. J. Phys. 1980, 58, 1200–1211. [Google Scholar] [CrossRef] [Green Version]
- Stephens, P.J.; Devlin, F.J.; Chabalowski, C.F.; Frisch, M.J. Ab Initio Calculation of Vibrational Absorption and Circular Dichroism Spectra Using Density Functional Force Fields. J. Phys. Chem. 1994, 98, 11623–11627. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple Selection Criteria for Drug-like Chemical Matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- O’Donnell, F.; Smyth, T.; Ramachandran, V.; Smyth, W. A study of the antimicrobial activity of selected synthetic and naturally occurring quinolines. Int. J. Antimicrob. Agents 2010, 35, 30–38. [Google Scholar] [CrossRef] [Green Version]
- Perveen, S.; Kumari, D.; Singh, K.; Sharma, R. Tuberculosis drug discovery: Progression and future interventions in the wake of emerging resistance. Eur. J. Med. Chem. 2022, 229, 114066. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Cryst. 2016, B72, 171–179. [Google Scholar] [CrossRef]


| Compound | 3a | 3b | 4a | 4b | CIP 2 | VAN 2 | FCZ 2 |
|---|---|---|---|---|---|---|---|
| Microorganism | MIC 1 [µg/mL] | ||||||
| Gram-positive bacteria | |||||||
| S. aureus ATCC 25923 | 1000 | 7.8 | 1000 | 1000 | 0.49 | 0.98 | - |
| S. epidermidis ATCC 12228 | 0.49 | 7.8 | 3.9 | 0.98 | 0.49 | 0.98 | - |
| M. luteus ATCC 10240 | 500 | 250 | 1000 | 62.5 | 0.98 | 0.12 | - |
| B. subtilis ATCC 6633 | 0.49 | 31.3 | 125 | 31.3 | 0.03 | 0.24 | - |
| B. cereus ATCC 10876 | 250 | 125 | 125 | 1000 | 0.12 | 0.98 | - |
| Gram-negative bacteria | |||||||
| E. coli ATCC 25922 | 1000 | 1000 | >1000 | >1000 | 0.004 | - | - |
| K. pneumoniae ATCC 13883 | 1000 | >1000 | >1000 | >1000 | 0.12 | - | - |
| P. mirabilis ATCC 12453 | >1000 | 1000 | >1000 | >1000 | 0.03 | - | - |
| Yeast | |||||||
| C. parapsilosis ATCC 22019 | 250 | 250 | 250 | 250 | - | - | 1.95 |
| Compd. | MIC 1 [µg/mL] | ||
|---|---|---|---|
| H37Rv 2 | Spec. 192 | Spec. 210 | |
| 3a | 6.2 | 6.2 | 6.2 |
| 3b | 25 | 25 | 25 |
| 4a | 3.1 | 12.5 | 3.1 |
| 4b | 25 | 25 | 25 |
| INH 3 | 12.5 | 12.5 | 25 |
| PZA 3 | 25 | 25 | >400 |
| 3a | 3b | 4a | 4b | |
|---|---|---|---|---|
| Chemical formula | C17H13N5O3 | C17H13N5O2S | C17H12N4O4 | 2(C17H12N4O3S)·H2O |
| Mr | 335.32 | 351.38 | 336.31 | 722.75 |
| Crystal system, space group | Monoclinic, P21 | Triclinic, P-1 | Monoclinic, P21/c | Monoclinic, C2/c |
| a, b, c (Å) | 7.9576 (1), 6.8200 (1), 14.7329 (3) | 9.3981 (2), 16.2901 (4), 21.5857 (6) | 9.8950 (4), 16.6918 (7), 10.2708 (5) | 27.8383 (15), 6.6147 (4), 18.6149 (10) |
| α, β, γ (°) | 90, 101.1060 (4), 90 | 78.623 (1), 81.641 (1), 75.018 (1) | 90, 114.976 (1), 90 | 90, 112.197 (1), 90 |
| V (Å3) | 784.59 (2) | 3114.04 (13) | 1537.74 (12) | 3173.8 (3) |
| Z | 2 | 8 | 4 | 4 |
| Crystal size (mm) | 0.34 × 0.18 × 0.17 | 0.57 × 0.24 × 0.10 | 0.23 × 0.18 × 0.16 | 0.45 × 0.42 × 0.32 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 8763, 2625, 2622 | 60,640, 12,299, 11,734 | 16,684, 3038, 2922 | 16,666, 3125, 3114 |
| Rint | 0.016 | 0.025 | 0.021 | 0.025 |
| (sin θ/λ)max (Å−1) | 0.618 | 0.619 | 0.618 | 0.618 |
| R[F2 > 2σ(F2)], wR(F2), S | 0.023, 0.063, 1.06 | 0.052, 0.128, 1.10 | 0.032, 0.084, 1.04 | 0.031, 0.083, 1.07 |
| No. of parameters | 233 | 925 | 229 | 234 |
| No. of restraints | 1 | 0 | 0 | 0 |
| Δmax, Δmin (e Å−3) | 0.19, −0.12 | 2.56, −0.85 | 0.29, −0.24 | 0.42, −0.28 |
| Absolute structure | Flack (1983) | – | – | – |
| Absolute structure parameter | 0.14 (3) | – | – | – |
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N4—H4A⋯N2 i | 0.86 (3) | 2.57 (2) | 3.2842 (19) | 141 (2) |
| N4—H4B⋯N42 ii | 0.93 (3) | 2.24 (2) | 3.0384 (18) | 143 (2) |
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N4B—H4BA⋯N2A | 0.86 (3) | 2.25 (3) | 3.041 (3) | 153 (3) |
| N4D—H4DA⋯O33B | 0.87 (4) | 2.49 (4) | 3.125 (3) | 130 (3) |
| N4A—H4AB⋯N2B | 0.86 (3) | 2.33 (3) | 3.107 (3) | 150 (3) |
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N3—H3⋯O33 i | 0.862 (17) | 2.278 (17) | 3.1072 (13) | 161.6 (17) |
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1W—H1W⋯O4 i | 0.81 (2) | 2.14 (2) | 2.9372 (13) | 169 (2) |
| N3—H3⋯O1W | 0.88 | 2.39 | 3.1280 (16) | 142 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gobis, K.; Szczesio, M.; Olczak, A.; Korona-Głowniak, I.; Augustynowicz-Kopeć, E.; Mazernt-Politowicz, I.; Ziembicka, D.; Główka, M.L. Differences in the Structure and Antimicrobial Activity of Hydrazones Derived from Methyl 4-Phenylpicolinimidate. Materials 2022, 15, 3085. https://doi.org/10.3390/ma15093085
Gobis K, Szczesio M, Olczak A, Korona-Głowniak I, Augustynowicz-Kopeć E, Mazernt-Politowicz I, Ziembicka D, Główka ML. Differences in the Structure and Antimicrobial Activity of Hydrazones Derived from Methyl 4-Phenylpicolinimidate. Materials. 2022; 15(9):3085. https://doi.org/10.3390/ma15093085
Chicago/Turabian StyleGobis, Katarzyna, Małgorzata Szczesio, Andrzej Olczak, Izabela Korona-Głowniak, Ewa Augustynowicz-Kopeć, Ida Mazernt-Politowicz, Dagmara Ziembicka, and Marek L. Główka. 2022. "Differences in the Structure and Antimicrobial Activity of Hydrazones Derived from Methyl 4-Phenylpicolinimidate" Materials 15, no. 9: 3085. https://doi.org/10.3390/ma15093085
APA StyleGobis, K., Szczesio, M., Olczak, A., Korona-Głowniak, I., Augustynowicz-Kopeć, E., Mazernt-Politowicz, I., Ziembicka, D., & Główka, M. L. (2022). Differences in the Structure and Antimicrobial Activity of Hydrazones Derived from Methyl 4-Phenylpicolinimidate. Materials, 15(9), 3085. https://doi.org/10.3390/ma15093085









