Perna canaliculus as an Ecological Material in the Removal of o-Cresol Pollutants from Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
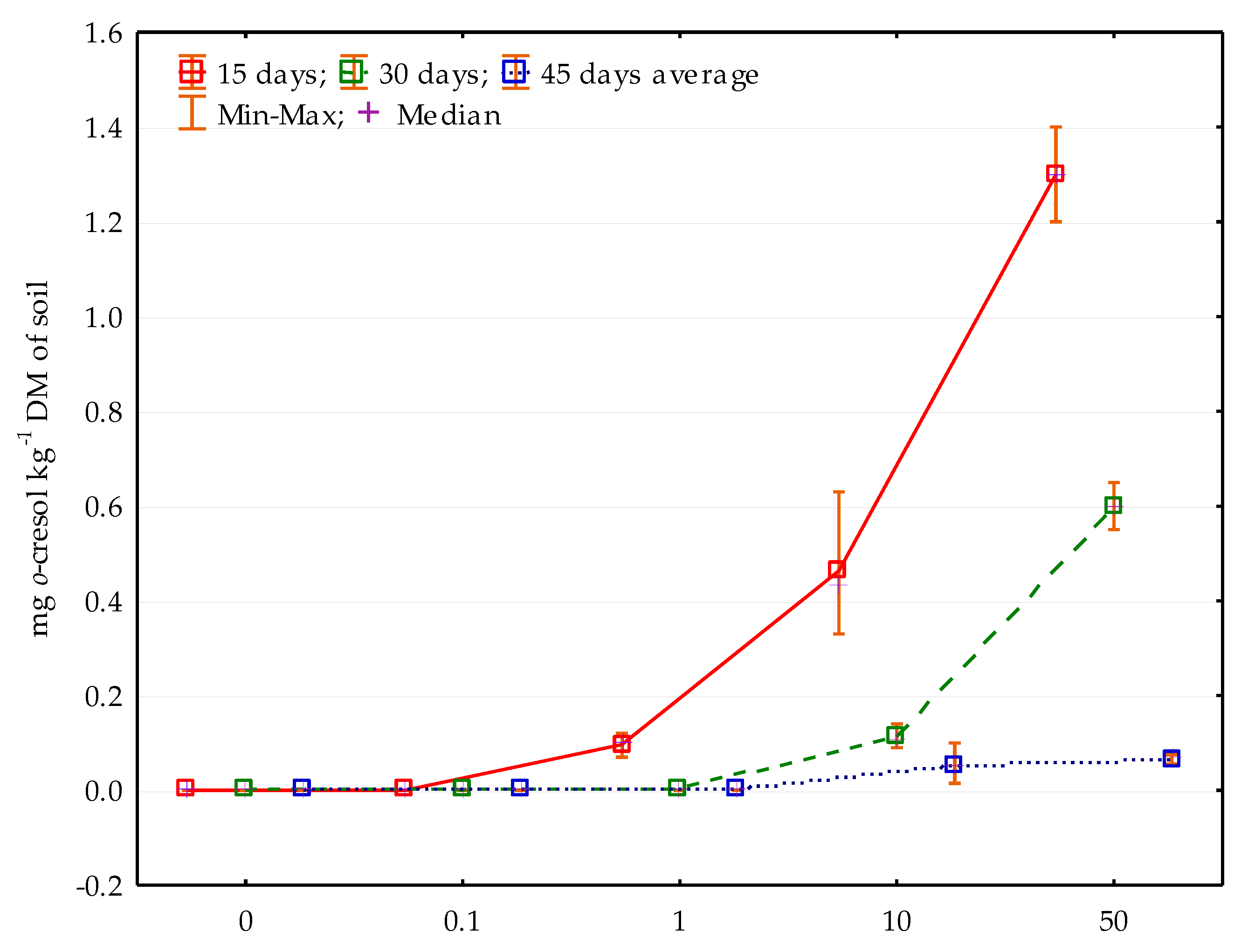
2.2. Characteristics and Determination of o-Cresol Residues in the Soil
2.3. Characteristics of Perna Canaliculus Mussel Meal
2.4. Determination of Soil Microorganisms
2.5. DNA Isolation and Bioinformatic Analysis of Bacterial Taxa
2.6. Determination of Soil Enzyme Activity
2.7. Statistical Data Analysis and Methodology of Calculations
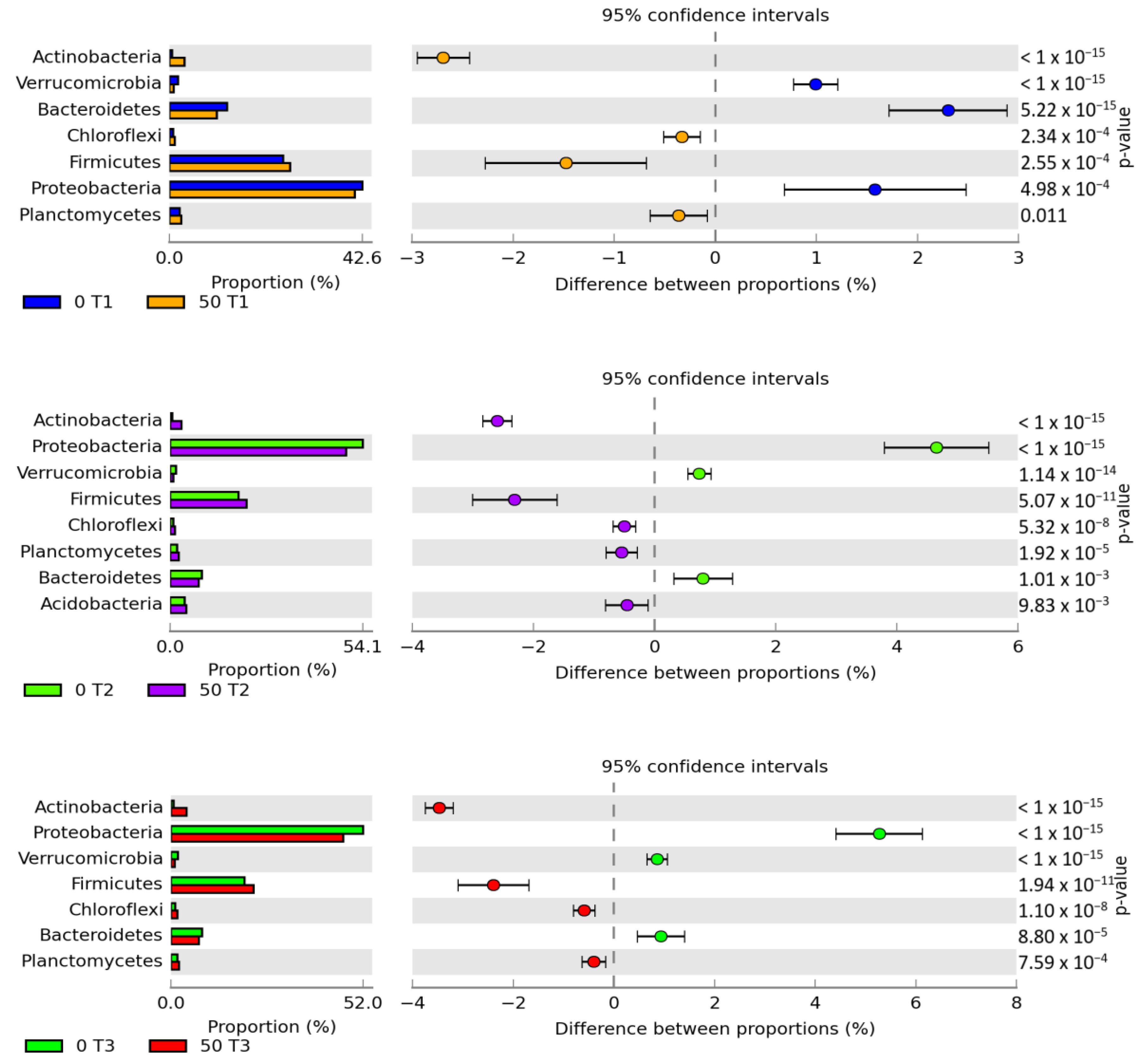
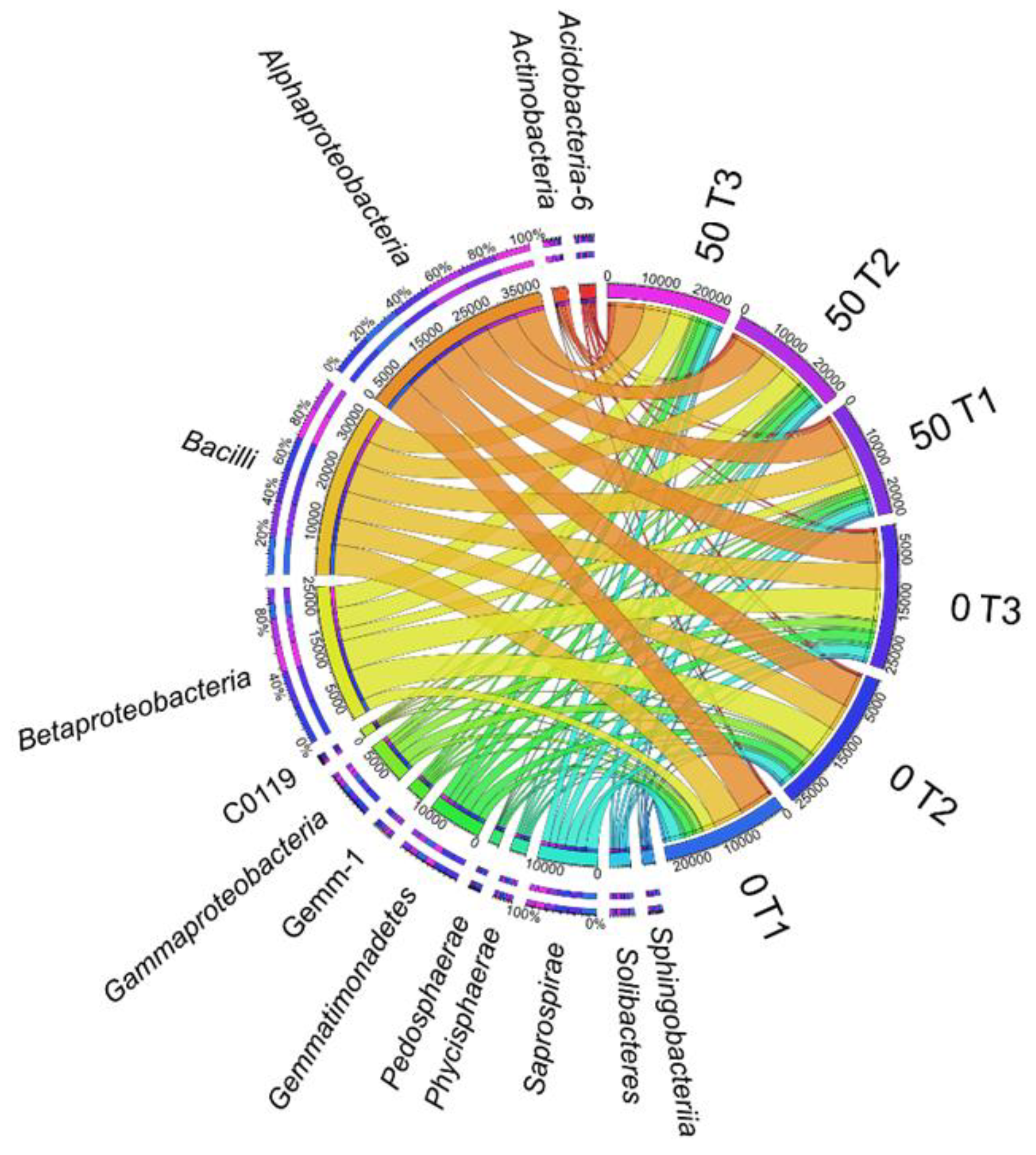
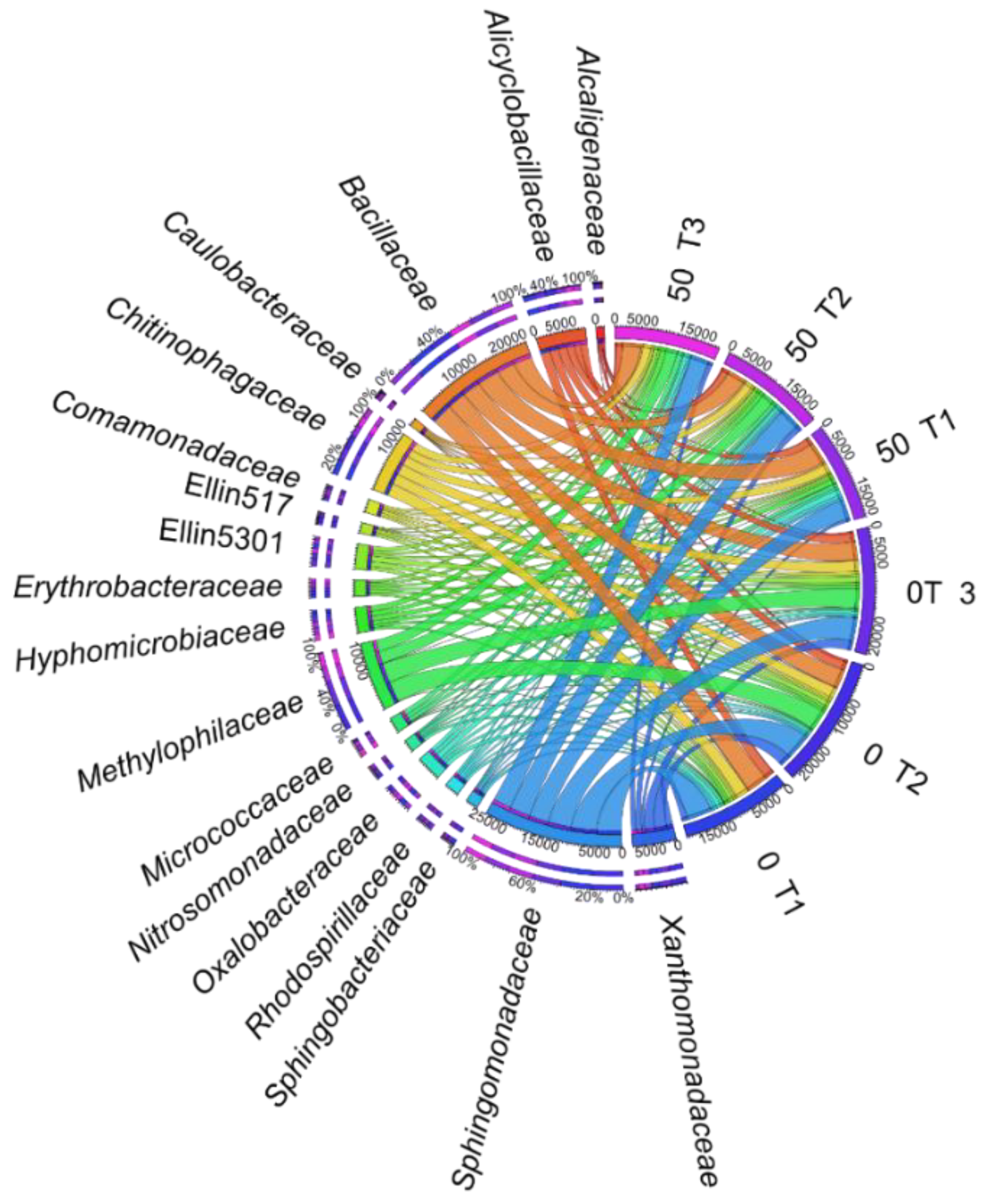
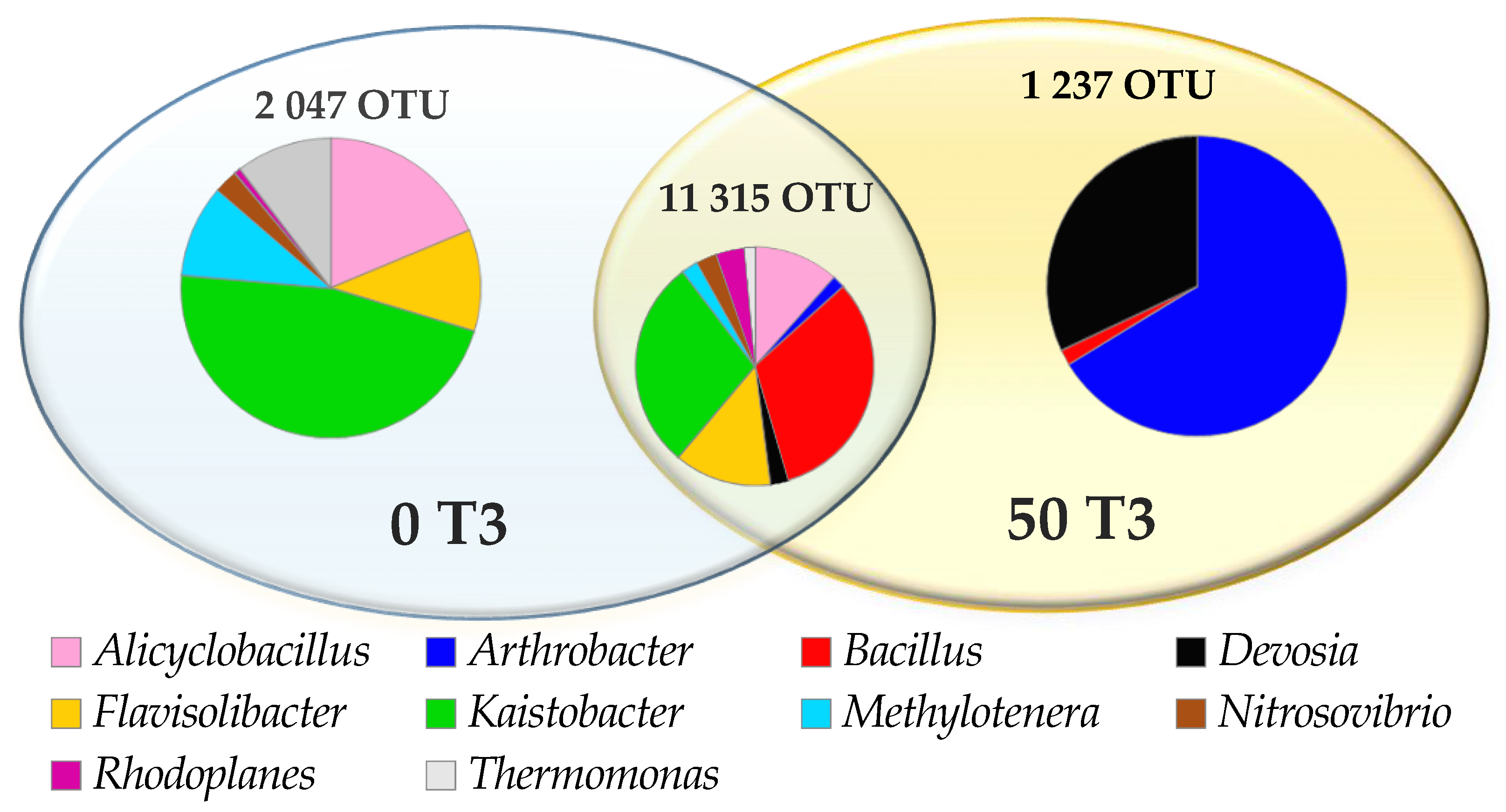
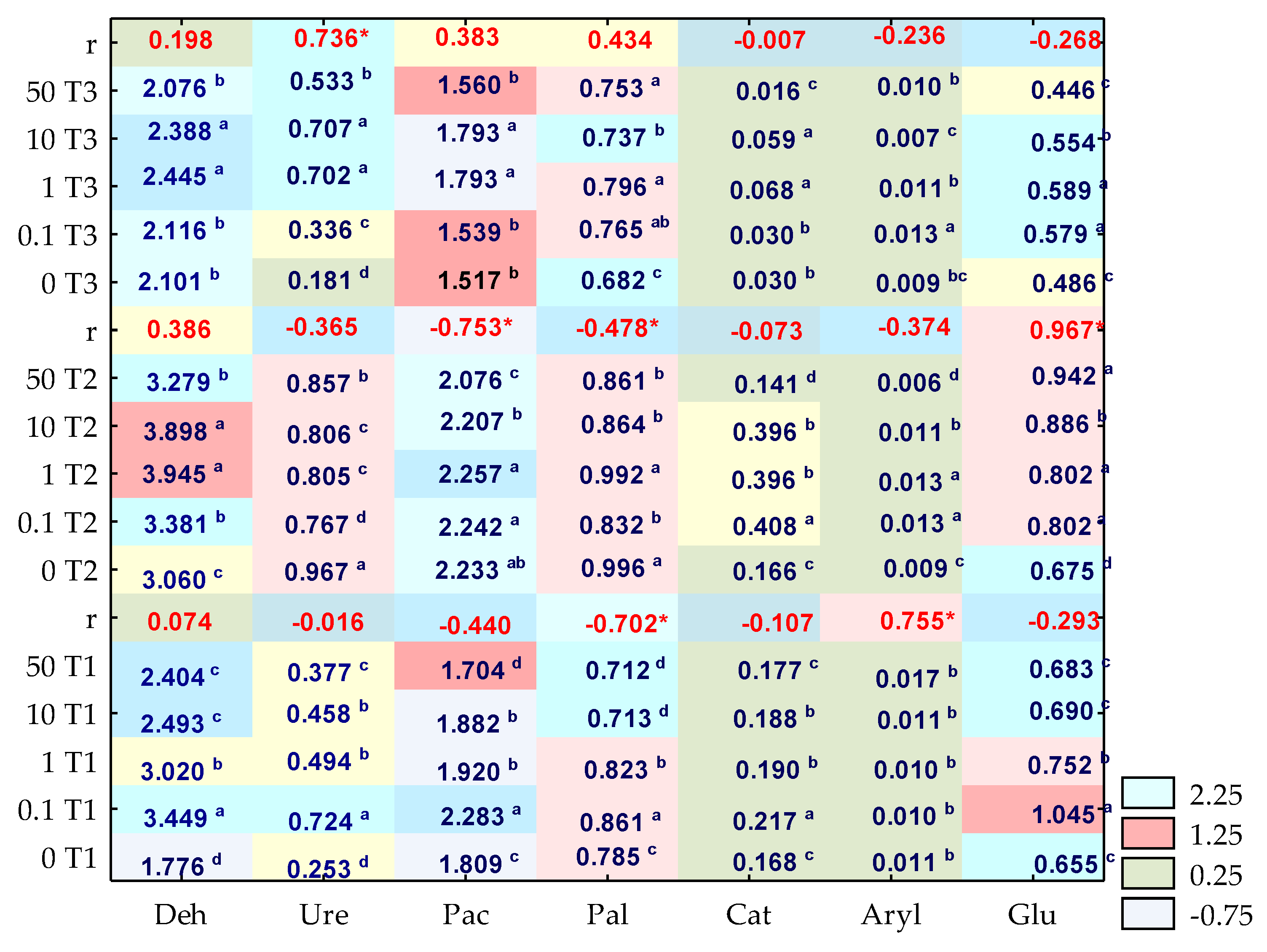
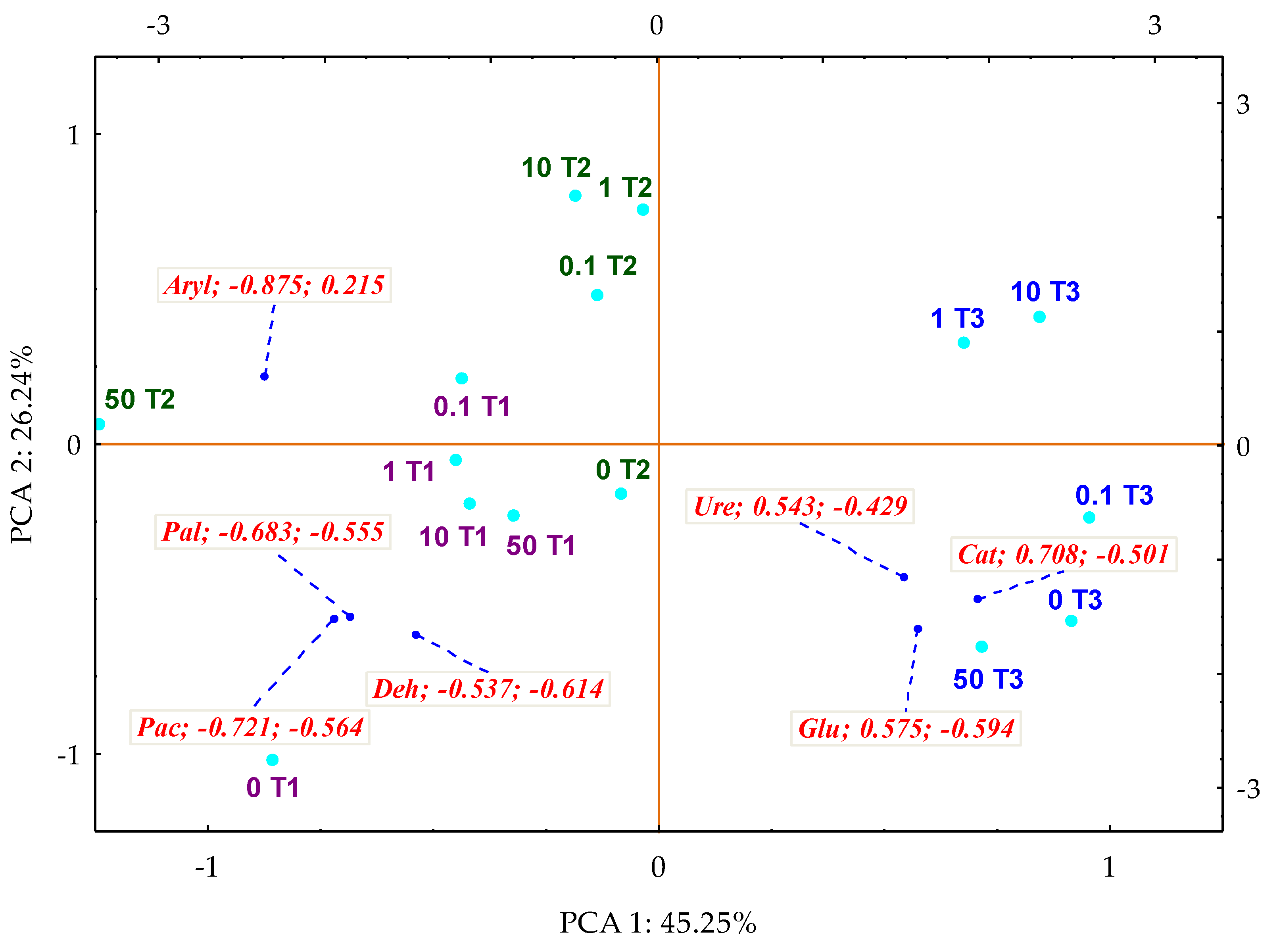
3. Results
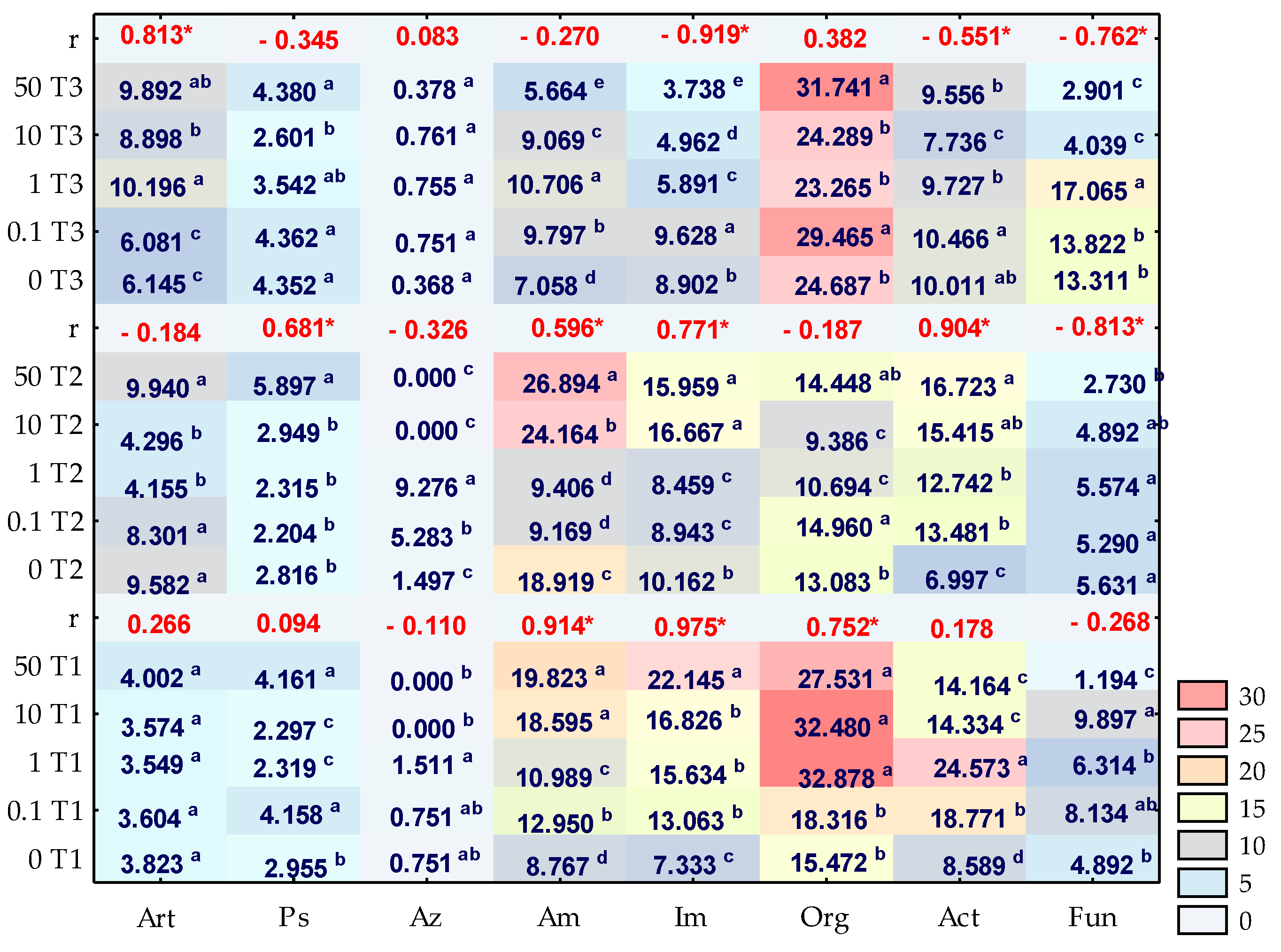
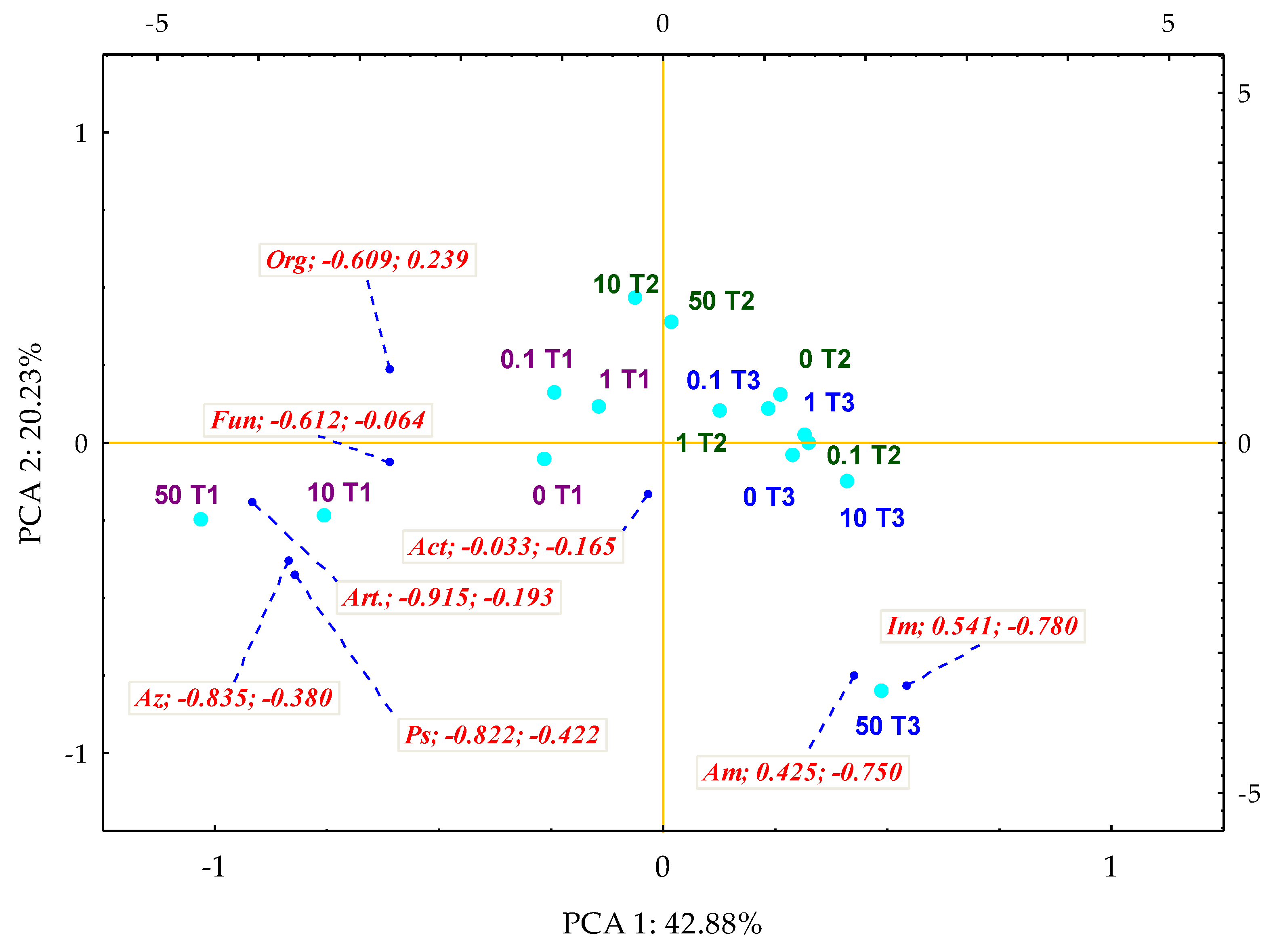
3.1. Counts and Diversity of Microorganisms
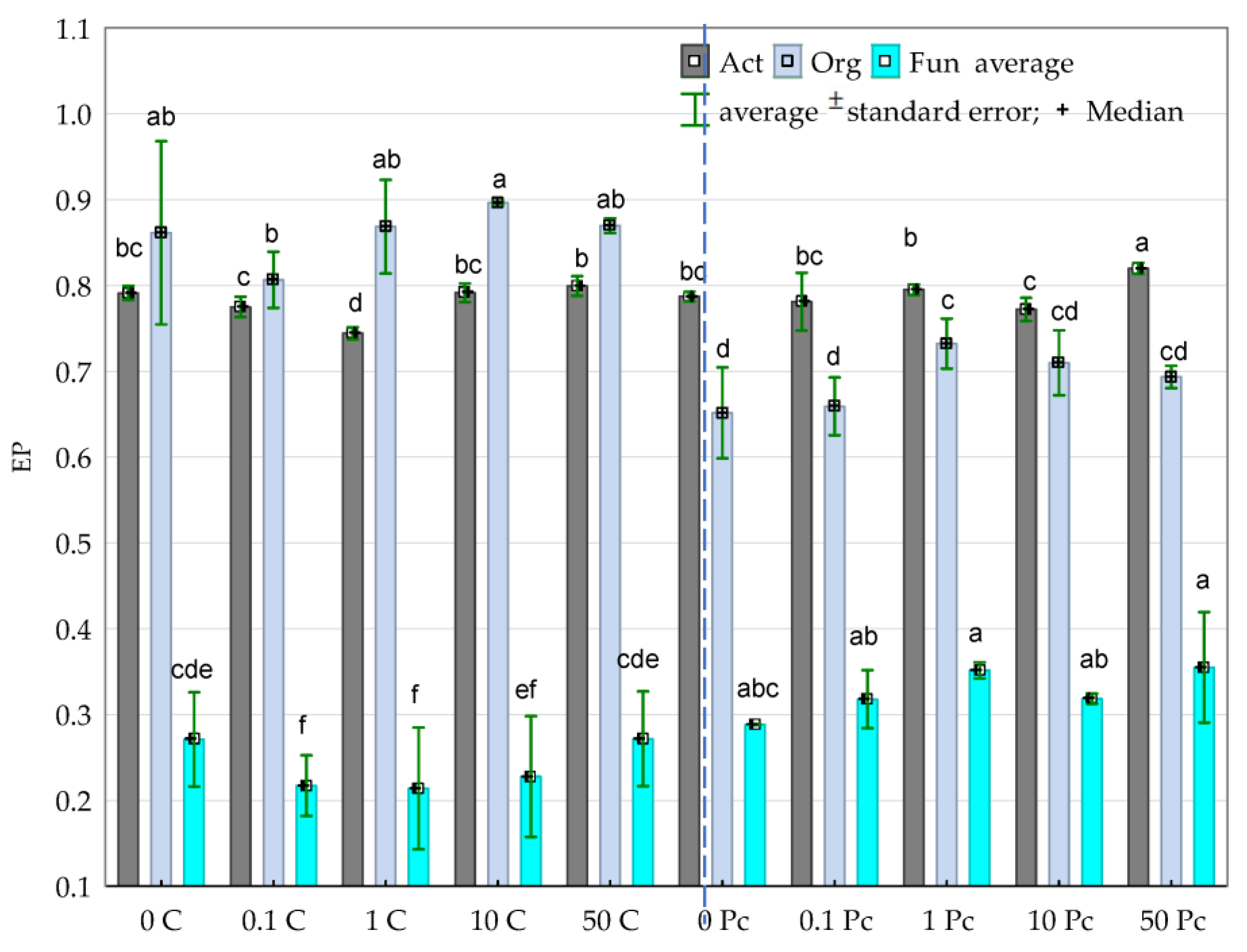
3.2. Enzyme Activity
4. Discussion
4.1. Counts and Diversity of Bacteria
4.2. Soil Enzymes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Villegas, L.G.C.; Mashhadi, N.; Chen, M.; Mukherjee, D.; Taylor, K.E.; Biswas, N. A short review of techniques for phenol removal from wastewater. Curr. Pollut. Rep. 2016, 2, 157–167. [Google Scholar] [CrossRef]
- Bennett, S.J.; Page, H.A. Implications of climate change for the petrochemical industry: Mitigation measures and feedstock transitions. In Handbook of Climate Change Mitigation and Adaptation; Springer International Publishing: Cham, Switzerland, 2017; pp. 383–426. [Google Scholar] [CrossRef]
- Zhu, X.; Wu, X.; Yao, J.; Wang, F.; Liu, W.; Luo, Y.; Jiang, X. Toxic effects of binary toxicants of cresol frother and Cu (II) on soil microorganisms. Int. Biodeterior. Biodegrad. 2018, 128, 155–163. [Google Scholar] [CrossRef]
- Dehmani, Y.; Abouarnadasse, S. Study of the adsorbent properties of nickel oxide for phenol depollution. Arab. J. Chem. 2020, 13, 5312–5325. [Google Scholar] [CrossRef]
- Zhang, N.; Li, Z.; Xiao, Y.; Pan, Z.; Jia, P.; Feng, G.; Bao, C.; Zhou, Y.; Hua, L. Lignin-based phenolic resin modified with whisker silicon and its application. J. Biores. Biopr. 2020, 5, 67–77. [Google Scholar] [CrossRef]
- Pradeep, N.V.; Anupama, S.; Navya, K.; Shalini, H.N.; Idris, M.; Hampannavar, U.S. Biological removal of phenol from wastewaters: A mini review. Appl. Water Sci. 2015, 5, 105–112. [Google Scholar] [CrossRef]
- Mahdavianpoura, M.; Moussavia, G.; Farrokhi, M. Biodegradation and COD removal of p-cresol in a denitrification baffled reactor: Performance evaluation and microbial community. Process Biochem. 2018, 69, 153–160. [Google Scholar] [CrossRef]
- Milia, S.; Cappai, G.; Perra, M.; Carucci, A. Biological treatment of nitrogen-rich refinery wastewater by partial nitritation (SHARON) process. Environ. Technol. 2012, 33, 1477–1483. [Google Scholar] [CrossRef]
- Wang, W.; Kannan, P.; Xue, J.; Kannan, K. Synthetic phenolic antioxidants, including butylated hydroxytoluene (BHT), in resin-based dental sealants. Environ. Res. 2016, 151, 339–343. [Google Scholar] [CrossRef]
- Chen, M.; Xu, P.; Zeng, G.; Yang, C.; Huang, D.; Zhang, J. Bioremediation of soils contaminated with polycyclic aromatic hydrocarbons, petroleum, pesticides, chlorophenols and heavy metals by composting: Applications, microbes and future research needs. Biotechnol. Adv. 2015, 33, 745–755. [Google Scholar] [CrossRef]
- Baldé, C.P.; Forti, V.; Gray, V.; Kuehr, R.; Stegmann, P. The Global E-Waste Monitor—2017; United Nations University (UNU), International Telecommunication Union (ITU) and International Solid Waste Association: Bonn, Germany; Geneva, Switzerland; Vienna, Austria, 2017; pp. 1–116. [Google Scholar]
- Michałowicz, J.; Duda, W. Phenols—Sources and toxicity. Pol. J. Environ. Stud. 2007, 16, 347–362. [Google Scholar]
- Li, Y.; Wang, Z.; Hu, Z.; Xu, B.; Li, Y.; Pu, W.; Zhao, J. A review of in situ upgrading technology for heavy crude oil. Petroleum 2021, 7, 117–122. [Google Scholar] [CrossRef]
- Schmidt, S.; Kirby, G.W. Dioxygenative cleavage of C-methylated hydroquinones and 2,6-dichlorohydroquinone by Pseudomonas sp. HH35. Biochim. Biophys. Acta. 2001, 1568, 83–89. [Google Scholar] [CrossRef]
- Riegert, U.; Burger, S.; Stolz, A. Altering catalytic properties of 3-chlorocatechol—Oxidizing extradiol dioxygenase from Sphingomonas xenophaga BN6 by random mutagenesis. J. Bacteriol. 2001, 183, 2322–2330. [Google Scholar] [CrossRef]
- Singh, S.; Malhotra, S.; Mukherjee, P.; Mishra, R.; Farooqi, F.; Sharma, R.S.; Mishra, V. Peroxidases from an invasive Mesquite species for management and restoration of fertility of phenolic-contaminated soil. J. Environ. Manag. 2020, 256, 109908. [Google Scholar] [CrossRef]
- Laplante, K.L.; Sarkisian, S.A.; Woodmansee, S.; Rowley, D.C.; Seeram, N.P. Effects of cranberry extracts on growth and biofilm production of Escherichia coli and Staphylococcus species. Phytother Res. 2012, 26, 1371–1374. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, J.; Pan, D.; Ge, X.; Jin, X.; Chen, S.; Wu, F. p-Coumaric can alter the composition of cucumber rhizosphere microbial communities and induce negative plant-microbial interactions. Biol. Fertil. Soils. 2018, 54, 363–372. [Google Scholar] [CrossRef]
- Noszczyńska, M.; Piotrowska-Seget, Z. Bisphenols: Application, occurrence, safety, and Biodegradation mediated by bacterial communities in wastewater treatment plants and rivers. Chemosphere 2018, 201, 214–223. [Google Scholar] [CrossRef]
- Setlhare, B.; Kumar, A.; Mokoena, M.P.; Pillay, B.; Olaniran, A.O. Phenol hydroxylase from Pseudomonas sp. KZNSA: Purification, characterization and prediction of three-dimensional structure. Int. J. Biol. Macromol. 2020, 146, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Gordeziani, M.S.; Varazi, T.G.; Pruidze, M.V. Structuralefunctional organization of cytochrome P450 containing monooxygenase and some aspects of modelling. Ann. Agrar. Sci. 2016, 14, 82–94. [Google Scholar] [CrossRef]
- Heinaru, E.; Truu, J.; Stottmeister, U.; Heinaru, A. Three types of phenol and p-cresol catabolism in phenol- and p-cresol-degrading bacteria isolated from river water continuously polluted with phenolic compounds. FEMS Microbiol. Ecol. 2000, 31, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Zaborowska, M. Krezole a drobnoustroje środowiska glebowego. Post. Mikrobiol. 2017, 56, 7–17. [Google Scholar]
- Heider, J.; Fuchs, G. Microbial anaerobic aromatic metabolism. Anaerobe 1997, 3, 1–22. [Google Scholar] [CrossRef]
- Johannes, J.; Bluschke, A.; Jehmlich, N.; Bergen, M.; Boll, M. Purification and characterization of active-site components of the putative p-cresol Methylhydroxylase Membrane Complex from Geobacter metallireducens. J. Bacteriol. 2008, 190, 6493–6500. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bielská, L.; Škulcová, L.; Neuwirthová, N.; Cornelissen, G.; Hale, S.E. Sorption, bioavailability and ecotoxic effects of hydrophobic organic compounds in biochar amended soils. Sci. Total Environ. 2018, 624, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Gao, H.; Song, X.; Deng, Y.; Zhang, Y. Adsorption of phenol on porous carbon from Toona sinensis leaves and its mechanism. Chem. Phys. Lett. 2020, 739, 137046. [Google Scholar] [CrossRef]
- Fu, Y.; Shen, Y.; Zhang, Z.; Ge, X.; Chen, M. Activated bio-chars derived from rice husk via one- and two-step KOH-catalyzed pyrolysis for phenol adsorption. Sci. Total Environ. 2019, 646, 1567–1577. [Google Scholar] [CrossRef]
- Dhanjai, A.; Sinha, A.; Wu, L.; Lu, X.; Chen, J.; Jain, R. Advances in sensing and biosensing of bisphenols: A review. Anal. Chim. Acta 2018, 998, 1–27. [Google Scholar] [CrossRef]
- Alkorta, I.; Epelde, L.; Garbisu, C. Environmental parameters altered by climate change affect the activity of soil microorganisms involved in bioremediation. FEMS Microbiol. Lett. 2017, 364, 1–9. [Google Scholar] [CrossRef]
- Singh, S.; Mishra, R.; Sharma, R.S.; Mishra, V. Phenol remediation by peroxidase from an invasive mesquite: Turning an environmental wound into wisdom. J. Hazard Mater. 2017, 334, 201–211. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef]
- FAO. Cultured aquatic species information programme. Perna canaliculus. Cultured aquatic species information programme. Text by kaspar, H. In FAO Fisheries and Aquaculture Department (Online); FAO: Rome, Italy, 2018; Updated 8 December. [Google Scholar]
- Caballero, G.; Garza, M.D.; Varela, M. The institutional foundations of economic performance of mussel production: The Spanish case of the Galician floating raft culture. Mar. Policy 2009, 33, 288–296. [Google Scholar] [CrossRef]
- Murphy, J.N.; Schneider, C.M.; Hawboldt, K.; Kerton, F.M. Hard to soft: Biogenic absorbent sponge-like material from waste mussel shells. Matter 2020, 3, 2029–2041. [Google Scholar] [CrossRef]
- Birnstiel, S.; Soares-Gomes, A.; da Gama, B.A.P. Depuration reduces microplastic content in wild and farmed mussels. Mar. Pollut. Bull. 2019, 140, 241–247. [Google Scholar] [CrossRef]
- Santos, F.S.; Neves, R.A.F.; Crapez, M.A.C.; Teixeira, V.L.; Krepsky, N. How does the brown mussel Perna respond to environmental pollution? A review on pollution biomarkers. J. Environ. Sci. 2022, 111, 412–428. [Google Scholar] [CrossRef]
- Lushchak, T.M.; Matviishyn, V.V.; Husak, J.M.; Storey, K.B. Storey. Pesticide toxicity: A mechanistic approach. EXCLI J. 2018, 17, 1101–1136. [Google Scholar] [PubMed]
- Conde-Cid, M.; Fernández-Calviño, D.; Núñez-Delgado, A.; Fernández-Sanjurjo, M.; Arias-Estévez, M.; Álvarez-Rodríguez, E. Influence of mussel shell, oak ash and pine bark on the adsorption and desorption of sulfonamides in agricultural soils. J. Environ. Manag. 2020, 261, 110221. [Google Scholar] [CrossRef]
- Romar-Gasalla, A.; Santas-Miguel, V.; Novoa-Munoz, J.C.; Arias- Estévez, J.C.; Alvarez- Rodríguez, E.; Núnez-Delgado, A.; Fernandez-Sanjurjo, M.J. Chromium and fluoride sorption/desorption on un-amended and waste-amended forest and vineyard soils and pyritic material. J. Environ. Manag. 2018, 222, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Kucharski, J.; Tomkiel, M.; Baćmaga, M.; Borowik, A.; Wyszkowska, J. Enzyme activity and microorganisms diversity in soil contaminated with the Boreal 58 WG herbicide. J. Environ. Sci. Health B. 2016, 51, 446–454. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Borowik, A.; Olszewski, J.; Kucharski, J. Soil bacterial community and soil enzyme activity depending on the cultivation of Triticum aestivum, Brassica napus, and Pisum sativum ssp. arvense. Diversity 2019, 11, 246. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Kucharski, M.; Kucharski, J. The role of Dactylis glomerata and diesel oil in the formation of microbiome and soil enzyme activity. Sensors 2020, 20, 3362. [Google Scholar] [CrossRef]
- IUSS Working Group WRB. World Reference Base for Soil Resources 2014, Update 2015. In International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; World Soil Resources Reports No. 106; FAO: Rome, Italy, 2015. [Google Scholar]
- Borowik, A.; Wyszkowska, J.; Wyszkowski, M. Resistance of aerobic microorganisms and soil enzyme response to soil contamination with Ekodiesel Ultra fuel. Environ. Sc. Pollut. Res. 2017, 24, 24346–24363. [Google Scholar] [CrossRef]
- ISO. 10390: Soil Quality-Determination of pH; International Organization for Standardization: Geneve, Switzerland, 2005. [Google Scholar]
- Klute, A. Methods of Soil Analysis; Agronomy Monograph 9; Soil Science Society of America, Inc.: Madison, WI, USA; American Society of Agronomy, Inc.: Madison, WI, USA, 1996. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total carbon, organic carbon, and organic matter. In Method of Soil Analysis: Chemical Methods; Sparks, D.L., Ed.; American Society of Agronomy: Madison, WI, USA, 1996; pp. 1201–1229. [Google Scholar]
- Egner, H.; Riehm, H.; Domingo, W.R. Untersuchun-gen über die chemische Bodenanalyse als Grundlage für die Beurteilung des Nährstoffzustandes der Böden. II. Chemische Extractionsmethoden zur Phospor- und Kaliumbestimmung. Ann. Royal Agricult. Coll. Swed. 1960, 26, 199–215. [Google Scholar]
- Schlichting, E.; Blume, H.P.; Stahr, K. Bodenkundliches Praktikum. Pareys Studientexte 81; Blackwell Wissenschafts-Verlag: Berlin, Germany, 1995. [Google Scholar]
- Mikami, D.; Kurihara, H.; Ono, M.; Kim, S.M.; Takahashi, K. Inhibition of algal bromophenols and their related phenols against glucose 6-phosphate dehydrogenase. Fitoterapia 2016, 108, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Renewable Fuels Association (RFA). The Impact of Accidental Ethanol Releases on the Environment. Renewable Fuels Association, Updated 29 May. Available online: https://ethanolrfa.org/producers/safety/ (accessed on 25 May 2021).
- Journal of Laws No. 1, Item 1395. Regulation of the Minister of the Environment of September 1, 2016 on the Way of Assessing the Pollution of the Earth’s Surface. 2016. Available online: https://isap.sejm.gov.pl/ (accessed on 25 May 2021).
- ATSDR, 2017. Substance Priority List. Atlanta, GA, USA: Agency for Toxic Substances and Disease Registry. Available online: http://www.atsdr.cdc.gov/spl/ (accessed on 25 May 2021).
- Aquaculture New Zealand. New Zealand Aquaculture: A Sector overview with key facts and statistics. In Proceedings of the New Zealand Aquaculture Conference, Blenheim, New Zealand, 18–19 September 2019; pp. 1–21. [Google Scholar]
- Guy, S.; Gaw, S.; Pearson, A.J.; Golovkoc, O.; Lechermann, M. Spatial variability in Polonium-210 and Lead-210 activity concentration in New Zealand shellfish and dose assessment. J. Environ. Radioactiv. 2020, 211, 106043. [Google Scholar] [CrossRef] [PubMed]
- Chandurvelan, R.; Marsden, I.D.; Gaw, S.; Glover, C.N. Waterborne cadmium impacts immunocytotoxic and cytogenotoxic endpoints in green-lipped mussel, Perna canaliculus. Aquat. Toxicol. 2013, 142–143, 283–293. [Google Scholar] [CrossRef]
- Zaborowska, M.; Kucharski, J.; Wyszkowska, J. Biochemical and microbiological activity of soil contaminated with o-cresol and biostimulated with Perna canaliculus mussel meal. Environ. Monit. Assess. 2018, 190, 602. [Google Scholar] [CrossRef] [PubMed]
- Núnez-Delgado, A.; Romar-Gasalla, A.; Santas-Miguel, V.; Fernandez-Sanjurjo, M.J.; Alvarez-Rodríguez, E.; Novoa-Munoz, J.C.; Arias-Estévez, M. Cadmium and Lead sorption/desorption on non-amended and by-product-amended soil samples and pyritic material. Water 2017, 9, 886. [Google Scholar] [CrossRef]
- Öhlinger, R. Dehydrogenase activity with the substrate TTC. In Methods in Soil Biology; Schinner, F., Ohlinger, R., Kandler, E., Margesin, R., Eds.; Springer: Berlin, Germany, 1996; pp. 241–243. [Google Scholar]
- Alef, K.; Nannipieri, P. Methods in Applied Soil Microbiology and Biochemistry, Enzyme Activities; Alef, K., Nannipieri, P., Eds.; Academic: London, UK, 1998; pp. 316–365. [Google Scholar]
- Dell Inc. Dell Statistica (Data Analysis Software System); Dell Inc.: Tulsa, OK, USA, 2016; Version 13.1. [Google Scholar]
- Krzywiński, M.I.; Schein, J.E.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. Gplots: Various R Programming Tools for Plotting Data. R Package Version 2.17.0. 2020. Available online: https://CRAN.R-project.org/package=gplots (accessed on 23 February 2020).
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2019; Available online: http://www.rstudio.com/ (accessed on 10 May 2021).
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 10 May 2021).
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Bünemann, E.K.; Bongiorno, G.; Bai, Z.; Creamer, R.E.; De Deyn, G.B.; de Goede, R.G.M.; Fleskens, L.; Geissen, V.; Kuyper, T.W.; Mäder, P.; et al. Soil quality—A critical review. Soil Biol. Biochem. 2018, 120, 105–125. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Boros-Lajszner, E.; Borowik, A.; Kucharski, J.; Baćmaga, M.; Tomkiel, M. Changes in the microbiological and biochemical properties of soil contaminated with zinc. J. Elementol. 2017, 22, 437–451. [Google Scholar] [CrossRef]
- Wu, H.; Wu, L.; Wang, J.; Zhu, Q.; Lin, S.; Xu, J.; Zheng, C.; Chen, J.; Qin, X.; Fang, C.; et al. Mixed phenolic acids mediated proliferation of pathogens Talaromyces helicus and Kosakonia sacchari in continuously monocultured Radix pseudostellariae rhizosphere soil. Front. Microbiol. 2016, 7, 335. [Google Scholar] [CrossRef] [PubMed]
- Siczek, A.; Frąc, M.; Gryta, A.; Kalembasa, S.; Kalembasa, D. Variation in soil microbial population and enzyme activities under faba bean as affected by pentachlorophenol. Appl. Soil Ecol. 2020, 150, 103466. [Google Scholar] [CrossRef]
- Zwetsloot, M.J.; Ucros, J.M.; Wickings, K.; Wilhelm, R.C.; Sparks, J.; Buckley, D.H.; Bauerle, T.L. Prevalent root-derived phenolics drive shifts in microbial community composition and prime decomposition in forest soil. Soil Biol. Biochem. 2020, 145, 107797. [Google Scholar] [CrossRef]
- Rongsayamanont, C.; Khongkhaem, P.; Luepromchai, E.; Khan, E. Inhibitory effect of phenol on wastewater ammonification. Bioresour. Technol. 2020, 309, 123312. [Google Scholar] [CrossRef]
- Wu, B.; He, C.; Yuan, S.; Hu, Z.; Wang, W. Hydrogen enrichment as a bioaugmentation tool to alleviate ammonia inhibition on anaerobic digestion of phenol-containing wastewater. Bioresour. Technol. 2019, 276, 97–102. [Google Scholar] [CrossRef]
- Wei, X.; Gilevska, T.; Wetzig, F.; Dorer, C.; Richnow, H.H.; Vogt, C. Characterization of phenol and cresol biodegradation by compound specific stable isotope analysis. Environ. Pollut. 2016, 210, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Du, D.; Wei, Z.; Xiaobo, Z.; Guojun, C. Phenol degradation and genotypic analysis of dioxygenase genes in bacteria isolated from sediments. Braz. J. Microbiol. 2017, 48, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Goswami, M.; Shivaraman, N.; Singh, R.P. Microbial metabolism of 2-chlorophenol, phenol and o-cresol by Rhodococcus erythropolis M1 in co-culture with Pseudomonas fluorescens P1. Microbiol. Res. 2005, 160, 101–109. [Google Scholar] [CrossRef]
- Zaborowska, M.; Wyszkowska, J.; Borowik, A.; Kucharski, J. Soil Microbiome Response to Contamination with Bisphenol A, Bisphenol F and Bisphenol S. Int. J. Mol. Sci. 2020, 21, 3529. [Google Scholar] [CrossRef]
- Panigrahy, N.; Barik, M.; Sahoob, R.K.; Sahoo, N.K. Metabolic profile analysis and kinetics of p-cresol biodegradation by an indigenous Pseudomonas citronellolis NS1 isolated from coke oven wastewater. Int. Biodeterior. Biodegrad. 2020, 147, 104837. [Google Scholar] [CrossRef]
- Atagana, H.I. Biodegradation of phenol, o-cresol, m-cresol and p-cresol by indigenous soil fungi in soil contaminated with creosote. World J. Microbiol. Biotechnol. 2004, 20, 851–858. [Google Scholar] [CrossRef]
- Keweloh, H.; Diefenbach, R.; Rehm, H.J. Increase of phenol tolerance of Escherichia coli by alterations of the fatty acid composition of the membrane lipids. Arch. Microbiol. 1991, 157, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Guzik, U.; Greń, I.; Wojcieszyńska, D.; Łabużek, S. Dioksygenazy—Główne enzymy degradacji związków aromatycznych. Biotechnologia 2008, 3, 71–88. [Google Scholar]
- Borowik, A.; Wyszkowska, J.; Kucharski, J. Impact of various grass species on soil bacteriobiome. Diversity 2020, 12, 212. [Google Scholar] [CrossRef]
- Li, Z.; Fu, J.; Zhou, R.; Wang, D. Effects of phenolic acids from ginseng rhizosphere on soil fungi structure, richness and diversity in consecutive monoculturing of ginseng. Saudi J. Biol. Sci. 2018, 25, 1788–1794. [Google Scholar] [CrossRef] [PubMed]
- Moridani, M.Y.; Siraki, A.; Chevaldina, T.; Scobie, H.; Obrie, N.P.J. Quantitative structure toxicity relationship for catechols in isolated rat hepatocytes. Chem. Biol. Interact. 2004, 147, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Grienke, U.; Silke, J.; Tasdemir, D. Bioactive compounds from marine mussels and their effects on human health. Food Chem. 2014, 142, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Srisunont, C.; Babel, S. Uptake, release, and absorption of nutrients into the marine environment by the green mussel (Perna viridis). Mar. Pollut. Bull. 2015, 97, 285–293. [Google Scholar] [CrossRef]
- Bakker, M.G.; Chaparro, J.M.; Manter, D.K.; Vivanco, J.M. Impacts of bulk soil microbial community structure on rhizosphere microbiomes of Zea mays. Plant Soil. 2015, 392, 115–126. [Google Scholar] [CrossRef]
- Salam, M.; Varma, A. Bacterial community structure in soils contaminated with electronic waste pollutants from Delhi NCR, India. Electron. J. Biotechn. 2019, 41, 72–80. [Google Scholar] [CrossRef]
- Patil, J.S.; Anil, A.C. Efficiency of copper and cupronickel substratum to resist development of diatom biofilms. Int. Biodeterior. Biodegrad. 2015, 105, 203–214. [Google Scholar] [CrossRef]
- Morrissey, E.M.; Mau, R.L.; Schwartz, E.; McHugh, T.A.; Dijkstra, P.; Koch, B.J.; Marks, J.C.; Hungate, B.A. Bacterial carbon use plasticity, phylogenetic diversity and the priming of soil organic matter. ISME J. 2017, 11, 1890–1899. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Chen, S.; Ren, Y.; Wei, C. Interaction and biodegradation evaluate of m-cresol and quinoline in co-exist system. Int. Biodeterior. Biodegrad. 2014, 86, 252–257. [Google Scholar] [CrossRef]
- Briganti, F.; Pessione, E.; Giunta, C.; Scozzafav, A. Purification, biochemical properties and substrate specificity of a catechol 1,2-dioxygenase from a phenol degrading Acinetobacter radioresistens. FEBS Lett. 1997, 416, 61–64. [Google Scholar] [CrossRef]
- Travkin, V.M.; Jadan, A.P.; Briganti, F.; Scozzafav, A.; Golovleva, A.L. Characterization of an intradiol dioxygenase involved in the biodégradation of the chlorophenoxy herbicides 2,4-D and 2,4,5-T. FEBS Lett. 1997, 407, 69–72. [Google Scholar] [CrossRef]
- Casciello, C.; Tonin, F.; Berini, F.; Fasoli, E.; Marinelli, F.; Pollegioni, L.; Rosini, E. A valuable peroxidase activity from the novel species Nonomuraea gerenzanensis growing on alkali lignin. Biotechnol. Rep. 2017, 13, 49–57. [Google Scholar] [CrossRef]
- Min, K.; Freeman, C.; Kang, H.; Choi, S.U. The regulation by phenolic compounds of soil organic matter dynamics under a changing environment. BioMed Res. Int. 2015, 2015, 825098. [Google Scholar] [CrossRef]
- Hoostal, M.J.; Bidart-Bouzat, M.G.; Bouzat, J.L. Local adaptation of microbial communities to heavy metal stress in polluted sediments of Lake Erie. FEMS Microbiol. Ecol. 2008, 65, 156–168. [Google Scholar] [CrossRef]
- Szaleniec, M.; Borowski, T.; Schühle, K.; Witko, M.; Heider, J. Ab inito modeling of ethylbenzene dehydrogenase reaction mechanism. J. Am. Chem. Soc. 2010, 132, 6014–6024. [Google Scholar] [CrossRef]
- Daudzai, Z.; Treesubsuntorn, C.; Thiravetyan, P. Inoculated Clitoria ternatea with Bacillus cereus ERBP for enhancing gaseous ethylbenzene phytoremediation: Plant metabolites and expression of ethylbenzene degradation genes. Ecotox. Environ. Saf. 2018, 164, 50–60. [Google Scholar] [CrossRef]
- Aguiar, B.; Vieira, J.; Cunha, A.E.; Vieira, C.P. No evidence for Fabaceae Gametophytic self-incompatibility being determined by Rosaceae, Solanaceae, and Plantaginaceae S-RNase lineage genes. BMC Plant Biol. 2015, 15, 129. [Google Scholar] [CrossRef]
- Perotti, E.B.R. Impact of hydroquinone used as a redox effector model on potential denitrification, microbial activity and redox condition of a cultivable soil. Rev. Argent. Microbiol. 2015, 47, 212–218. [Google Scholar] [CrossRef]
- Herter, S.; Schmidt, M.; Thompson, M.L.; Mikolasch, A.; Schauer, F. Study of enzymatic properties of phenol oxidase from nitrogen-fixing Azotobacter chroococcum. AMB Express 2011, 1, 14. [Google Scholar] [CrossRef]
- Mustafa, S.; Perveen, S.; Khan, A. Synthesis, enzyme inhibition and anticancer investigations of unsymmetrical 1,3-disubstituted urease. J. Serb. Chem. Soc. 2014, 79, 1–10. [Google Scholar] [CrossRef]
- Cantarella, H.; Otto, R.; Soares, J.R.; Silva, B.A.G. Agronomic efficiency of NBPT as a urease inhibitor: A review. J. Adv. Res. 2018, 13, 19–27. [Google Scholar] [CrossRef]
- Vassiliou, S.; Kosikowska, P.; Grabowiecka, A.; Yiotakis, A.; Kafarski, P.; Berlicki, L. Computer-aided optimization of phosphinic inhibitors of bacterial ureases. J. Med. Chem. 2010, 53, 5597–5606. [Google Scholar] [CrossRef] [PubMed]
- Zaborska, W.; Krajewska, B.; Kot, M.; Karcz, W. Quinone-induced inhibition of urease. Elucidation of its mechanisms by probing thiol groups of the enzyme. Bioorg. Chem. 2007, 35, 233–242. [Google Scholar] [CrossRef]
- Krajewska, B.; Zaborska, W. Double mode of inhibition-inducing interactions of 1,4-naphthoquinone with urease: Arylation versus oxidation of enzyme thiols. Bioorg. Med. Chem. 2007, 15, 4144–4151. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Chen, H.; He, H.; Ma, C. Assays for alkaline phosphatase activity: Progress and prospects. TrAC Trend Anal. Chem. 2019, 113, 32–43. [Google Scholar] [CrossRef]
- Margalef, O.; Sardans, J.; Fernández-Martínez, M.; Molowny-Horas, R.; Janssens, I.A.; Ciais, P.; Goll, D.; Richter, A.; Obersteiner, M.; Asensio, D.; et al. Global patterns of phosphatase activity in natural soils. Sci. Rep. 2017, 7, 1337–1350. [Google Scholar] [CrossRef] [PubMed]
- Bonting, C.F.C.; Schneider, S.; Schmidtberg, G.; Fuchs, G. Anaerobic degradation of m-cresol via methyl oxidation to 3-hydroxybenzoate by a denitrifying bacterium. Arch. Microbiol. 1995, 164, 63–69. [Google Scholar] [CrossRef]
- Franchi, O.; Rosenkranz, F.; Chamy, R. Key microbial populations involved in anaerobic degradation of phenol and p-cresol using different inocula. Electron. J. Biotechnol. 2018, 35, 33–38. [Google Scholar] [CrossRef]
- Liebeg, E.W.; Cutright, T.J. The investigation of enhanced bioremediation through the addition of macro and micro nutrients in a PAH contaminated soil. Int. Biodeter. Biodegr. 1999, 44, 55–64. [Google Scholar] [CrossRef]
- Shibata, A.; Yasushi, I.; Arata, K. Aerobic and anaerobic biodegradation of phenol derivatives in various paddy soils. Sci. Total Environ. 2006, 367, 979–987. [Google Scholar] [CrossRef] [PubMed]
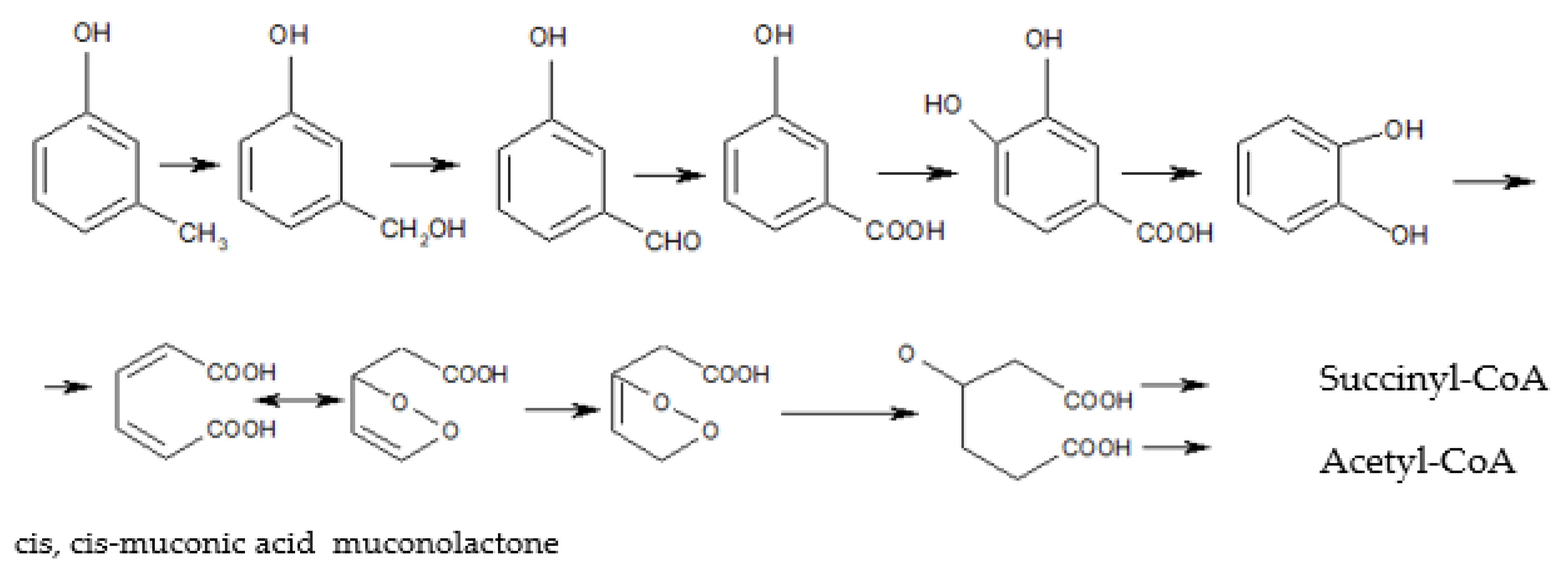



| Properties | Unit | Value | Reference |
|---|---|---|---|
| pHKCl | 7.0 | [46] | |
| HAC | mM (+) kg−1 DM of soil | 6.40 | [47] |
| EBC | 165.90 | [47] | |
| CEC | 172.30 | [47] | |
| BS | (%) | 96.29 | [47] |
| Corg | g kg−1 DM of soil | 6.40 | [48] |
| Ke | mg kg−1 DM of soil | 180.00 | [49] |
| Cae | 2571.40 | [49] | |
| Nae | 20.00 | [49] | |
| Mge | 59.50 | [50] |
| Chemical Formula | Chemical Structure | Synonyms | Identification Numbers | |||
|---|---|---|---|---|---|---|
| NIOSH | EPA | RTECS | HSDB | |||
| C7H8O | 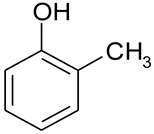 | 2-methylophenol 2-hydroxytoluene o-cresylic acid | G06300000 | F004 | U052 | 1813 |
| Molecular Weight | MP | BP | WS | PC | VP | BCF | ||
|---|---|---|---|---|---|---|---|---|
| 1 atm | 10 mm Hg | Log Octanol/Water | Log Koc | |||||
| 108.14 | 30.94 | 191 | 74.90 | 25.95 | 1.95 | 1.03 | 0.230 | 1.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaborowska, M.; Wyszkowska, J.; Borowik, A.; Kucharski, J. Perna canaliculus as an Ecological Material in the Removal of o-Cresol Pollutants from Soil. Materials 2021, 14, 6685. https://doi.org/10.3390/ma14216685
Zaborowska M, Wyszkowska J, Borowik A, Kucharski J. Perna canaliculus as an Ecological Material in the Removal of o-Cresol Pollutants from Soil. Materials. 2021; 14(21):6685. https://doi.org/10.3390/ma14216685
Chicago/Turabian StyleZaborowska, Magdalena, Jadwiga Wyszkowska, Agata Borowik, and Jan Kucharski. 2021. "Perna canaliculus as an Ecological Material in the Removal of o-Cresol Pollutants from Soil" Materials 14, no. 21: 6685. https://doi.org/10.3390/ma14216685
APA StyleZaborowska, M., Wyszkowska, J., Borowik, A., & Kucharski, J. (2021). Perna canaliculus as an Ecological Material in the Removal of o-Cresol Pollutants from Soil. Materials, 14(21), 6685. https://doi.org/10.3390/ma14216685









