A Methodology for Porcine Circovirus 2 (PCV-2) Quantification Based on Gold Nanoparticles
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Antibodies
2.3. Samples
2.4. DNA Extraction for qPCR Analysis
2.5. Instruments
2.6. Synthesis of Gold Nanoparticles and Formation of Self-Assembled Monolayers
2.7. Quantitative PCR (qPCR) for PCV
3. Results and Discussion
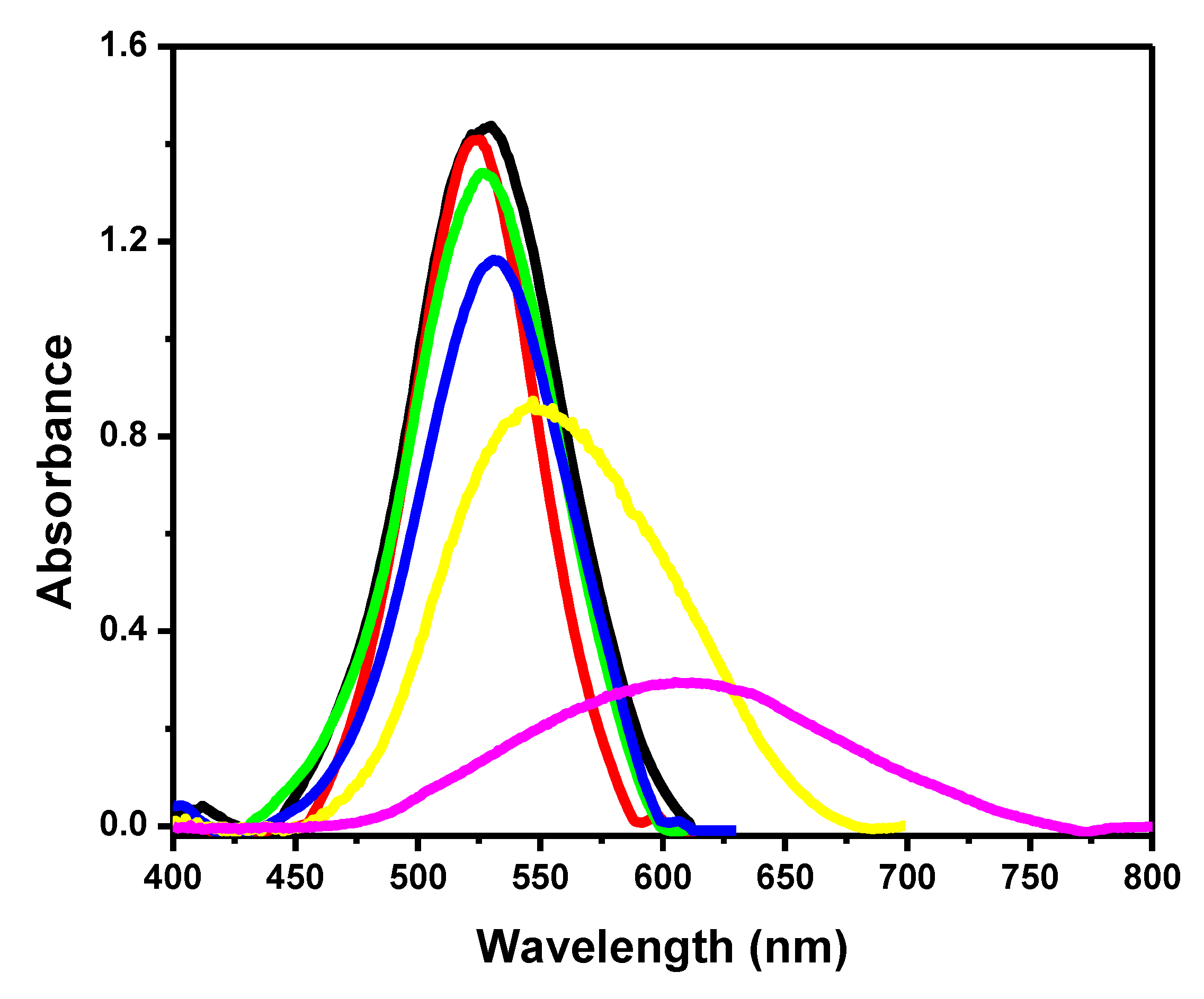
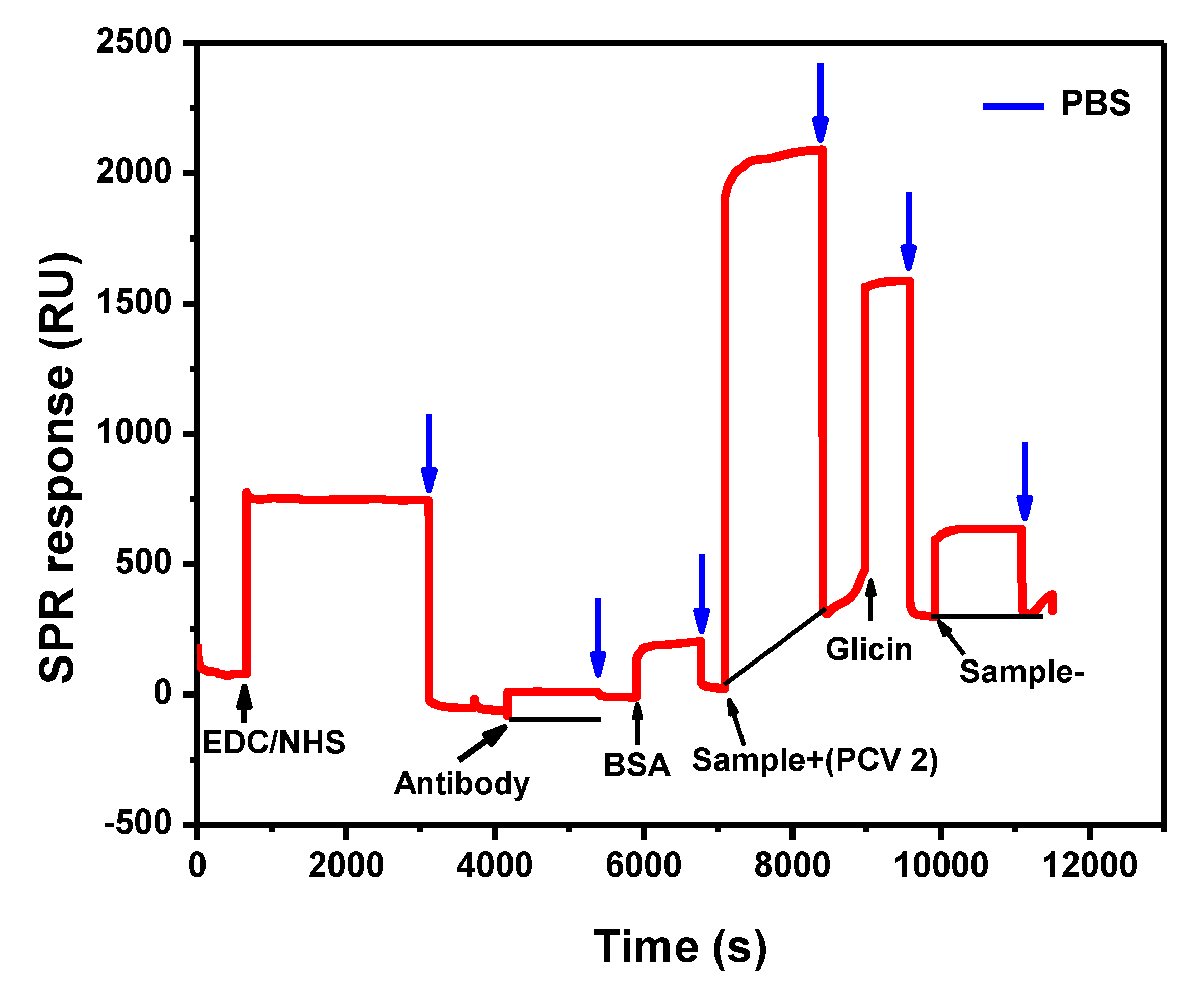
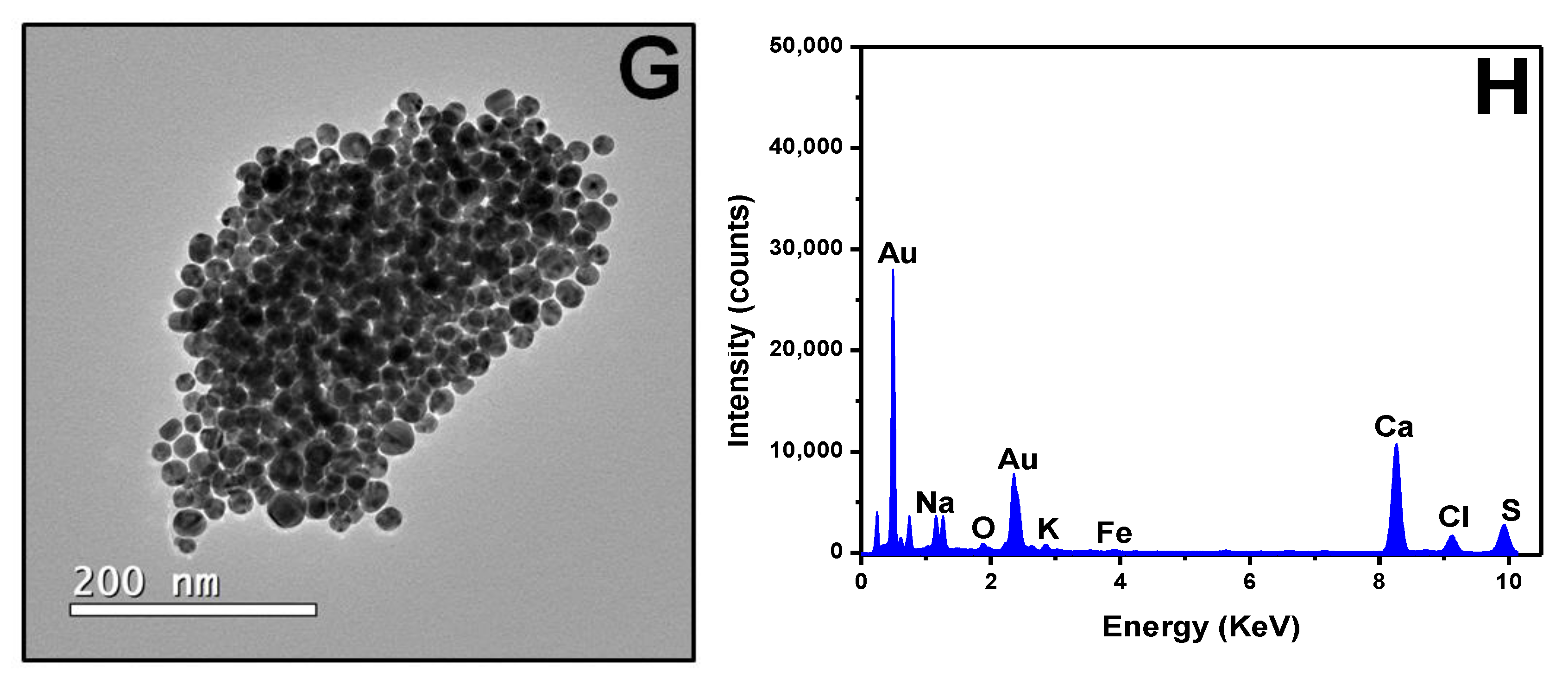
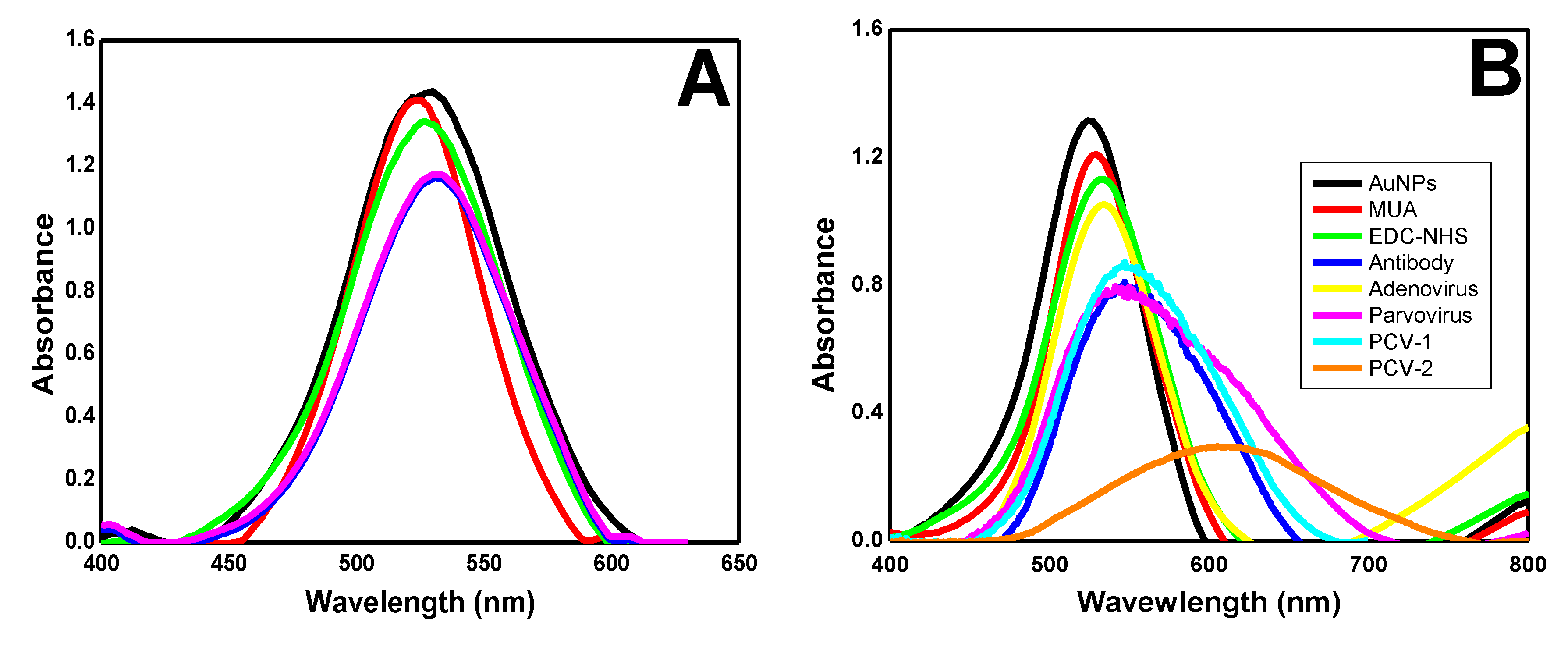
3.1. Biosensor Development
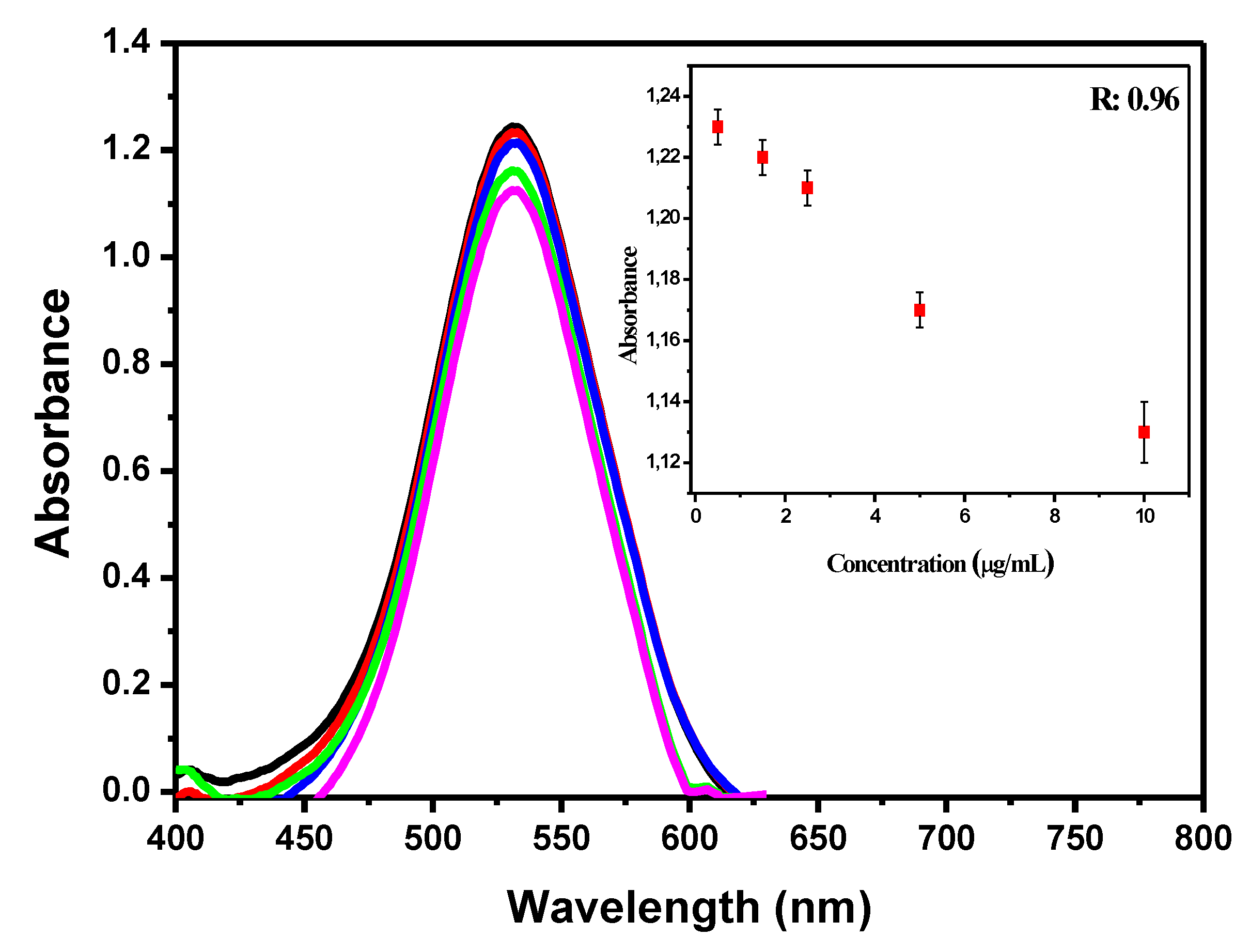
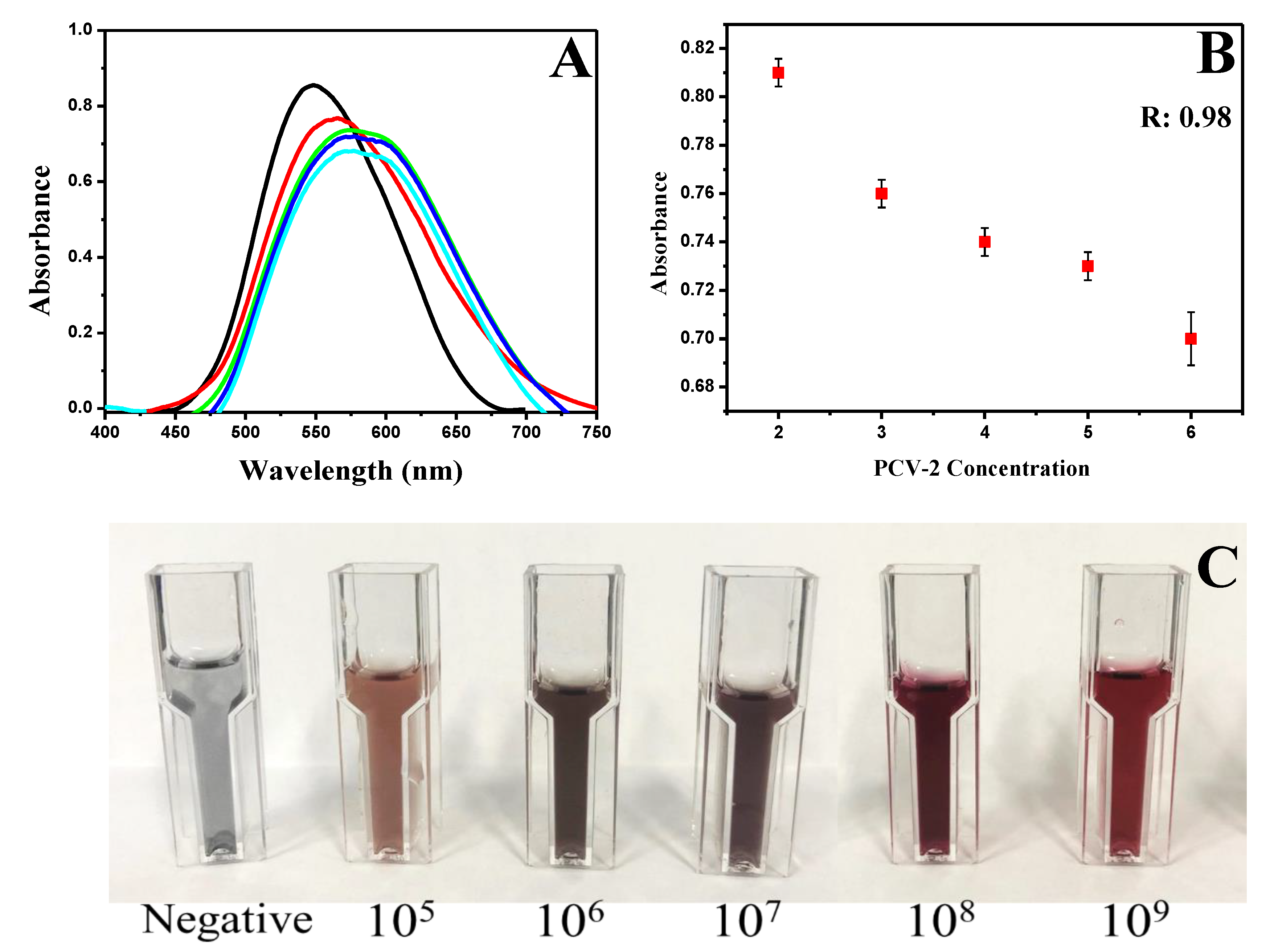
3.2. The Dynamic Range of Signal
3.3. Biosensor Characterization and Negative Control
3.4. Negative control
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Franzo, G.; Cortey, M.; Olvera, A.; Novosel, D.; Castro, A.M.M.G.; Biagini, P.; Segalés, J.; Drigo, M. Full-genome sequencing of porcine circovirus 3 field strains from Denmark, Italy and Spain demonstrates a high within-Europe genetic heterogeneity. Virol. J. 2015, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zanella, J.R.C. Situação atual da circovirose no Brasil. Embrapa Suíno e Aves. 2017, 18, 150–156. [Google Scholar]
- Gava, D.; Serrão, V.H.B.; Fernandes, L.T.; Cantão, M.E.; Ciacci-Zanella, J.R.; Morés, N.; Schaefer, R. Structure analysis of capsid protein of porcine circovirus type 2 from pigs with systemic disease. Braz. J. Microb. 2018, 49, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Zanella, J.R.C.; Morés, N.; De Barcellos, D.E.S.N. Principais ameaças sanitárias endêmicas da cadeia produtiva de suínos no Brasil. Pesquisa Agropecuária Brasileira 2016, 51, 443–453. [Google Scholar] [CrossRef]
- Opriessing, T.; Xiao, C.T.; Halbur, P.G.; Gerber, P.F.; Matzinger, S.R.; Meng, X.J. A commercial porcine circovirus (PCV) type 2a-based vaccine reduces PCV 2d viremia and shedding and prevents PCV2d transmission to naïve pigs under experimental conditions. Vaccine 2017, 35, 248–254. [Google Scholar] [CrossRef]
- Wei, R.; Xie, J.; Theuns, S.; Nauwynck, H.J. Changes on the viral capsid surface during the evolution of porcine circovirus type 2 (PCV2) from 2009 till 2018 may lead to a better receptor binding. Virus Evol. 2019, 5, vez026. [Google Scholar] [CrossRef]
- Correa-Fiz, F.; Franzo, G.; Llorens, A.; Segalés, J.; Kekarainen, T. Porcine circovirus 2 (PCV-2) genetic variability under natural infection scenario reveals a complex network of viral quasispecies. Sci. Rep. 2018, 8, 15469. [Google Scholar] [CrossRef]
- Cruz, T.F.; Magro, A.J.; De Castro, A.M.; Pedraza-Ordoñez, F.J.; Tsunemi, M.H.; Perahia, D.; Araujo, J.P. In vitro and in silico studies reveal capsid-mutant Porcine circovirus 2b with novel cytopathogenic and structural characteristics. Virus Res. 2018, 251, 22–33. [Google Scholar] [CrossRef]
- Mukherjee, P.; Milton, A.A.P.; Karam, A.; Chakraborty, A.K.; Das, S.; Puro, K.; Sanjukta, R.; Ghatak, S.; Shakuntala, I.; Laha, R.G.; et al. Porcine circovirus2 (PCV2): A review of disease profileand control. Indian J. Comp. Microbiol. Immunol. Infect. Dis. 2018, 39, 38–50. [Google Scholar] [CrossRef]
- Patterson, R.; Nevel, A.; Diaz, A.V.; Martineau, H.M.; Demmers, T.; Browne, C.; Mavrommatis, B.; Werling, D. Exposure to environmental stressors result in increased viral load and further reduction of production parameters in pigs experimentally infected with PCV2b. Vet. Microbiol. 2015, 177, 261–269. [Google Scholar] [CrossRef]
- Olvera, A.; Sibila, M.; Calsamiglia, M.; Segalés, J.; Domingo, M. Comparison of porcine circovirus type 2 load in serum quantified by a real time PCR in postweaning multisystemic wasting syndrome and porcine dermatitis and nephropathy syndrome naturally affected pigs. J. Virol. Methods 2004, 117, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Henriques, A.M.; Duarte, M.; Barros, S.C.; Fagulha, T.; Ramos, F.; Luís, T.; Fevereiro, M. Development and validation of a real-time PCR for the detection and quantification of porcine circovirus type 2. VirusDisease 2018, 29, 355–361. [Google Scholar] [CrossRef] [PubMed]
- López-Lorenzo, G.; Díaz-Cao, J.M.; Prieto, A.; López-Novo, C.; López, C.M.; Díaz, P.; Rodríguez-Vega, V.; Díez-Baños, P.; Fernández, G. Environmental distribution of Porcine Circovirus Type 2 (PCV2) in swine herds with natural infection. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cruz, T.F.; Kanashiro, T.M.; De Castro, A.M.; Baldin, C.M.; Richtzenhain, L.J.; Araujo, J.P., Jr. A double-antibody sandwich ELISA based on the porcine circovirus type 2 (PCV2) propagated in cell culture for antibody detection. Pesquisa Veterinária Brasileira 2016, 36, 1171–1177. [Google Scholar] [CrossRef]
- De Souza, C.D.; Nogueira, B.R.; Rostelato, M.E.C. Review of the methodologies used in the synthesis gold nanoparticles by chemical reduction. J. Alloy. Compd. 2019, 798, 714–740. [Google Scholar] [CrossRef]
- Ming, M.A.; Mi, S.; Yue, L.; Jim, L.Y.; Fang, L.; Hua, L.; Bin, Q.; Yu, L.Z.; Nan, C.G. Progress of visual viosensor based on gold nanoparticles. Chin. J. Anal. Chem. 2018, 46, 1–10. [Google Scholar]
- Wang, H.; Rao, H.; Luo, M.; Xue, X.; Xue, Z.; Lu, X. Noble metal nanoparticles growth-based colorimetric strategies: From monocolorimetric to multicolorimetric sensors. Co-ord. Chem. Rev. 2019, 398, 113003. [Google Scholar] [CrossRef]
- Basso, C.R.; Tozato, C.C.; Crulhas, B.P.; Castro, G.R.; Junior, J.P.A.; Pedrosa, V.A. An easy way to detect dengue virus using nanoparticle-antibody conjugates. Virology 2018, 513, 85–90. [Google Scholar] [CrossRef]
- Cordeiro, M.; Carlos, F.F.; Pedrosa, P.; Lopez, A.; Baptista, P.V. Gold Nanoparticles for Diagnostics: Advances towards Points of Care. Diagnostics 2016, 6, 43. [Google Scholar] [CrossRef]
- Vines, J.B.; Yoon, J.-H.; Ryu, N.-E.; Lim, N.-J.; Park, H. Gold Nanoparticles for Photothermal Cancer Therapy. Front. Chem. 2019, 7, 167. [Google Scholar] [CrossRef]
- Basso, C.R.; Tozato, C.D.C.; Ribeiro, M.C.M.; Júnior, J.P.A.; Pedrosa, V.A. A immunosensor for the diagnosis of canine distemper virus infection using SPR and EIS. Anal. Methods 2013, 5, 5089. [Google Scholar] [CrossRef]
- Ahmed, S.R.; Kim, J.; Tran, V.T.; Suzuki, T.; Neethirajan, S.; Lee, J.; Park, E.Y. In situ self-assembly of gold nanoparticles on hydrophilic and hydrophobic substrates for influenza virus-sensing platform. Sci. Rep. 2017, 7, 44495. [Google Scholar] [CrossRef] [PubMed]
- Lohse, S.E.; Abadeer, N.S.; Zoloty, M.; White, J.C.; Newman, L.A.; Murphy, C.J. Nanomaterial probes in the environment: Gold nanoparticles soil retention and environment stability as a function of surface chemistry. ACS Sustain. Chem. Eng. 2017, 5, 11451–11458. [Google Scholar] [CrossRef]
- Zhang, L.; Salmain, M.; Liedberg, B.; Boujday, S. Naked Eye Immunosensing of Food Biotoxins Using Gold Nanoparticle-Antibody Bioconjugates. ACS Appl. Nano Mater. 2019, 2, 4150–4158. [Google Scholar] [CrossRef]
- Liu, G.; Lu, M.; Huang, X.; Li, T.; Xu, D. Application of Gold-Nanoparticle Colorimetric Sensing to Rapid Food Safety Screening. Sensors 2018, 18, 4166. [Google Scholar] [CrossRef]
- Winkler, T.E.; Stevenson, F.O.; Kim, E.; Kang, M.; Payne, G.F.; Kelly, D.L.; Ghodssi, R. The Role of Microsystems Integration Towards Point-of-Care Clozapine Treatment Monitoring in Schizophrenia. IEEE Sens. Lett. 2018, 2, 1–4. [Google Scholar] [CrossRef]
- Enomoto, J.; Kageyama, T.; Myasnikova, D.; Onishi, K.; Kobayashi, Y.; Taruno, Y.; Kanai, T.; Fukuda, J. Gold cleaning methods for preparation of cell culture surfaces for self-assembled monolayers of zwitterionic oligopeptides. J. Biosci. Bioeng. 2018, 125, 606–612. [Google Scholar] [CrossRef]
- Ladekjær-Mikkelsen, A.-S.; Nielsen, J.; Stadejek, T.; Storgaard, T.; Krakowka, S.; Ellis, J.; McNeilly, F.; Allan, G.; Bøtner, A. Reproduction of postweaning multisystemic wasting syndrome (PMWS) in immunostimulated and non-immunostimulated 3-week-old piglets experimentally infected with porcine circovirus type 2 (PCV2). Vet. Microbiol. 2002, 89, 97–114. [Google Scholar] [CrossRef]
- Basso, C.R.; Tozato, C.C.; Junior, J.P.A.; Pedrosa, V.A. A fast and highly sensitive method for the detection of canine distemper virus by the naked eye. Anal. Methods 2015, 7, 2264–2267. [Google Scholar] [CrossRef]
- Verdoodt, N.; Basso, C.R.; Rossi, B.F.; Pedrosa, V.A. Development of a rapid and sensitive immunosensor for the detection of bacteria. Food Chem. 2017, 221, 1792–1796. [Google Scholar] [CrossRef]
- Wang, C.; Irudayaraj, J. Gold Nanorod Probes for the Detection of Multiple Pathogens. Small 2008, 4, 2204–2208. [Google Scholar] [CrossRef] [PubMed]
- Damborský, P.; Švitel, J.; Katrlík, J.; Vitel, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar] [PubMed]
- Reth, M. Matching cellular dimensions with molecular sizes. Nat. Immunol. 2013, 14, 765–767. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Yu, K.; Chang, J.; Steeber, U.A.; Ocola, L.E.; Chen, J. Direct Growth of Vertically-oriented Graphene for Field-Effect Transistor Biosensor. Sci. Rep. 2013, 3, 1696. [Google Scholar] [CrossRef]
- Ions, R.; Narayanan, M.; Browning, M.; Gaillard, E.A.; Stiefel, G.; Tang, J.W. Case presentation: Persistent adenovirus B3 infections associated with bronchiolitis obliterans treated with cidofovir in a child with mosaic tetrasomy 9p. BMC Infect. Dis. 2018, 18, 529. [Google Scholar] [CrossRef]
- Chirambo-Kalolekesha, M.; Kaile, T.; Mwaba, F.; Daka, V.; Simakando, M.; Kowa, S. Seroprevalence of parvovirus B19 in blood donors: The risks and challenges of blood transfusion in Zambia in the era of HIV/AIDS at the Kitwe Central Hospital, blood bank. Afr. Health Sci. 2018, 18, 496–502. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basso, C.R.; Cruz, T.F.; Silva, B.L.; Pedrosa, V.A.; Araújo Junior, J.P. A Methodology for Porcine Circovirus 2 (PCV-2) Quantification Based on Gold Nanoparticles. Materials 2020, 13, 1087. https://doi.org/10.3390/ma13051087
Basso CR, Cruz TF, Silva BL, Pedrosa VA, Araújo Junior JP. A Methodology for Porcine Circovirus 2 (PCV-2) Quantification Based on Gold Nanoparticles. Materials. 2020; 13(5):1087. https://doi.org/10.3390/ma13051087
Chicago/Turabian StyleBasso, Caroline R., Taís F. Cruz, Bruna L. Silva, Valber A. Pedrosa, and João P. Araújo Junior. 2020. "A Methodology for Porcine Circovirus 2 (PCV-2) Quantification Based on Gold Nanoparticles" Materials 13, no. 5: 1087. https://doi.org/10.3390/ma13051087
APA StyleBasso, C. R., Cruz, T. F., Silva, B. L., Pedrosa, V. A., & Araújo Junior, J. P. (2020). A Methodology for Porcine Circovirus 2 (PCV-2) Quantification Based on Gold Nanoparticles. Materials, 13(5), 1087. https://doi.org/10.3390/ma13051087





