1. Introduction
With the rapid development of civilization, the demand for plastic products has increased significantly. However, plastic waste has become a worldwide environmental issue because plastic materials are durable and difficult to degrade [
1,
2,
3]. As a result, bioplastics, which are made from biological substances, have attracted significant interest as an alternative for conventional plastics made from petroleum [
4,
5]. Polyhydroxyalkanoate (PHA) polymers are a type of green bioplastics that are produced naturally by bacteria and utilized by the cells for energy and carbon storage [
6]. PHAs have been used commercially for a wide range of applications due to their excellent versatility, biocompatibility, and biodegradability Their application areas range from food packaging and agricultural films to biomedical fields including use as drug carriers, tissue engineering scaffolds, etc. [
7,
8,
9].
To fulfill the requirements for various applications, fabrication of PHAs with a variety of properties, such as physical, thermal, and mechanical properties, becomes a new challenge. PHAs with different properties are achieved by tuning the monomers and configurations [
10,
11,
12]. It is known that approximately 150 types of PHA monomers can be used to constitute different PHA copolymers [
13]. These combinations of single monomers can provide PHA polymers with diverse and flexible properties. In addition, the polymer properties can also be tuned by modifying chemical configurations, i.e., the size, shape, and branching of polymers [
14,
15]. Therefore, the design and synthesis of novel PHAs typically need trial and error experimentation to obtain the structure–property–performance relationships. This traditional approach requires enormous lab and labor investments, and progress is typically slow.
As data science has rapidly developed in the last decade, machine learning (ML) became a promising tool for data analytics and predictions. ML is part of data science and it is a technology for processing a large amount of data. It learns from the existing data, finds data patterns, and provides solutions [
16]. It is faster, cheaper, and more flexible than experimentation. A neural network is an algorithm of ML. Inspired by the neural network of the human brain, an artificial neural network can learn from past data and generate a response [
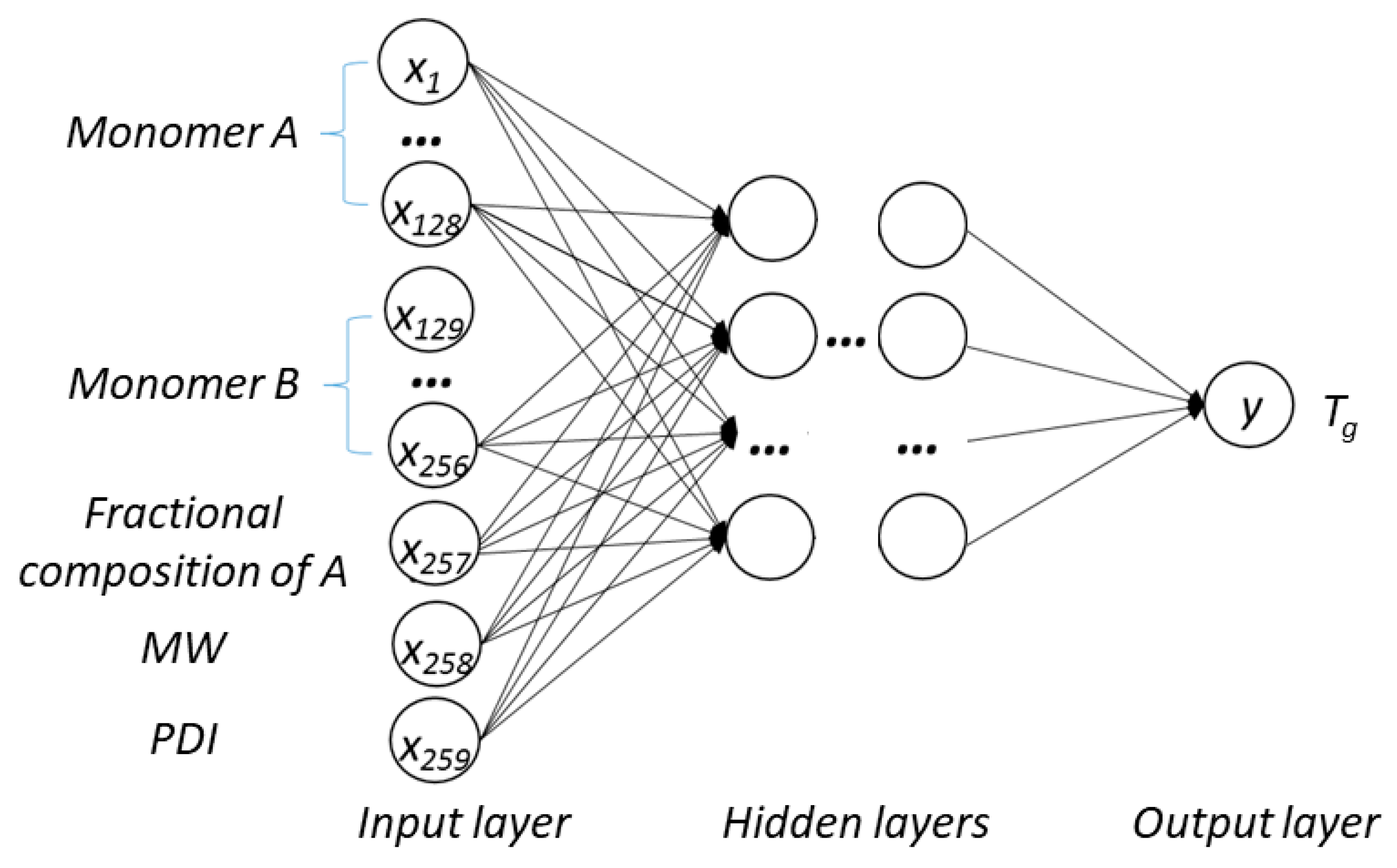
17]. The structure of a typical artificial neural network is usually composed of three essential types of layers, i.e., the input layer, hidden layer(s), and output layer. They are used for receiving, processing, and exporting information, respectively. Each neuron is connected with an assigned weight and the weight sum is calculated by a transfer function [
18]. Deep learning is a class of ML that allows multiple hidden layers for data processing [
19]. A deep neural network (DNN) combines an artificial neural network with deep learning and is capable of providing a better solution to problems in cognitive learning such as speech and image recognition [
20]. So far, DNN models have been successfully applied to learn and predict a range of properties of diverse types of materials, including metals, ceramics, and macromolecular materials [
21,
22,
23].
Due to the increasing demand for plastic materials and rising awareness of the plastic waste crisis, PHAs have been intensively explored since their first discovery in 1926 [
24]. With tremendous historical experimental efforts focused on PHA synthesis and characterization, a large amount of data is accessible from published sources. Therefore, this allows data to be extracted from the literature and used to build a data-based ML model to investigate the connections between structures and properties of PHAs and to extract design rules and useful chemical trends.
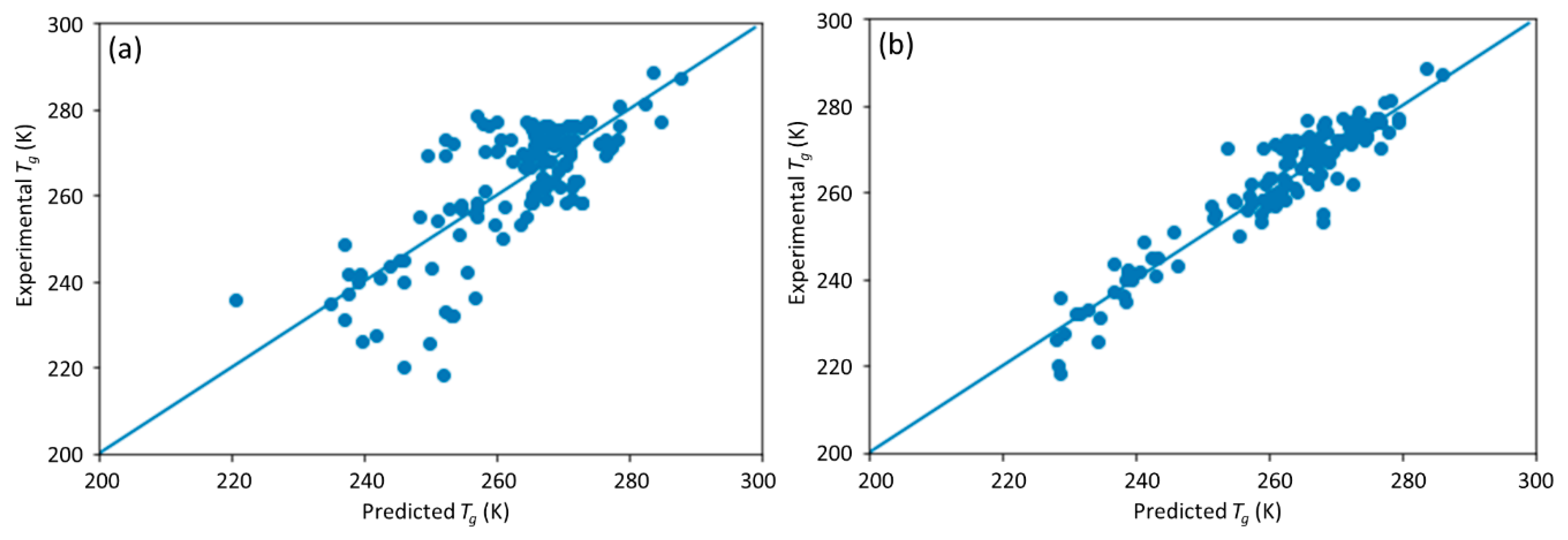
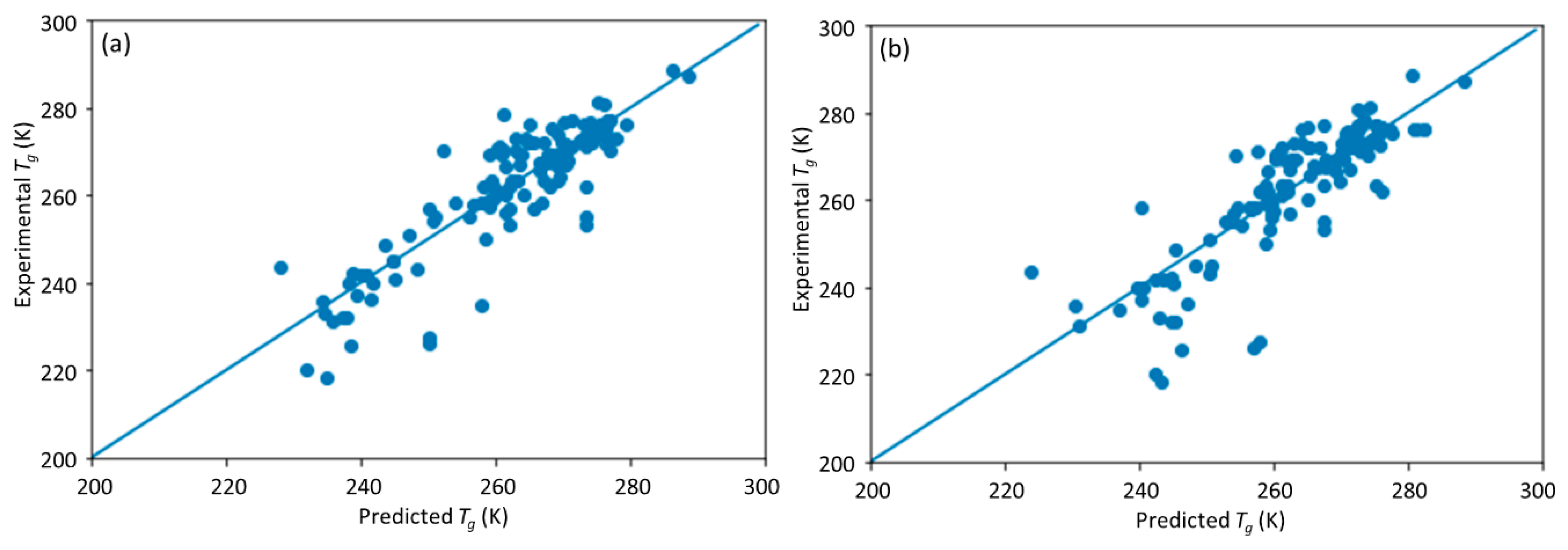
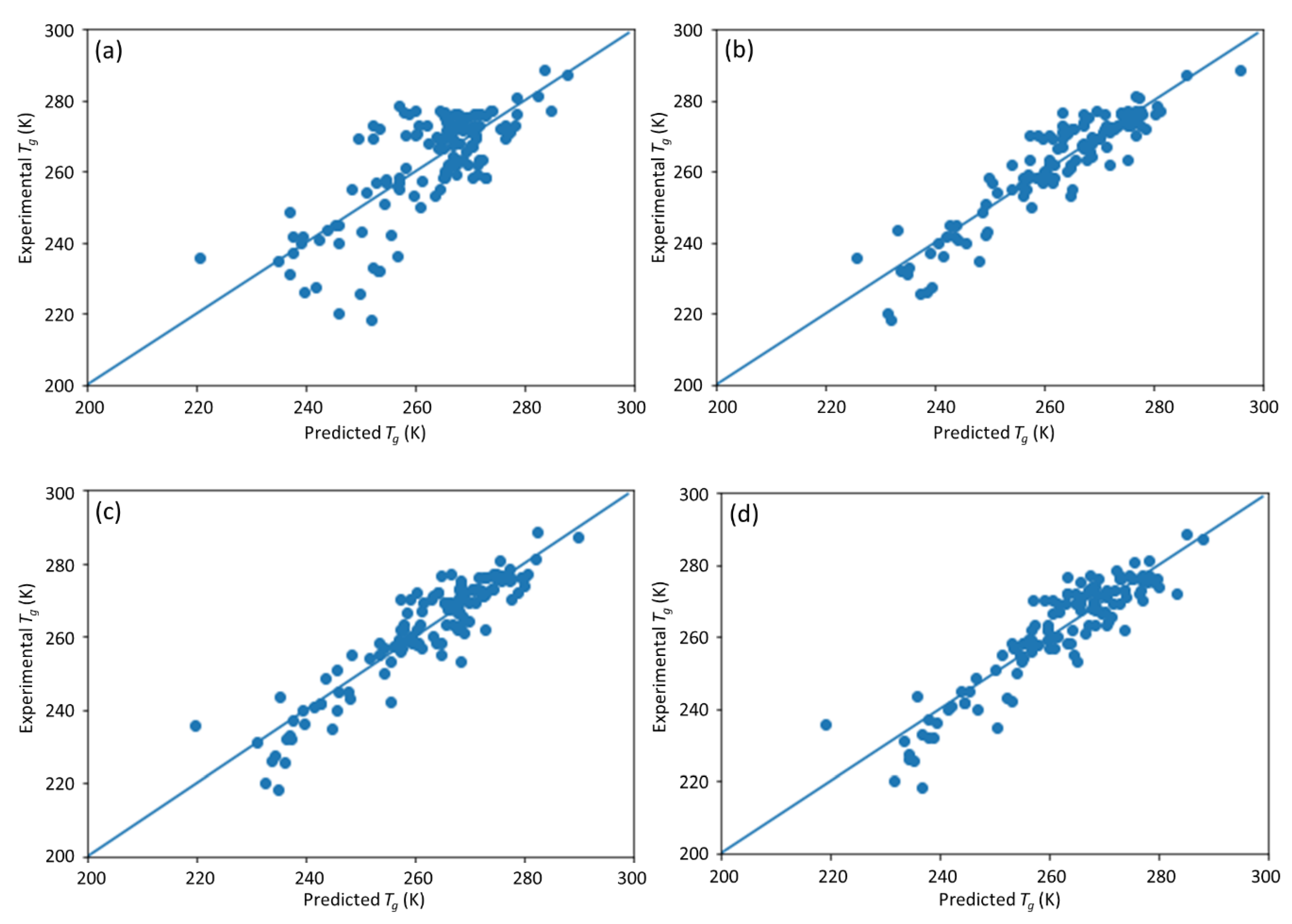
This study is a follow-up to [
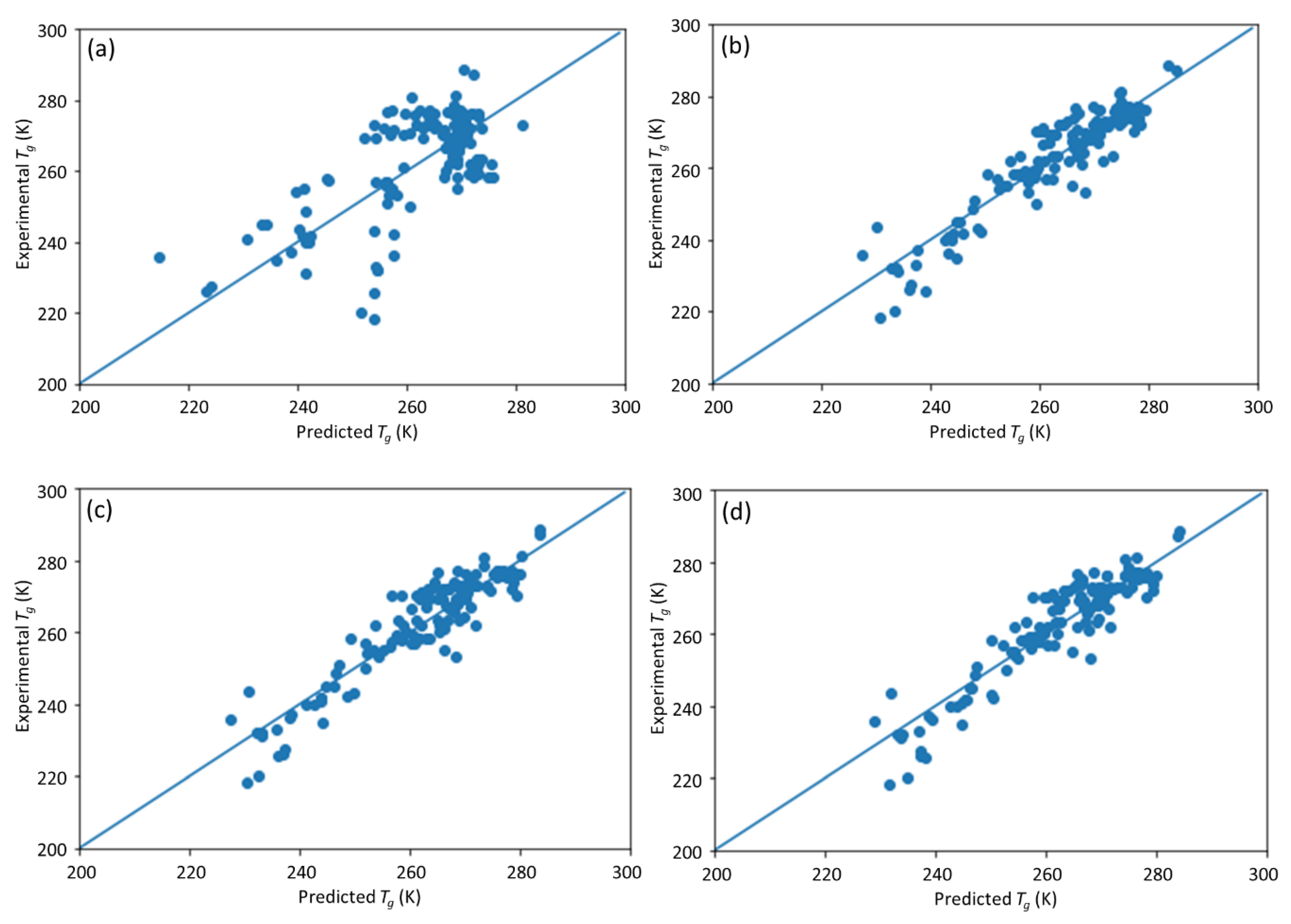
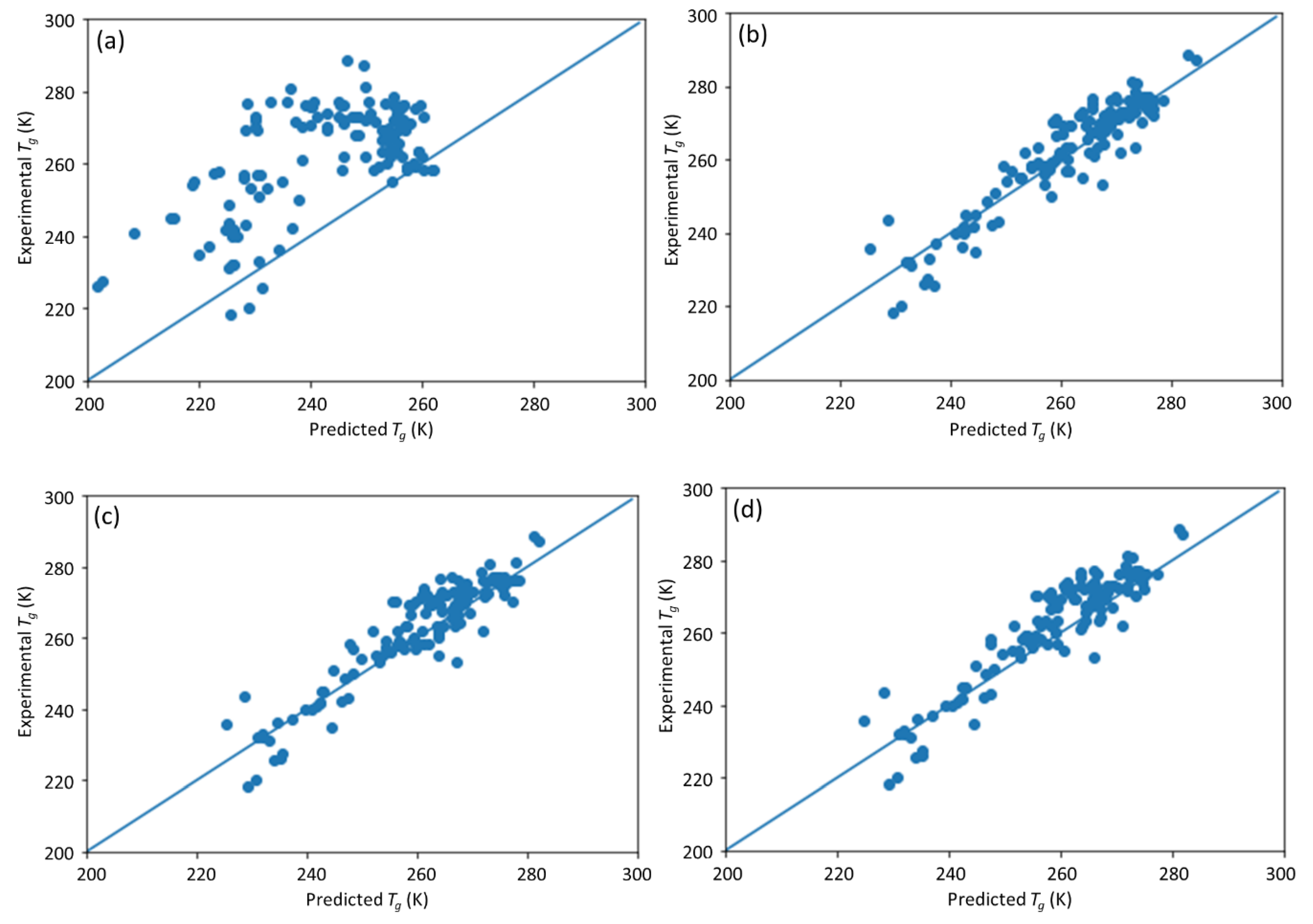
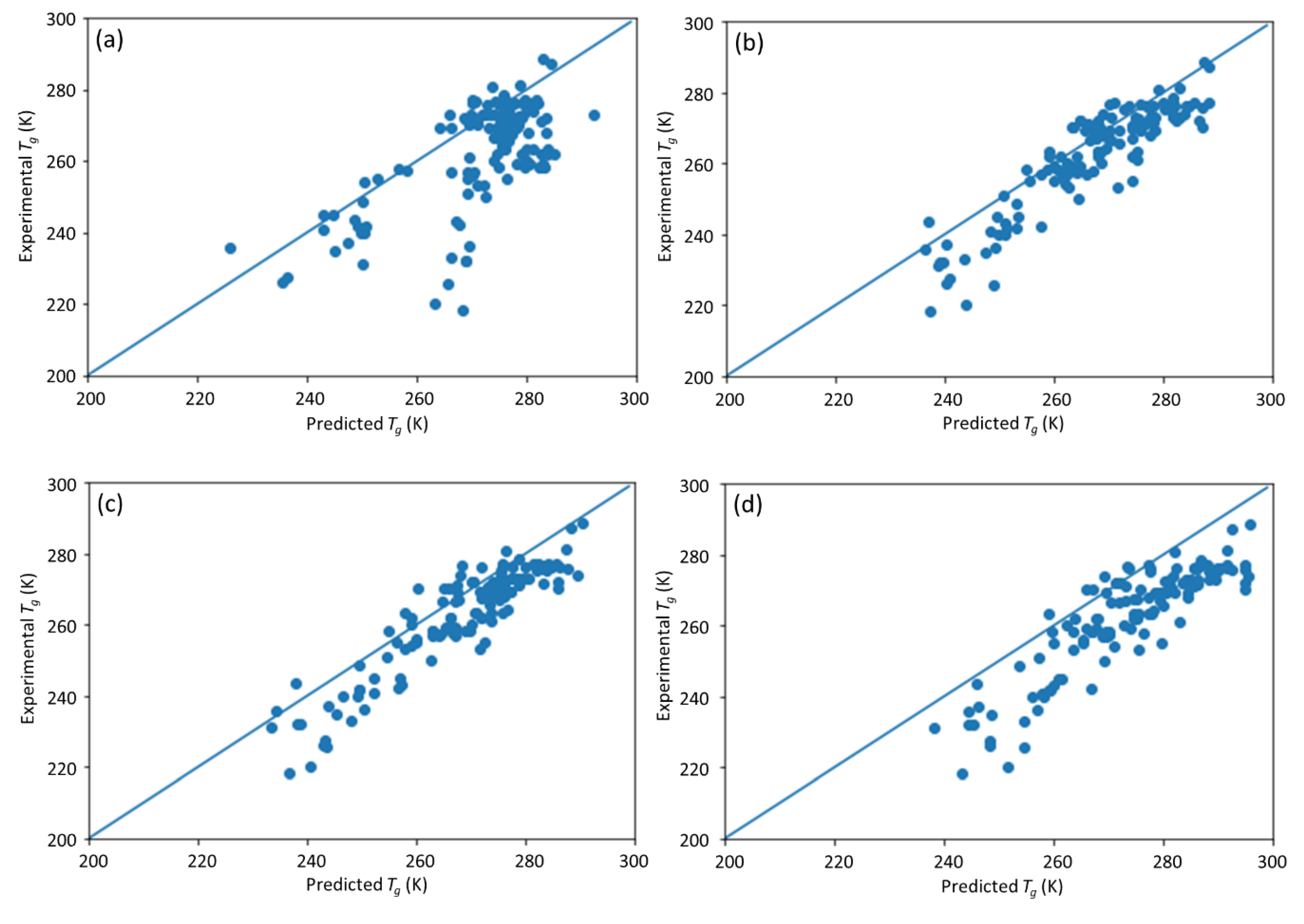
25], in which an ML model incorporating descriptors extracted from the quantitative structure–property relationship (QSPR) for glass transition temperature (
Tg) prediction was used. It describes the development of a DNN-based model for
Tg predictions of PHA homo- and copolymers. We used
Tg as the predicted property of this model because
Tg is an important thermal property for polymers turning from a rigid state to a rubbery state.
Tg has been extensively studied and its value depends on the structure of polymers, e.g., molecular weight and branching network [
26,
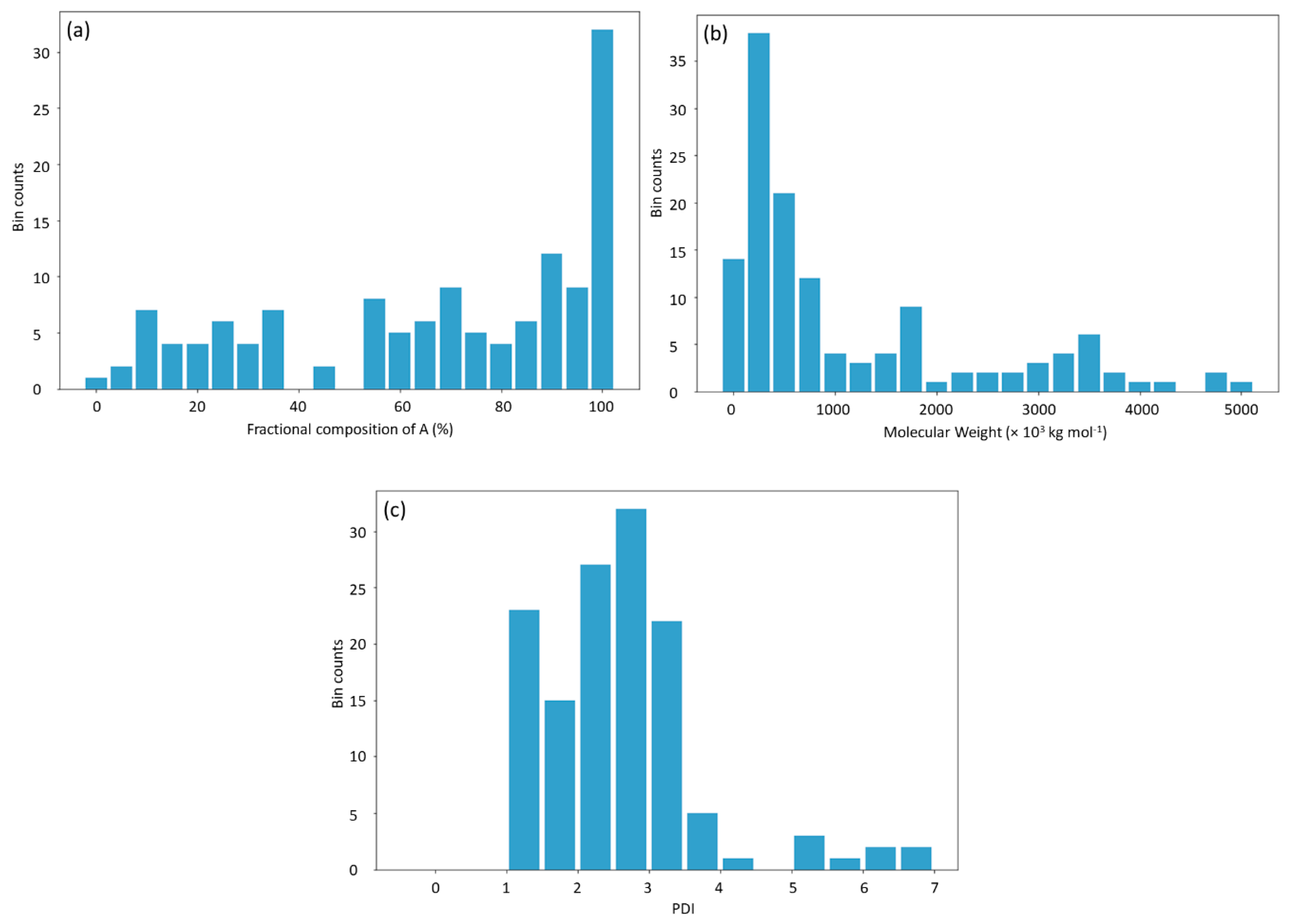
27]. The database consists of 133 data points obtained from published experiments [
25]. The DNN-based model was used to investigate the connections between the inputs, i.e., structure of PHA polymers, and the output, i.e.,
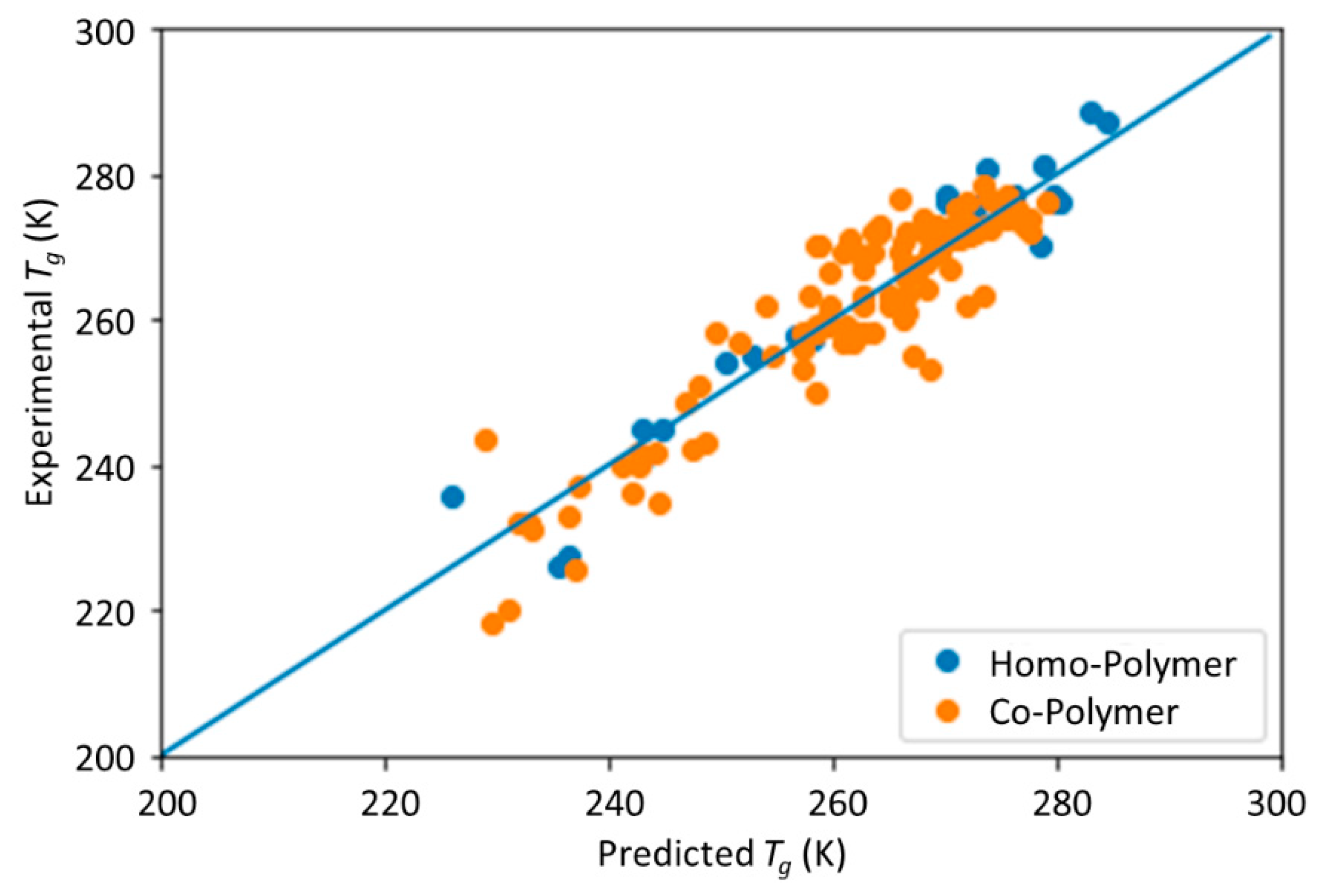
Tg. The trained model can provide guidance for material design of PHA polymers before conducting experiments, which can significantly reduce experimental trials, and, therefore, save time and expense.
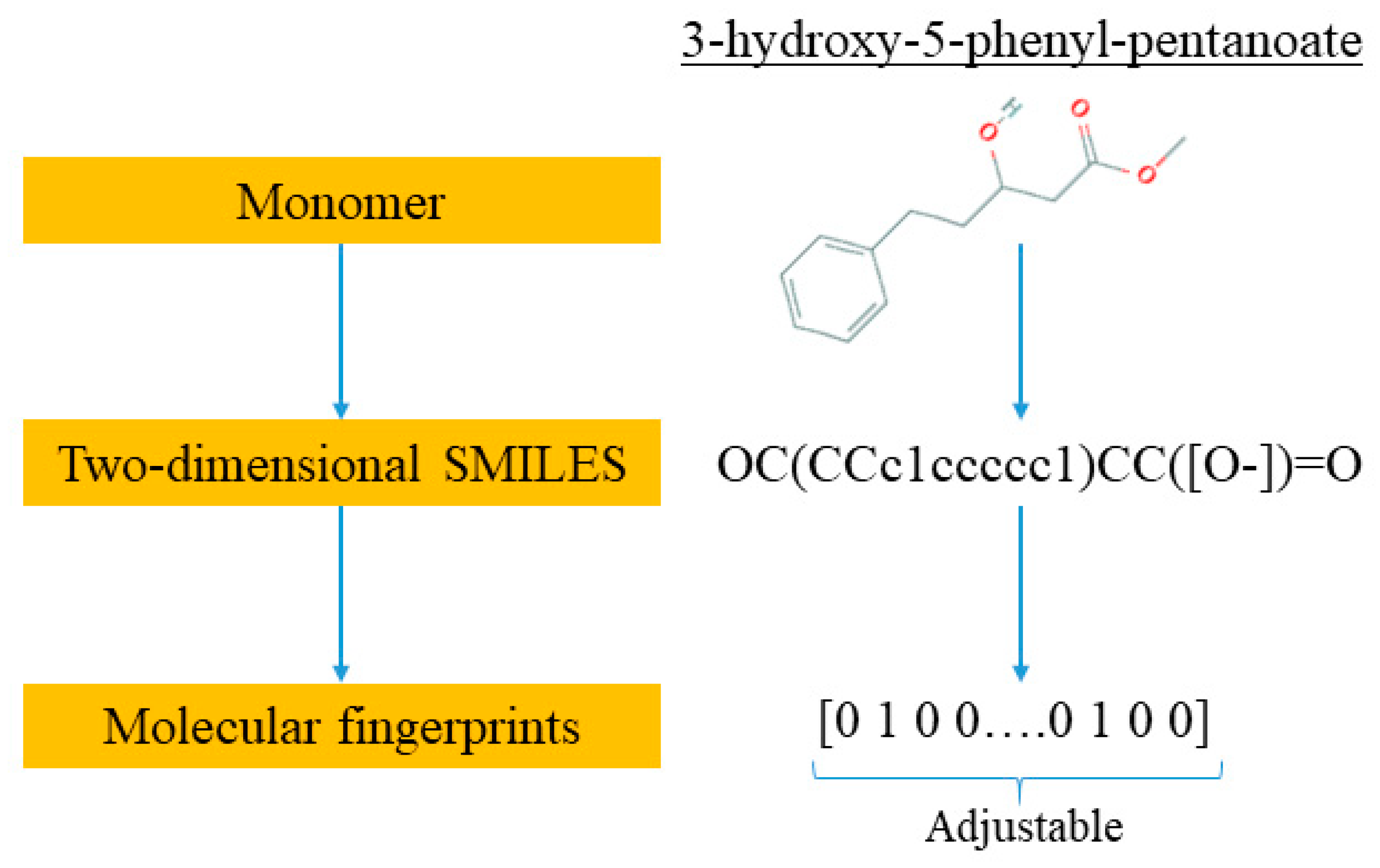
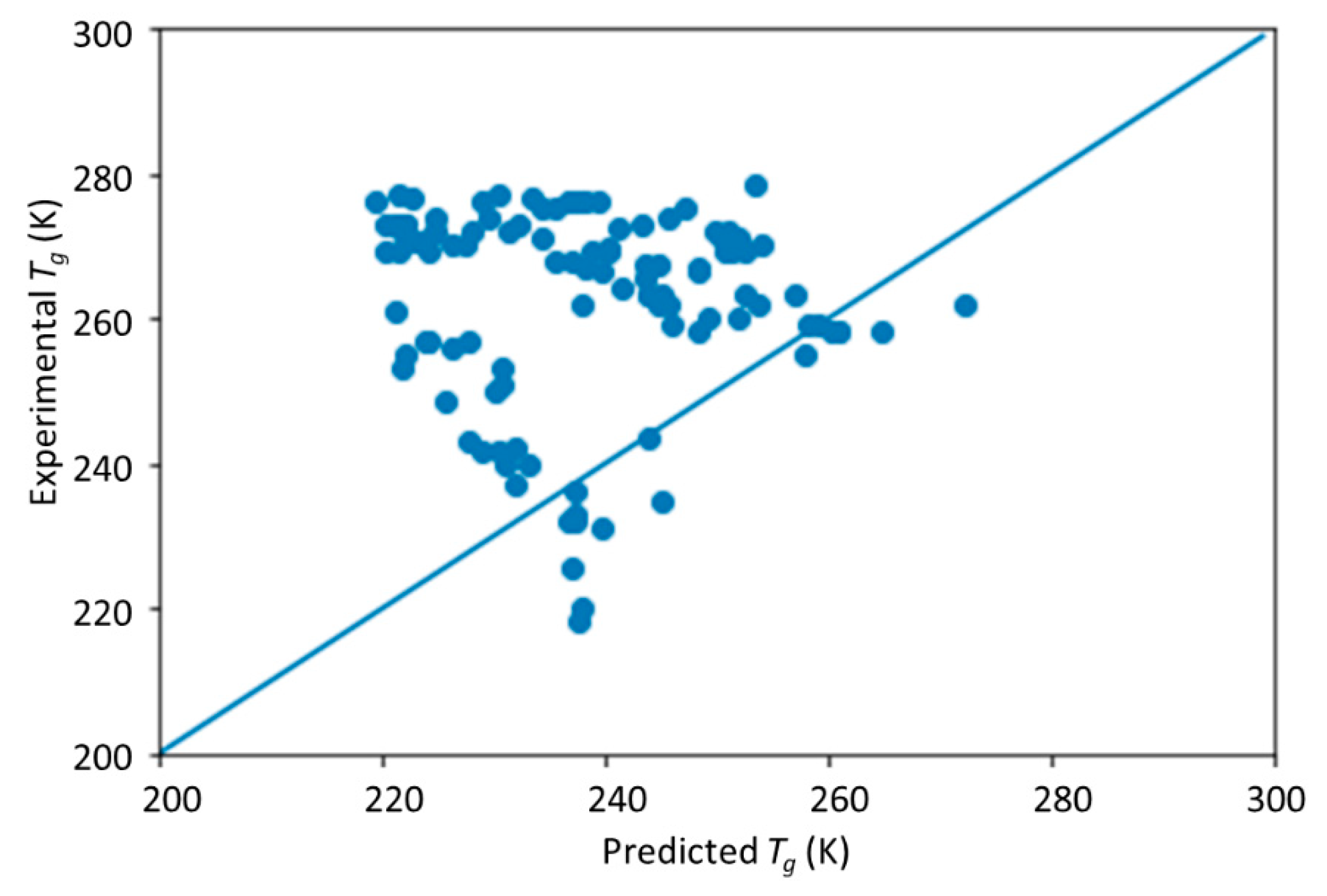
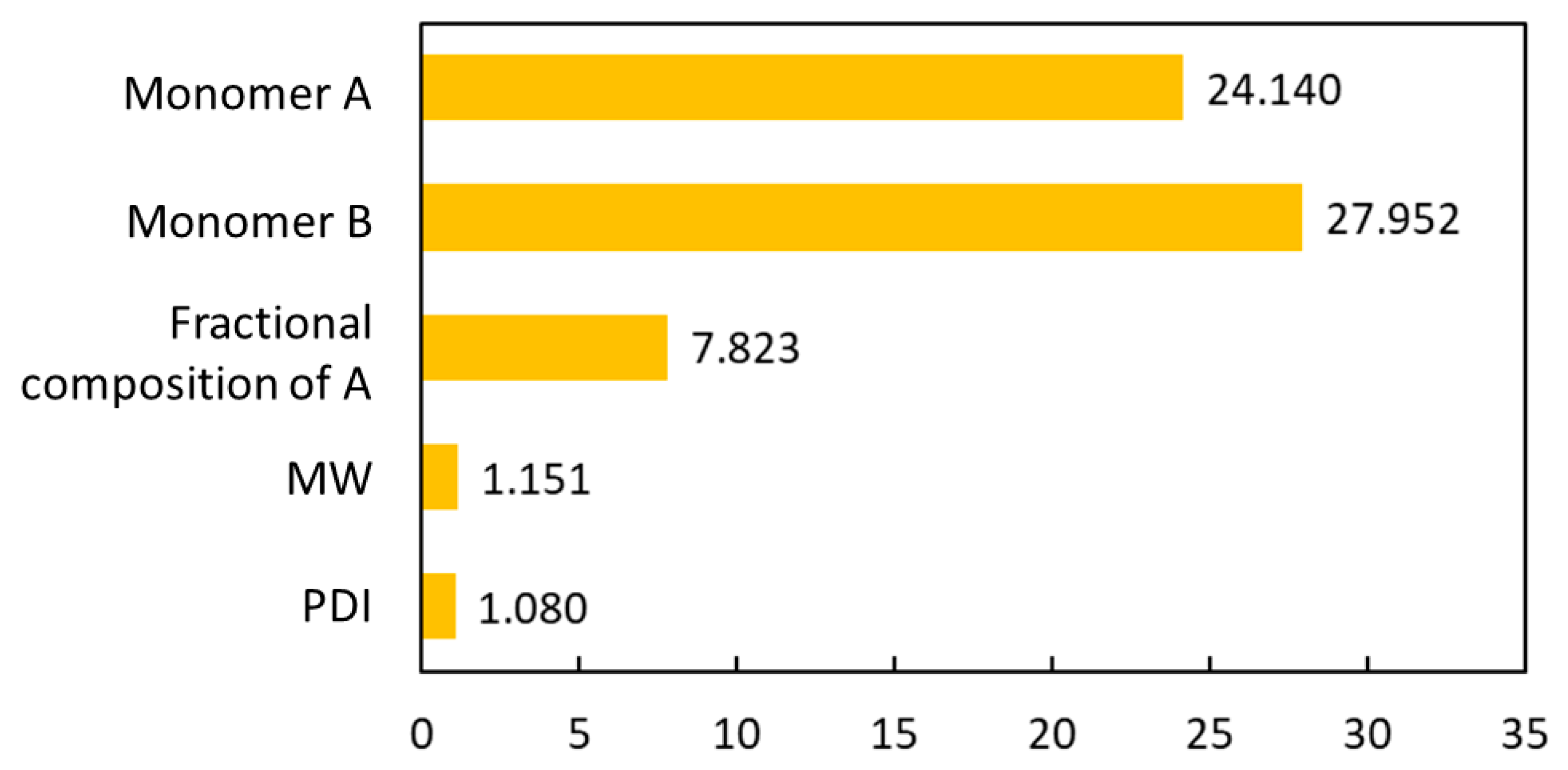
The novelty of this work is the prediction of the glass transition temperature using a DNN model that is simple to implement and still achieves accurate results. Compared with prior ML model approach (i.e., [
25]), this model does not require the assistance of an expert to obtain the chemical descriptors from QSPR; instead, it uses standard molecular fingerprints to convert the polymers into machine-readable language. This is not only much easy to encode the copolymers but also avoids possible cognitive bias in QSPR by different professionals. This ML model framework can also be easily extended to a broader range of polymers beyond PHA copolymers.