Applications of Bionano Sensor for Extracellular Vesicles Analysis
Abstract
1. Introduction
2. Surface Plasmon Resonance-Based Analysis Methods for EV Detection
3. Colorimetric/Fluorescence-Based Analysis Methods for EV Detection
4. Electrochemical-Based Analysis Methods for EV Detection
5. Raman-Based Analysis Methods for EV Detection
6. Conclusions and Future Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorák, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta Gen. Subj. 2012, 1820, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Cutler, E.G.; Cho, H. Therapeutic nanoplatforms and delivery strategies for neurological disorders. Nano Converg. 2018, 5, 35. [Google Scholar] [CrossRef]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.-F.; Jacob, H.J.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 1–14. [Google Scholar] [CrossRef]
- Johnsen, K.B.; Gudbergsson, J.M.; Andresen, T.L.; Simonsen, J.B. What is the blood concentration of extracellular vesicles? Implications for the use of extracellular vesicles as blood-borne biomarkers of cancer. Biochim. Biophys. Acta Bioenerg. 2018, 1871, 109–116. [Google Scholar] [CrossRef]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer—Implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef]
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear Export of MicroRNA Precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef]
- Lai, E.C. Micro RNAs are Complementary to 3′ UTR Sequence Motifs that Mediate Negative Post-Transcriptional Regulation. Nat. Genet. 2002, 30, 363. [Google Scholar] [CrossRef]
- Xu, H.; Ye, B.-C. Advances in biosensing technologies for analysis of cancer-derived exosomes. TrAC Trends Anal. Chem. 2020, 123, 115773. [Google Scholar] [CrossRef]
- Ibsen, S.D.; Wright, J.; Lewis, J.M.; Kim, S.; Ko, S.-Y.; Ong, J.; Manouchehri, S.; Vyas, A.; Akers, J.; Chen, C.C.; et al. Rapid Isolation and Detection of Exosomes and Associated Biomarkers from Plasma. ACS Nano 2017, 11, 6641–6651. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Xu, R.; Ji, H.; Tauro, B.J.; Simpson, R.J. A Protocol for Exosome Isolation and Characterization: Evaluation of Ultracentrifugation, Density-Gradient Separation, and Immunoaffinity Capture Methods. In Proteomic Profiling; Springer: Berlin/Heidelberg, Germany, 2015; pp. 179–209. [Google Scholar]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernández, A.F.; Gammon, S.T.; Kaye, J.; le Bleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Zacharias, W.; Gercel-Taylor, C. Exosome Isolation for Proteomic Analyses and RNA Profiling. In Serum/Plasma Proteomics; Springer: Berlin/Heidelberg, Germany, 2011; pp. 235–246. [Google Scholar]
- Gangoda, L.; Liem, M.; Ang, C.-S.; Keerthikumar, S.; Adda, C.G.; Parker, B.S.; Mathivanan, S. Proteomic Profiling of Exosomes Secreted by Breast Cancer Cells with Varying Metastatic Potential. Proteomics 2017, 17, 1600370. [Google Scholar] [CrossRef]
- Yu, J.; Lin, Y.; Xiong, X.; Li, K.; Yao, Z.; Dong, H.; Jiang, Z.; Yu, D.; Yeung, S.-C.J.; Zhang, H. Detection of Exosomal PD-L1 RNA in Saliva of Patients With Periodontitis. Front. Genet. 2019, 10, 202. [Google Scholar] [CrossRef]
- Merdalimova, A.; Chernyshev, V.; Nozdriukhin, D.; Rudakovskaya, P.; Gorin, D.A.; Yashchenok, A.M. Identification and Analysis of Exosomes by Surface-Enhanced Raman Spectroscopy. Appl. Sci. 2019, 9, 1135. [Google Scholar] [CrossRef]
- Bu, J.; Shim, J.-E.; Lee, T.H.; Cho, Y.-H. Multi-Modal liquid biopsy platform for cancer screening: Screening both cancer-associated rare cells and cancer cell-derived vesicles on the fabric filters for a reliable liquid biopsy analysis. Nano Converg. 2019, 6, 1–8. [Google Scholar] [CrossRef]
- Im, H.; Lee, K.; Weissleder, R.; Lee, H.; Castro, C.M. Novel nanosensing technologies for exosome detection and profiling. Lab Chip 2017, 17, 2892–2898. [Google Scholar] [CrossRef]
- Shao, B.; Xiao, Z. Recent achievements in exosomal biomarkers detection by nanomaterials-based optical biosensors—A review. Anal. Chim. Acta 2020, 1114, 74–84. [Google Scholar] [CrossRef]
- Singh, P. SPR Biosensors: Historical Perspectives and Current Challenges. Sens. Actuators B Chem. 2016, 229, 110–130. [Google Scholar] [CrossRef]
- Kim, D.; Choi, E.; Lee, C.; Choi, Y.; Kim, H.; Yu, T.; Piao, Y. Highly sensitive and selective visual detection of Cr (VI) ions based on etching of silver-coated gold nanorods. Nano Converg. 2019, 6, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Chun, H.J.; Kim, J.-H.; Yoon, H.; Yoon, H.C. A non-spectroscopic optical biosensor for the detection of pathogenic Salmonella Typhimurium based on a stem-loop DNA probe and retro-reflective signaling. Nano Converg. 2019, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Dong, B.; Lee, C. Progress of infrared guided-wave nanophotonic sensors and devices. Nano Converg. 2020, 7, 12–34. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Kim, B.-C.; Oh, B.-K.; Choi, J.-W. Highly sensitive localized surface plasmon resonance immunosensor for label-free detection of HIV-1. Nanomedicine 2013, 9, 1018–1026. [Google Scholar] [CrossRef]
- Gong, C.; Leite, M. Noble Metal Alloys for Plasmonics. ACS Photonics 2016, 3, 507–513. [Google Scholar] [CrossRef]
- Haes, A.J.; Zou, S.; Schatz, G.C.; van Duyne, R.P. Nanoscale Optical Biosensor: Short Range Distance Dependence of the Localized Surface Plasmon Resonance of Noble Metal Nanoparticles. J. Phys. Chem. B 2004, 108, 6961–6968. [Google Scholar] [CrossRef]
- Sugawa, K.; Tahara, H.; Yamashita, A.; Otsuki, J.; Sagara, T.; Harumoto, T.; Yanagida, S. Refractive Index Susceptibility of the Plasmonic Palladium Nanoparticle: Potential as the Third Plasmonic Sensing Material. ACS Nano 2015, 9, 1895–1904. [Google Scholar] [CrossRef]
- Haes, A.J.; Zou, S.; Schatz, G.C.; van Duyne, R.P. A Nanoscale Optical Biosensor: The Long Range Distance Dependence of the Localized Surface Plasmon Resonance of Noble Metal Nanoparticles. J. Phys. Chem. B 2004, 108, 109–116. [Google Scholar] [CrossRef]
- Yang, L.; Lee, J.-H.; Rathnam, C.; Hou, Y.; Choi, J.-W.; Lee, K.-B. Dual-Enhanced Raman Scattering-Based Characterization of Stem Cell Differentiation Using Graphene-Plasmonic Hybrid Nanoarray. Nano Lett. 2019, 19, 8138–8148. [Google Scholar] [CrossRef]
- Jain, P.K.; Huang, X.; El-Sayed, I.H.; El-Sayed, M.A. Review of Some Interesting Surface Plasmon Resonance-enhanced Properties of Noble Metal Nanoparticles and Their Applications to Biosystems. Plasmonics 2007, 2, 107–118. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface Plasmon Resonance: A Versatile Technique for Biosensor Applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef] [PubMed]
- Brolo, A.G. Plasmonics for future biosensors. Nat. Photonics 2012, 6, 709–713. [Google Scholar] [CrossRef]
- Belotelov, V.; Akimov, I.; Pohl, M.; Kalish, A.; Kasture, S.; Vengurlekar, A.; Gopal, A.; Kotov, V.; Yakovlev, D.; Zvezdin, A. Intensity magnetooptical effect in magnetoplasmonic crystals. J. Phys. Conference Ser. 2011, 303, 012038. [Google Scholar] [CrossRef]
- Rizal, C.; Belotelov, V. Sensitivity comparison of surface plasmon resonance (SPR) and magneto-optic SPR biosensors. Eur. Phys. J. Plus 2019, 134, 435. [Google Scholar] [CrossRef]
- Leung, A.; Shankar, P.; Mutharasan, R. A review of fiber-optic biosensors. Sens. Actuators B Chem. 2007, 125, 688–703. [Google Scholar] [CrossRef]
- Jing, J.-Y.; Wang, Q.; Zhao, W.-M.; Wang, B.-T. Long-Range surface plasmon resonance and its sensing applications: A review. Opt. Lasers Eng. 2019, 112, 103–118. [Google Scholar] [CrossRef]
- Vlček, J.; Pištora, J.; Lesňák, M. Design of Plasmonic-Waveguiding Structures for Sensor Applications. Nanomaterials 2019, 9, 1227. [Google Scholar] [CrossRef]
- Shpacovitch, V.; Hergenröder, R. Optical and surface plasmonic approaches to characterize extracellular vesicles. A review. Anal. Chim. Acta 2018, 1005, 1–15. [Google Scholar] [CrossRef]
- Qiu, G.; Thakur, A.; Xu, C.; Ng, S.P.; Lee, Y.J.; Wu, C.M.L. Detection of Glioma-Derived Exosomes with the Biotinylated Antibody-Functionalized Titanium Nitride Plasmonic Biosensor. Adv. Funct. Mater. 2018, 29, 1806761. [Google Scholar] [CrossRef]
- Wang, Q.; Zou, L.; Yang, X.; Liu, X.; Nie, W.; Zheng, Y.; Cheng, Q.; Wang, K. Direct quantification of cancerous exosomes via surface plasmon resonance with dual gold nanoparticle-assisted signal amplification. Biosens. Bioelectron. 2019, 135, 129–136. [Google Scholar] [CrossRef]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-Free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Qiu, G.; Ng, S.P.; Guan, J.; Yue, J.; Lee, Y.J.; Wu, C.M.L. Direct detection of two different tumor-derived extracellular vesicles by SAM-AuNIs LSPR biosensor. Biosens. Bioelectron. 2017, 94, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Bathini, S.; Raju, D.; Badilescu, S.; Kumar, A.; Ouellette, R.J.; Ghosh, A.; Packirisamy, M. Nano-Bio Interactions of Extracellular Vesicles with Gold Nanoislands for Early Cancer Diagnosis. Reserch 2018, 2018, 3917986. [Google Scholar] [CrossRef] [PubMed]
- Raghu, D.; Christodoulides, J.A.; Christophersen, M.; Liu, J.L.; Anderson, G.P.; Robitaille, M.C.; Byers, J.M.; Raphael, M.P. Nanoplasmonic pillars engineered for single exosome detection. PLoS ONE 2018, 13, e0202773. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Lu, Y.; He, L.; Pang, J.; Yang, F.; Liu, Y. Colorimetric sensor array based on gold nanoparticles: Design principles and recent advances. TrAC Trends Anal. Chem. 2020, 122, 115754. [Google Scholar] [CrossRef]
- Aldewachi, H.; Chalati, T.; Woodroofe, M.N.; Bricklebank, N.; Sharrack, B.; Gardiner, P. Gold nanoparticle-based colorimetric biosensors. Nanoscale 2018, 10, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Chen, T.; Chen, G.; Weng, Y.; Zeng, L.; Liao, Y.; Chen, W.; Lan, J.; Zhang, J.; Chen, J.-H. A nature-inspired colorimetric and fluorescent dual-modal biosensor for exosomes detection. Talanta 2020, 214, 120851. [Google Scholar] [CrossRef]
- Cheng, N.; Du, D.; Wang, X.; Liu, N.; Xu, W.; Luo, Y.; Lin, Y. Recent Advances in Biosensors for Detecting Cancer-Derived Exosomes. Trends Biotechnol. 2019, 37, 1236–1254. [Google Scholar] [CrossRef]
- Sabela, M.; Bechelany, M.; Janot, J.M.; Balme, S.; Bisetty, K. A Review of Gold and Silver Nanoparticle-Based Colorimetric Sensing Assays. Adv. Eng. Mater. 2017, 19, 1700270. [Google Scholar] [CrossRef]
- Chen, Z.; Cheng, S.-B.; Cao, P.; Qiu, Q.-F.; Chen, Y.; Xu, Y.; Xie, M.; Huang, W.-H. Detection of exosomes by ZnO nanowires coated three-dimensional scaffold chip device. Biosens. Bioelectron. 2018, 122, 211–216. [Google Scholar] [CrossRef]
- Zhang, P.; Zhou, X.; He, M.; Shang, Y.; Tetlow, A.L.; Godwin, A.K.; Zeng, Y. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat. Biomed. Eng. 2019, 3, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-M.; Liu, J.-W.; Adkins, G.; Shen, W.; Trinh, M.P.; Duan, L.-Y.; Jiang, J.-H.; Zhong, W. Enhancement of the Intrinsic Peroxidase-Like Activity of Graphitic Carbon Nitride Nanosheets by ssDNAs and Its Application for Detection of Exosomes. Anal. Chem. 2017, 89, 12327–12333. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Liu, M.; Wang, L.; Yan, A.; He, W.; Chen, M.; Lan, J.; Xu, J.; Guan, L.; Chen, J. A visible and colorimetric aptasensor based on DNA-capped single-walled carbon nanotubes for detection of exosomes. Biosens. Bioelectron. 2017, 92, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xu, Y.; Lu, Y.; Xing, W. Isolation and Visible Detection of Tumor-Derived exosomes from Plasma. Anal. Chem. 2018, 90, 14207–14215. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Shi, M.; Liu, Y.; Wan, S.; Cui, C.; Zhang, L.; Tan, W. Aptamer/AuNP Biosensor for Colorimetric Profiling of Exosomal Proteins. Angew. Chem. Int. Ed. 2017, 56, 11916–11920. [Google Scholar] [CrossRef]
- Liu, W.; Li, J.; Wu, Y.; Xing, S.; Lai, Y.; Zhang, G. Target-Induced proximity ligation triggers recombinase polymerase amplification and transcription-mediated amplification to detect tumor-derived exosomes in nasopharyngeal carcinoma with high sensitivity. Biosens. Bioelectron. 2018, 102, 204–210. [Google Scholar] [CrossRef]
- Sposito, A.J.; Kurdekar, A.; Zhao, J.; Hewlett, I. Application of nanotechnology in biosensors for enhancing pathogen detection. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2018, 10, e1512. [Google Scholar] [CrossRef]
- Jin, D.; Yang, F.; Zhang, Y.L.; Liu, L.; Zhou, Y.; Wang, F.; Zhang, G.-J. ExoAPP: Exosome-Oriented, Aptamer Nanoprobe-Enabled Surface Proteins Profiling and Detection. Anal. Chem. 2018, 90, 14402–14411. [Google Scholar] [CrossRef]
- Chen, X.; Lan, J.; Liu, Y.; Li, L.; Yan, L.; Xia, Y.; Wu, F.; Li, C.; Li, S.; Chen, J.-H. A paper-supported aptasensor based on upconversion luminescence resonance energy transfer for the accessible determination of exosomes. Biosens. Bioelectron. 2018, 102, 582–588. [Google Scholar] [CrossRef]
- Lee, J.-H.; Choi, J.-H.; Chueng, S.-T.D.; Pongkulapa, T.; Yang, L.; Cho, H.-Y.; Choi, J.-W.; Lee, K.-B. Nondestructive Characterization of Stem Cell Neurogenesis by a Magneto-Plasmonic Nanomaterial-Based Exosomal miRNA Detection. ACS Nano 2019, 13, 8793–8803. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, F.; Zhang, H.; Zhang, Y.; Liu, M.; Liu, Y. Universal Ti3C2 MXenes Based Self-Standard Ratiometric Fluorescence Resonance Energy Transfer Platform for Highly Sensitive Detection of exosomes. Anal. Chem. 2018, 90, 12737–12744. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Wang, J.; Yin, B.-C.; Ye, B.-C. Quantification of Exosome Based on a Copper-Mediated Signal Amplification Strategy. Anal. Chem. 2018, 90, 8072–8079. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Du, D.; Lin, Y. Graphene-Like 2D nanomaterial-based biointerfaces for biosensing applications. Biosens. Bioelectron. 2017, 89, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Saidur, M.; Aziz, A.A.; Basirun, W. Recent advances in DNA-based electrochemical biosensors for heavy metal ion detection: A review. Biosens. Bioelectron. 2017, 90, 125–139. [Google Scholar] [CrossRef]
- Shao, Y.; Wang, J.; Wu, H.; Liu, J.; Aksay, I.A.; Lin, Y. Graphene Based Electrochemical Sensors and Biosensors: A Review. Electroanalysis 2010, 22, 1027–1036. [Google Scholar] [CrossRef]
- Wang, J. Carbon-Nanotube Based Electrochemical Biosensors: A Review. Electroanalysis 2005, 17, 7–14. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Huang, K.-J.; Wu, X. Recent advances in transition-metal dichalcogenides based electrochemical biosensors: A review. Biosens. Bioelectron. 2017, 97, 305–316. [Google Scholar] [CrossRef]
- Hu, T.; Zhang, L.; Wen, W.; Zhang, X.; Wang, S. Enzyme catalytic amplification of miRNA-155 detection with graphene quantum dot-based electrochemical biosensor. Biosens. Bioelectron. 2016, 77, 451–456. [Google Scholar] [CrossRef]
- Beitollahi, H.; Ivari, S.G.; Torkzadeh-Mahani, M. Application of antibody–nanogold–ionic liquid–carbon paste electrode for sensitive electrochemical immunoassay of thyroid-stimulating hormone. Biosens. Bioelectron. 2018, 110, 97–102. [Google Scholar] [CrossRef]
- Figueroa-Miranda, G.; Feng, L.; Shiu, S.C.-C.; Dirkzwager, R.M.; Cheung, Y.-W.; Tanner, J.A.; Schöning, M.J.; Offenhäusser, A.; Mayer, D. Aptamer-Based electrochemical biosensor for highly sensitive and selective malaria detection with adjustable dynamic response range and reusability. Sens. Actuators B Chem. 2018, 255, 235–243. [Google Scholar] [CrossRef]
- Huang, Y.L.; Mo, S.; Gao, Z.F.; Chen, J.R.; Lei, J.L.; Luo, H.Q.; Li, N.B. Amperometric biosensor for microRNA based on the use of tetrahedral DNA nanostructure probes and guanine nanowire amplification. Microchim. Acta 2017, 481, 190–2604. [Google Scholar] [CrossRef]
- Ding, J.; Qin, W. Recent advances in potentiometric biosensors. TrAC Trends Anal. Chem. 2020, 124, 115803. [Google Scholar] [CrossRef]
- Doldán, X.; Fagúndez, P.; Cayota, A.; Laíz, J.; Tosar, J.P. Electrochemical Sandwich Immunosensor for Determination of Exosomes Based on Surface Marker-Mediated Signal Amplification. Anal. Chem. 2016, 88, 10466–10473. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, L.; Han, B.; Wang, Y.; Dai, Y.; Zhao, J. A catalytic molecule machine-driven biosensing method for amplified electrochemical detection of exosomes. Biosens. Bioelectron. 2019, 141, 111397. [Google Scholar] [CrossRef] [PubMed]
- Moura, S.L.; Martín, C.G.; Martí, M.; Pividori, M. Electrochemical immunosensing of nanovesicles as biomarkers for breast cancer. Biosens. Bioelectron. 2019, 150, 111882. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Li, R.; Zhang, F.; He, P. Magneto-Mediated Electrochemical Sensor for Simultaneous Analysis of Breast Cancer Exosomal Proteins. Anal. Chem. 2020, 92, 5404–5410. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, L.; Wan, S.; Cansiz, S.; Cui, C.; Liu, Y.; Cai, R.; Hong, C.-Y.; Teng, I.-T.; Shi, M.; et al. Aptasensor with Expanded Nucleotide Using DNA Nanotetrahedra for Electrochemical Detection of Cancerous Exosomes. ACS Nano 2017, 11, 3943–3949. [Google Scholar] [CrossRef]
- Huang, R.; He, L.; Xia, Y.; Xu, H.; Liu, C.; Xie, H.; Wang, S.; Peng, L.; Liu, Y.; Liu, Y.; et al. A Sensitive Aptasensor Based on a Hemin/G-Quadruplex-Assisted Signal Amplification Strategy for Electrochemical Detection of Gastric Cancer Exosomes. Small 2019, 15, e1900735. [Google Scholar] [CrossRef]
- Ding, S.-Y.; You, E.-M.; Tian, Z.-Q.; Moskovits, M. Electromagnetic theories of surface-enhanced Raman spectroscopy. Chem. Soc. Rev. 2017, 46, 4042–4076. [Google Scholar] [CrossRef]
- Dong, H.; Chen, H.; Jiang, J.; Zhang, H.; Cai, C.; Shen, Q. Highly Sensitive Electrochemical Detection of Tumor Exosomes Based on Aptamer Recognition-Induced Multi-DNA Release and Cyclic Enzymatic Amplification. Anal. Chem. 2018, 90, 4507–4513. [Google Scholar] [CrossRef]
- An, Y.; Jin, T.; Zhu, Y.; Zhang, F.; He, P. An ultrasensitive electrochemical aptasensor for the determination of tumor exosomes based on click chemistry. Biosens. Bioelectron. 2019, 142, 111503. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Sun, R.; He, P.; Zhang, X. Ultrasensitive Detection of Exosomes by Target-Triggered Three-Dimensional DNA Walking Machine and Exonuclease III-Assisted Electrochemical Ratiometric Biosensing. Anal. Chem. 2019, 91, 14773–14779. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Hou, T.; Huang, B.; Yang, L.; Li, F. Aptamer recognition-trigged label-free homogeneous electrochemical strategy for an ultrasensitive cancer-derived exosome assay. Chem. Commun. 2019, 55, 13705–13708. [Google Scholar] [CrossRef] [PubMed]
- Thind, A.; Wilson, C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles 2016, 5, 281. [Google Scholar] [CrossRef]
- Salehi, M.; Sharifi, M. Exosomal miRNAs as novel cancer biomarkers: Challenges and opportunities. J. Cell. Physiol. 2018, 233, 6370–6380. [Google Scholar] [CrossRef]
- Jin, X.; Chen, Y.; Chen, H.; Fei, S.; Chen, D.; Cai, X.; Liu, L.; Lin, B.; Su, H.; Zhao, L.; et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non–Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017, 23, 5311–5319. [Google Scholar] [CrossRef]
- Boriachek, K.; Umer, M.; Islam, N.; Gopalan, V.; Lam, A.K.-Y.; Nguyen, N.-T.; Shiddiky, M.J.A. An amplification-free electrochemical detection of exosomal miRNA-21 in serum samples. Analyst 2018, 143, 1662–1669. [Google Scholar] [CrossRef]
- Luo, L.; Wang, L.; Zeng, L.; Wang, Y.; Weng, Y.; Liao, Y.; Chen, T.; Xia, Y.; Zhang, J.; Chen, J.-H. A ratiometric electrochemical DNA biosensor for detection of exosomal MicroRNA. Talanta 2019, 207, 120298. [Google Scholar] [CrossRef]
- Guo, Q.; Yu, Y.; Zhang, H.; Cai, C.; Shen, Q. Electrochemical Sensing of Exosomal MicroRNA Based on Hybridization Chain Reaction Signal Amplification with Reduced False-Positive Signals. Anal. Chem. 2020, 92, 5302–5310. [Google Scholar] [CrossRef]
- Cheng, W.; Ma, J.; Cao, P.; Zhang, Y.; Xu, C.; Yi, Y.; Li, J. Enzyme-Free electrochemical biosensor based on double signal amplification strategy for the ultra-sensitive detection of exosomal microRNAs in biological samples. Talanta 2020, 121242. [Google Scholar] [CrossRef]
- Ding, S.-Y.; Yi, J.; Li, J.-F.; Ren, B.; Wu, D.-Y.; Selvam, R.P.P.; Tian, Z.-Q. Nanostructure-Based plasmon-enhanced Raman spectroscopy for surface analysis of materials. Nat. Rev. Mater. 2016, 1, 16021. [Google Scholar] [CrossRef]
- Mosier-Boss, P. Review of SERS Substrates for Chemical Sensing. Nanomaterials 2017, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- El-Said, W.A.; Yoon, J.; Choi, J.-W. Nanostructured surfaces for analysis of anticancer drug and cell diagnosis based on electrochemical and SERS tools. Nano Converg. 2018, 5, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Andrade, G.F.; Brolo, A.G. A review on recent advances in the applications of surface-enhanced Raman scattering in analytical chemistry. Anal. Chim. Acta 2019, 1097, 1–29. [Google Scholar] [CrossRef]
- Tian, Y.-F.; Ning, C.-F.; He, F.; Yin, B.-C.; Ye, B.-C. Highly sensitive detection of exosomes by SERS using gold nanostar@Raman reporter@nanoshell structures modified with a bivalent cholesterol-labeled DNA anchor. Analyst 2018, 143, 4915–4922. [Google Scholar] [CrossRef]
- Kwizera, E.A.; O’Connor, R.; Vinduska, V.; Williams, M.; Butch, E.R.; Snyder, S.E.; Chen, X.; Huang, X. Molecular Detection and Analysis of Exosomes Using Surface-Enhanced Raman Scattering Gold Nanorods and a Miniaturized Device. Theranostics 2018, 8, 2722. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, L.; Diefenbach, R.J.; Campbell, D.H.; Walsh, B.J.; Packer, N.H.; Wang, Y. Enabling Sensitive Phenotypic Profiling of Cancer-Derived Small Extracellular Vesicles Using Surface-Enhanced Raman Spectroscopy Nanotags. ACS Sens. 2020, 5, 764–771. [Google Scholar] [CrossRef]
- Pang, Y.; Shi, J.; Yang, X.; Wang, C.; Sun, Z.; Xiao, R. Personalized detection of circling exosomal PD-L1 based on Fe3O4@TiO2 isolation and SERS immunoassay. Biosens. Bioelectron. 2019, 148, 111800. [Google Scholar] [CrossRef]
- Wang, Z.; Zong, S.; Wang, Y.; Li, N.; Li, L.; Lu, J.; Wang, Z.; Chen, B.; Cui, Y. Screening and multiple detection of cancer exosomes using an SERS-based method. Nanoscale 2018, 10, 9053–9062. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, C.; Pei, Y.; Song, W.; Zhang, S. Preparation of a Novel Raman Probe and Its Application in the Detection of Circulating Tumor Cells and Exosomes. ACS Appl. Mater. Interfaces 2019, 11, 28671–28680. [Google Scholar] [CrossRef]
- Ma, D.; Huang, C.; Zheng, J.; Tang, J.; Li, J.; Yang, J.; Tan, W. Quantitative detection of exosomal microRNA extracted from human blood based on surface-enhanced Raman scattering. Biosens. Bioelectron. 2018, 101, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Wang, Z.; Zong, S.; Liu, Y.; Yang, K.; Li, N.; Wang, Z.; Li, L.; Tang, H.; Cui, Y. Hydrophobic Plasmonic Nanoacorn Array for a Label-Free and Uniform SERS-Based Biomolecular Assay. ACS Appl. Mater. Interfaces 2020. [Google Scholar] [CrossRef] [PubMed]
- Ning, C.-F.; Wang, L.; Tian, Y.-F.; Yin, B.-C.; Ye, B.-C. Multiple and sensitive SERS detection of cancer-related exosomes based on gold–silver bimetallic nanotrepangs. Analyst 2020, 145, 2795–2804. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.U.; Kim, W.H.; Lee, H.S.; Park, K.H.; Sim, S.J. Quantitative and Specific Detection of Exosomal miRNAs for Accurate Diagnosis of Breast Cancer Using a Surface-Enhanced Raman Scattering Sensor Based on Plasmonic Head-Flocked Gold Nanopillars. Small 2019, 15, e1804968. [Google Scholar] [CrossRef] [PubMed]
- Tsang, D.K.H.; Lieberthal, T.; Watts, C.; Dunlop, I.E.; Ramadan, S.; Hernández, A.E.D.R.; Klein, N. Chemically Functionalised Graphene FET Biosensor for the Label-free Sensing of Exosomes. Sci. Rep. 2019, 9, 13946. [Google Scholar] [CrossRef]
- Yu, Y.; Li, Y.-T.; Jin, D.; Yang, F.; Wu, D.; Xiao, M.-M.; Zhang, H.; Zhang, Z.; Zhang, G.-J. Electrical and Label-Free Quantification of Exosomes with a Reduced Graphene Oxide Field Effect Transistor Biosensor. Anal. Chem. 2019, 91, 10679–10686. [Google Scholar] [CrossRef]
- Etayash, H.; McGee, A.R.; Kaur, K.; Thundat, T. Nanomechanical sandwich assay for multiple cancer biomarkers in breast cancer cell-derived exosomes. Nanoscale 2016, 8, 15137–15141. [Google Scholar] [CrossRef]
- Chang, Y.-J.; Yang, W.-T.; Wu, J.-C. Isolation and detection of exosomes via AAO membrane and QCM measurement. Microelectron. Eng. 2019, 216, 111094. [Google Scholar] [CrossRef]
- Suthar, J.; Parsons, E.S.; Hoogenboom, B.W.; Williams, G.R.; Guldin, S. Acoustic Immunosensing of Exosomes Using a Quartz Crystal Microbalance with Dissipation Monitoring. Anal. Chem. 2020, 92, 4082–4093. [Google Scholar] [CrossRef]
- Zhu, F.; Li, D.; Ding, Q.; Lei, C.; Ren, L.; Ding, X.; Sun, X. 2D magnetic MoS2–Fe3O4 hybrid nanostructures for ultrasensitive exosome detection in GMR sensor. Biosens. Bioelectron. 2020, 147, 111787. [Google Scholar] [CrossRef]
- Ertsgaard, C.T.; Wittenberg, N.J.; Klemme, D.J.; Barik, A.; Shih, W.-C.; Oh, S.-H. Integrated Nanogap Platform for Sub-Volt Dielectrophoretic Trapping and Real-Time Raman Imaging of Biological Nanoparticles. Nano Lett. 2018, 18, 5946–5953. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Dutta, S.; Liu, Z.; Yu, X.; Mesgarzadeh, N.; Ji, F.; Bitan, G.; Xie, Y.-H. A Label-Free Platform for Identification of Exosomes from Different Sources. ACS Sens. 2019, 4, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Rabinowits, G.; Gercel-Taylor, C.; Day, J.M.; Taylor, U.D.; Kloecker, G.H. Exosomal MicroRNA: A Diagnostic Marker for Lung Cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating Exosomal microRNAs as Biomarkers of Colon Cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef]
- Cho, S.; Yang, H.C.; Rhee, W.J. Simultaneous multiplexed detection of exosomal microRNAs and surface proteins for prostate cancer diagnosis. Biosens. Bioelectron. 2019, 146, 111749. [Google Scholar] [CrossRef]
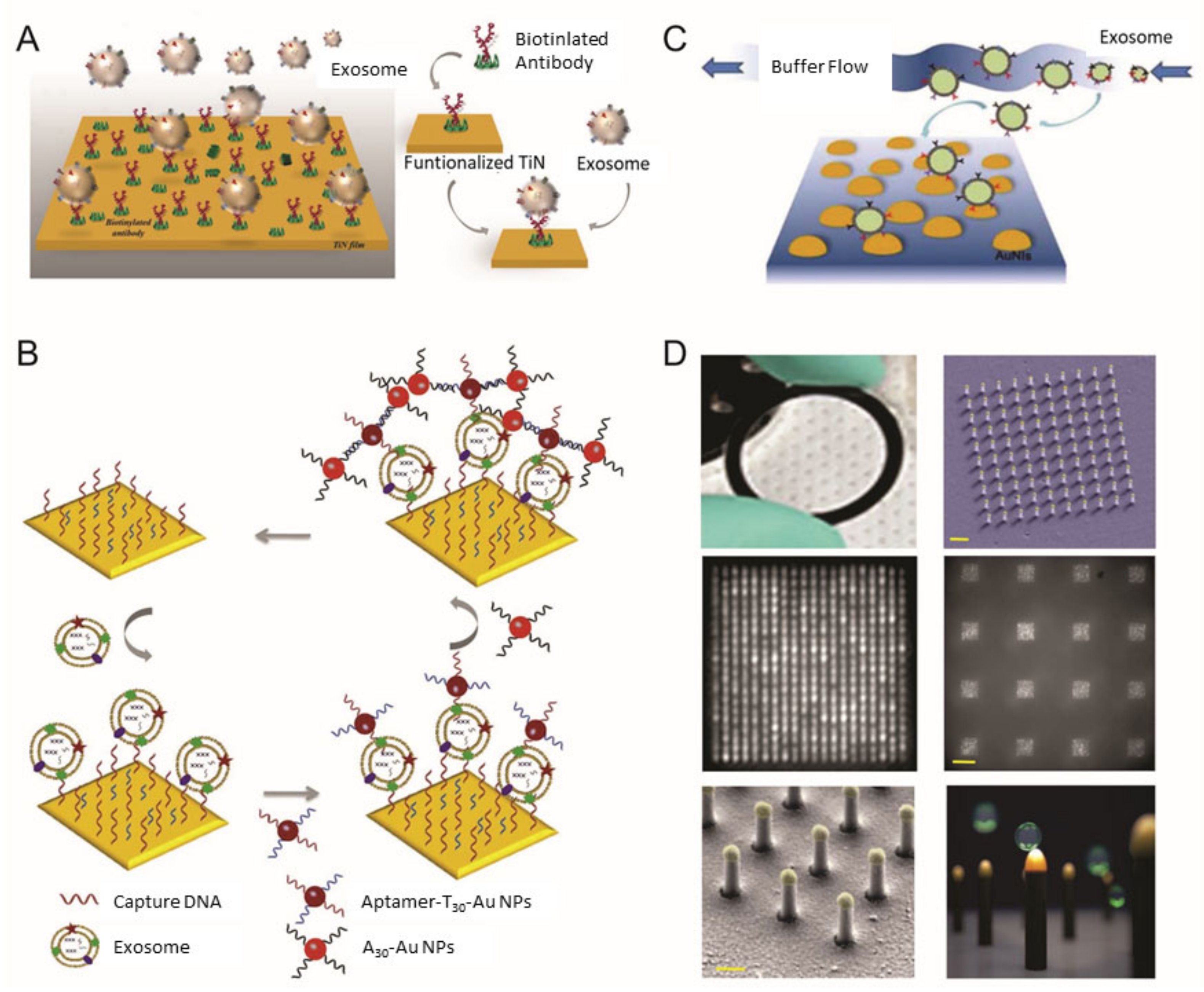
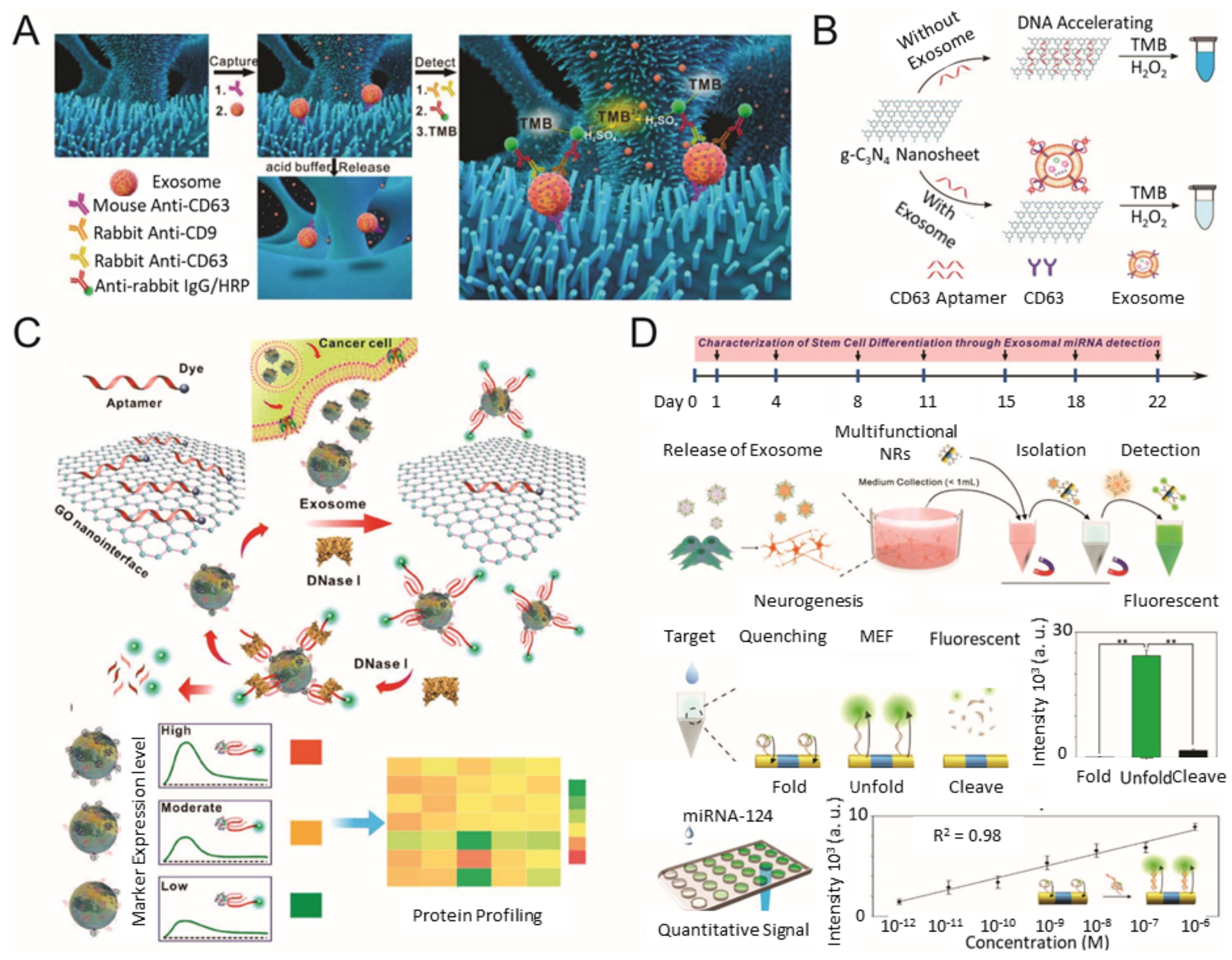
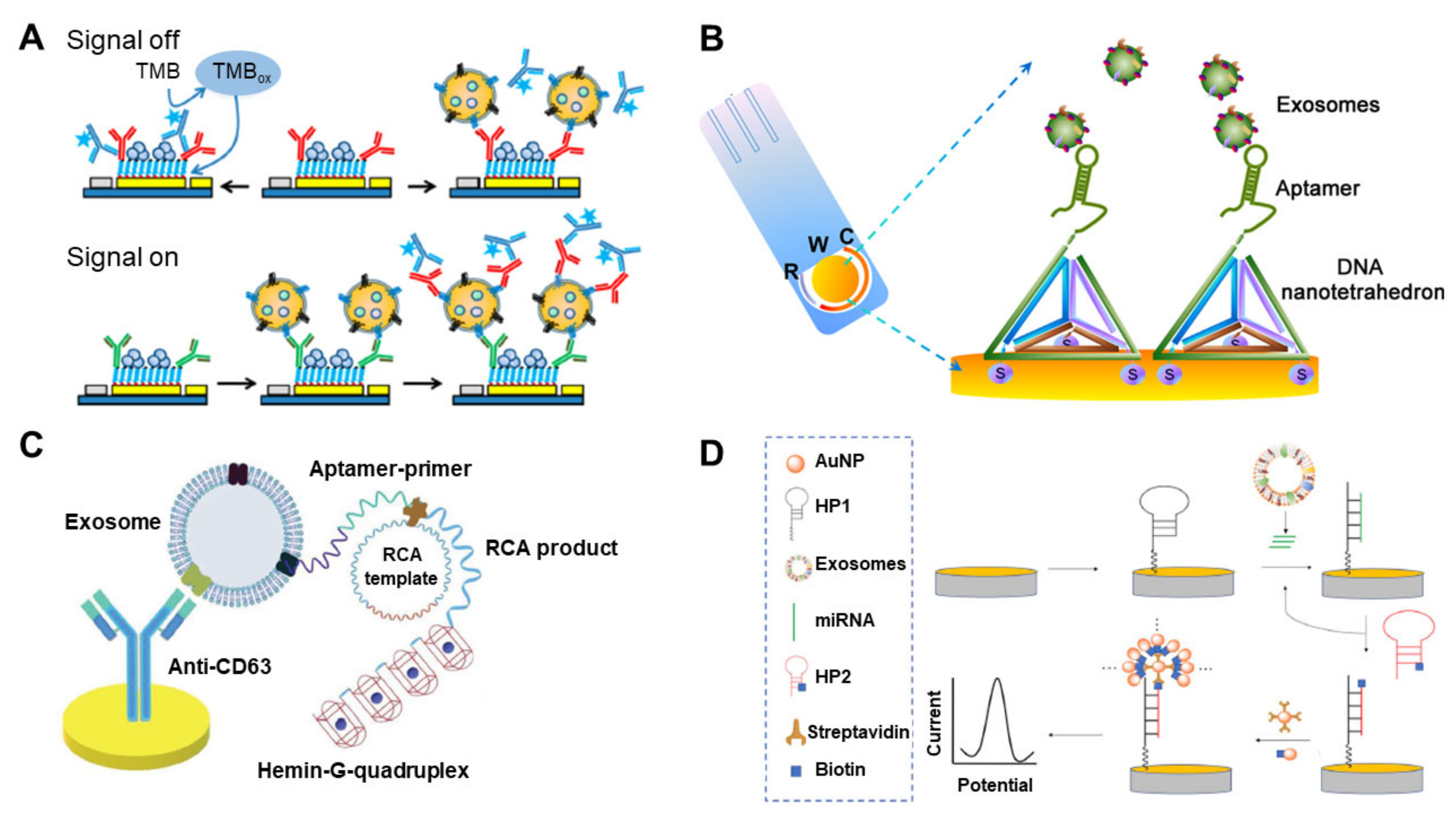
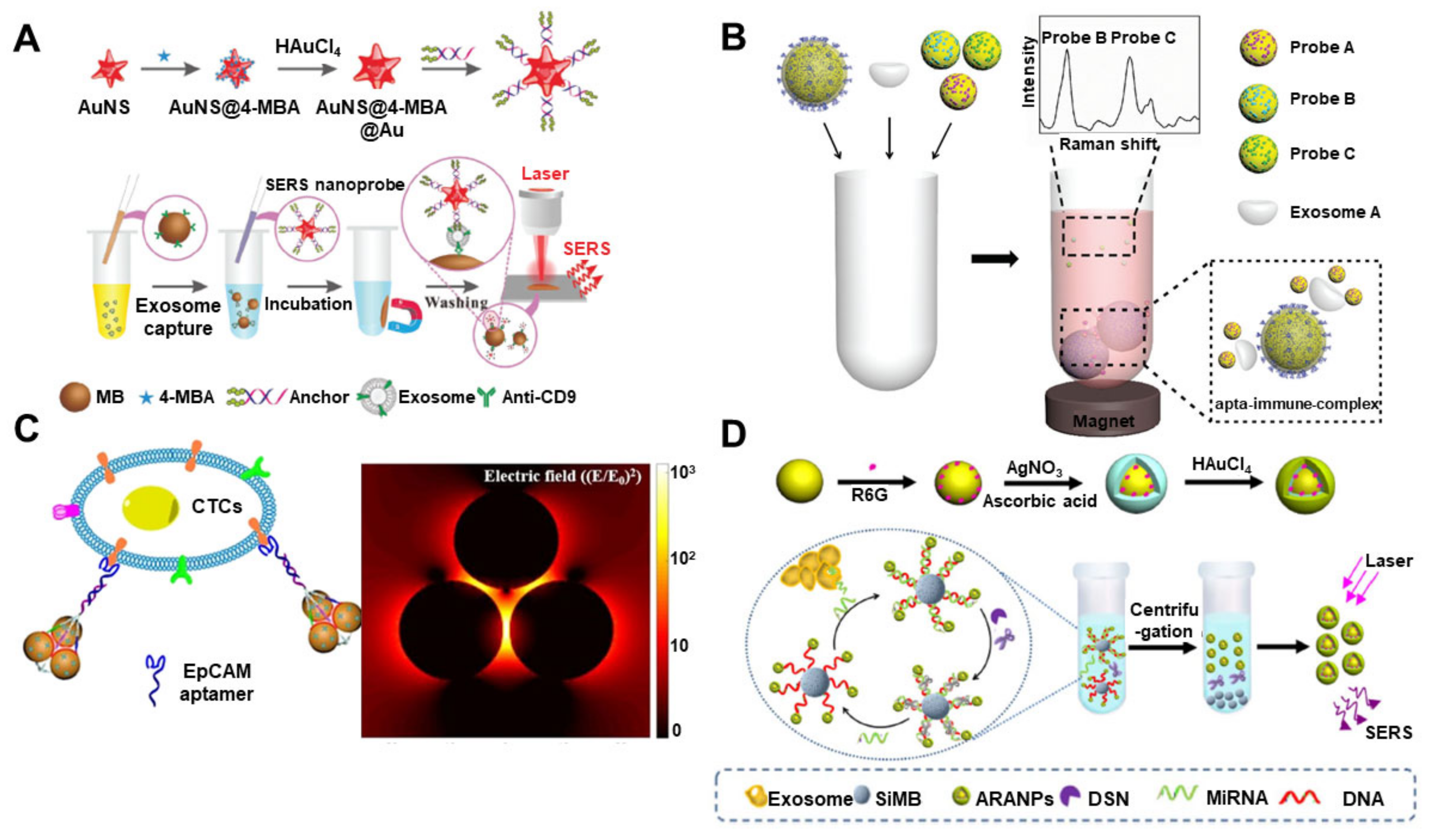
| Method | Working Principle | Target | Correlation Range | Detection Limit | Ref |
|---|---|---|---|---|---|
| SPR | TiN film functionalized by biotinylated anti-epidermal growth factor receptor variant-III (EGFRvIII) antibodies | Glioma cells (U251) | 0.005–500 µg/mL | 2.75 × 10−3 µg/mL | [41] |
| Dual signal amplification via two-step hybridization using aptamer functionalized Au NPs | MCF-7 breast cancer cells and MCF-10A normal breast cells | Not stated | 50 EVs/μL | [42] | |
| LSPR | Periodic nanohole arrays with gold layer functionalized antibodies | Ovarian cancer | 4.03 × 102–1.32 × 106 EVs μL | 4.03 × 102 EVs/μL | [45] |
| Self-assembled Au NIs without functionalization | A-549 and SH-SY5Y cells | 0.194–100 μg/mL | 0.194 μg/mL | [43] | |
| Au nanoplasmonic array functionalized with anti-CD63 antibody | Breast cancer | Not stated | 1 × 102 EVs/μL | [46] |
| Method | Working Principle | Target | Correlation Range | Detection Limit | Ref |
|---|---|---|---|---|---|
| Colorimetric | ZnO-nanowires-coated three-dimensional (3D) scaffold chip | Breast cancer (MCF-7) | 2.2 × 105–2.4 × 107 EVs/μL | 2.2 × 104 EVs/μL | [52] |
| microfluidic chip with self-assembled three-dimensional herringbone nanopatterns (nano–HB) | Ovarian cancer | 1 × 103–5 × 105 EVs/μL | 10 EVs/μL | [53] | |
| graphitic carbon nitride nanosheets (g-C3N4 NSs) | Breast cancer (MCF-7) | 0.19 × 107–3.38 × 107 EVs/μL | 1.352 × 103 EVs/μL | [54] | |
| aptamer-CD63 functionalized single-walled carbon nanotubes (s-SWCNTs) | Breast cancer (MCF-7) | 1.84 × 106–2.21 × 107 EVs/μL | 5.2 × 102 EVs/μL | [56] | |
| EpCAM aptamer modified Fe3O4 NPs | Prostate cancer | 0.4 × 105–6.0 × 105 EVs/μL | 3.58 × 103 EVs/μL | [57] | |
| Au NPs complexed with a panel of aptamers | Prostate cancer | 0–12.8 μg/mL | Not stated | [58] | |
| PLA–RPA–TMA assay | Nasopharyngeal carcinoma cell | 0.1–105 EVs/μL | 0.1 EVs/μL | [59] | |
| Fluorescence | Fluorescent labeled aptamer/GO nanoprobe | Prostate cancer | 1.6 × 102–1.6 × 105 EVs/μL | 1.6 x 102 EVs/μL | [65] |
| Cy3 labeled aptamer–CD63 and Ti3C2 MXenes complex | Melanoma (B16), Breast cancer (MCF-7), ovarian carcinoma (OVCAR-3), liver cancer (Hep G2) | 10–106 EVs/μL | 0.1 EVs/μL | [61] | |
| Aptamer modified UCNP and Au NRs | liver cancer (Hep G2) | 1.0 × 104–1.0 × 109 EVs/μL | 1.1 × 103 EVs/μL | [62] | |
| Multifunctional magneto-plasmonic NRs | Neurogenesis(miR-124) | 1 Pm–106 pM | 1 pM | [63] | |
| Copper-mediated fluorescent signal amplification | Cancer (Hep G2) | 7.5 × 104 to 1.5 × 107 EVs/μL | 4.8 × 104 EVs/μL | [64] |
| Method | Working Principle | Target | Correlation Range | Detection Limit | Ref |
|---|---|---|---|---|---|
| Electrochemical | Electrochemical sandwich immunosensor (amperometry) | Breast cancer (MCF-7) | 2 × 102–1 × 106 EVs/μL | 2 × 102/μL | [75] |
| Cascade toehold-mediated strand displacement reaction (CTSDR) (voltammetry) | Liver cancer (HepG2) | 1 × 103 to 5 × 105 EVs/μL | 1.72 × 102/μL | [76] | |
| Electrochemical immunosensor using magnetic bead (amperometry) | Breast cancer (MCF7, MDA-MB-231 and SK-BR-3) | 1 × 102–1 × 106 EVs/μL | 102/μL | [77] | |
| Electrochemical sensor based on graphene oxide-cucurbit modified carbon electrode (voltammetry) | Breast cancer (MCF-7, SK-BR-3, MDA-MB-231 and BT474) | 1.2 × 103–1.2 × 107 EVs/μL | 1.2 × 103/μL | [78] | |
| Electrochemical aptasensor using DNA nanotetrahedron and aptamer (voltammetry) | Liver cancer (HepG2) | 1 × 102–1 × 109 EVs/μL | 102/μL | [79] | |
| Electrochemical aptasensor by cyclic enzymatic amplification (voltammetry) | Prostate cancer and breast cancer (LNCaP and MCF-7) | 70 to 1 × 105 EVs/μL | 70/μL | [82] | |
| G-quadruplex circular template triggered rolling circle amplification (RCA) for electrochemical sensor (voltammetry) | Gastric cancer (GES-1 and SGC7901) | 0.954–7.8 × 103 EVs/μL | 0.954 /μL | [80] | |
| Electrochemical biosensor based on click chemistry of alkynyl-4-ONE (voltammetry) | Breast cancer (MCF-7) | 1.12 × 102–1.12 × 108 EVs/μL | 1.12 × 102/μL | [83] | |
| Electrochemical biosensor by coupling the DNA walking machine (voltammetry) | Breast cancer (MCF-7) | 13–1.0 × 107/μL | 13/μL | [84] | |
| Exo III-assisted cycling reaction for signal amplification (voltammetry) | Breast cancer (MCF-7) | 12–3.4 × 105 EVs/μL | 12/μL | [85] | |
| Electrochemical detection of exosomal miRNAs by using magnetic separation (voltammetry) | miR-21 | 1 pM–100 nM | 1 pM | [89] | |
| Electrochemical biosensor by Y-shaped locked nucleic acid (LNA) (voltammetry) | miR-21 | 10–70 fM | 2.3 fM | [90] | |
| Electrochemical assay of miR-122 by hybridization chain reaction (HCR) (voltammetry) | miR-122 | 1 × 102–1 × 1011 aM | 53 aM | [91] | |
| Enzyme-free electrochemical biosensor by the double signal amplification strategy (voltammetry) | miR-21 | 1 fM–200 pM | 0.4 fM | [81] |
| Method | Working Principle | Target | Correlation Range | Detection Limit | Ref |
|---|---|---|---|---|---|
| Raman | SERS-based immunosensor by gold nanostar@4-mercaptobenzoic acid@nanoshell structures | Liver cancer (HepG2) | 40–4 × 107 EVs/μL | 27/μL | [97] |
| Miniaturized affinity-based SERS-sensitive device by 3D printing | Breast cancer (MDA-MB-231, MDA-MB-468 and SK-BR-3) | 106–108 EVs/μL | 2×103/μL | [98] | |
| SERS-based EV profiling platform using surface proteins | Colorectal cancer and bladder cancer (SW480 and C3) | 2.3 × 103–2.3 × 108 EVs/μL | 2.3×103/μL | [99] | |
| SERS immunoassay using anti-PD-L1-functionalized Fe3O4@TiO2 nanoparticles | Lung cancer (A549) | 5–2 × 102 EVs/μL | 1/μL | [100] | |
| SERS-based multi-EV detection by MB@SiO2@Au nanoparticle | Breast cancer and prostate cancer (SK-BR-3, T84 and LNCaP) | 32–3.2 × 105 EVs/μL | 32/μL | [101] | |
| SERS-based aptasensor by Au nanoparticles and triangular pyramid DNA | Breast cancer and cervical cancer (MCF-7, Hela and HEK-293 T) | 1.0 × 103–1.0 × 107 EVs/μL | 1.0 × 103/μL | [102] | |
| SERS biosensor with hydrophobic assembled nanoacorn and Au@Ag nanocubes | Breast cancer (MCF7, MBA-MD-231) | 50–1.0 × 106 EVs/μL | 50/μL | [104] | |
| SERS aptasensor by gold–silver–silver core–shell–shell nanotrepangs | Breast cancer, prostate cancer and liver cancer (SK-BR-3, HepG2 and LNCaP) | 1–104 EVs/μL | 1/μL | [105] | |
| SERS-based sensor with duplex-specific nuclease (DSN) and Au@R6G@AgAu nanoparticles | miR-21 | 5 fM–20 pM | 5 fM | [103] | |
| Uniform plasmonic head-flocked gold nanopillar substrate for the signal enhancement of SERS | miR-21, 222, 200c | 1 aM–100 nM | 1 aM | [106] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, J.-H.; Lee, J.-H.; Choi, J.-W. Applications of Bionano Sensor for Extracellular Vesicles Analysis. Materials 2020, 13, 3677. https://doi.org/10.3390/ma13173677
Choi J-H, Lee J-H, Choi J-W. Applications of Bionano Sensor for Extracellular Vesicles Analysis. Materials. 2020; 13(17):3677. https://doi.org/10.3390/ma13173677
Chicago/Turabian StyleChoi, Jin-Ha, Jin-Ho Lee, and Jeong-Woo Choi. 2020. "Applications of Bionano Sensor for Extracellular Vesicles Analysis" Materials 13, no. 17: 3677. https://doi.org/10.3390/ma13173677
APA StyleChoi, J.-H., Lee, J.-H., & Choi, J.-W. (2020). Applications of Bionano Sensor for Extracellular Vesicles Analysis. Materials, 13(17), 3677. https://doi.org/10.3390/ma13173677







