Efficient Doxorubicin Loading to Isolated Dexosomes of Immature JAWSII Cells: Formulated and Characterized as the Bionanomaterial
Abstract
1. Introduction
2. Materials and Methods
2.1. Pretreated Solutions
2.2. Dexosome Production and Isolation
2.3. Evaluation of Dexosomal RNA and Protein Content
2.4. Confocal Studies after CellMask Membrane Staining
2.5. Loading of Dexosomes with Doxorubicin by Ultrasonication
2.6. NTA Analysis
2.7. DLS Analysis
2.8. SEM Analysis
2.9. In Vitro Cytotoxic Assay
2.10. Drug Release Analysis
3. Results
3.1. Evaluation of Dexosomal RNA and Protein Content
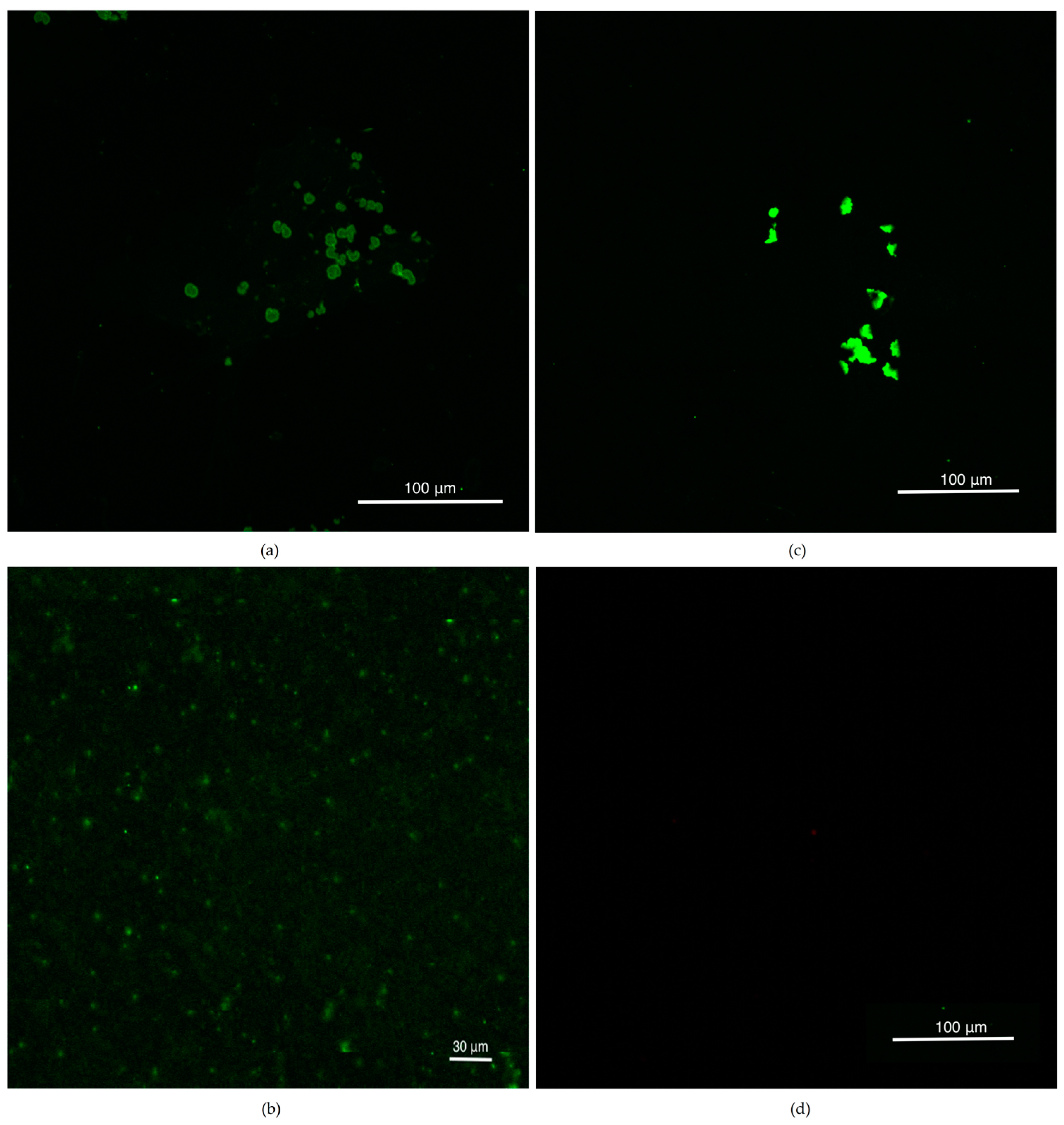
3.2. Confocal Studies after CellMask Membrane Staining
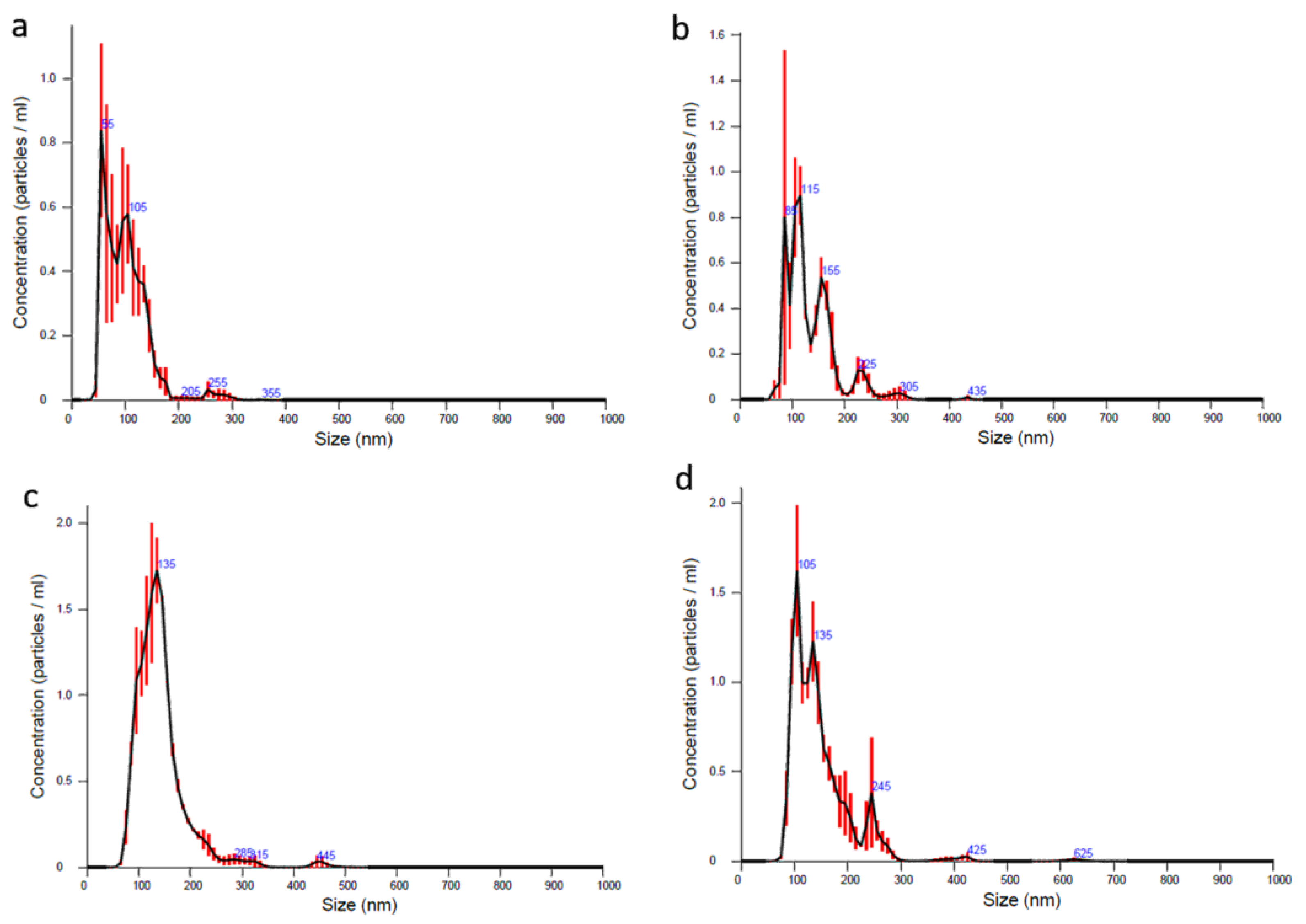
3.3. NTA Analysis
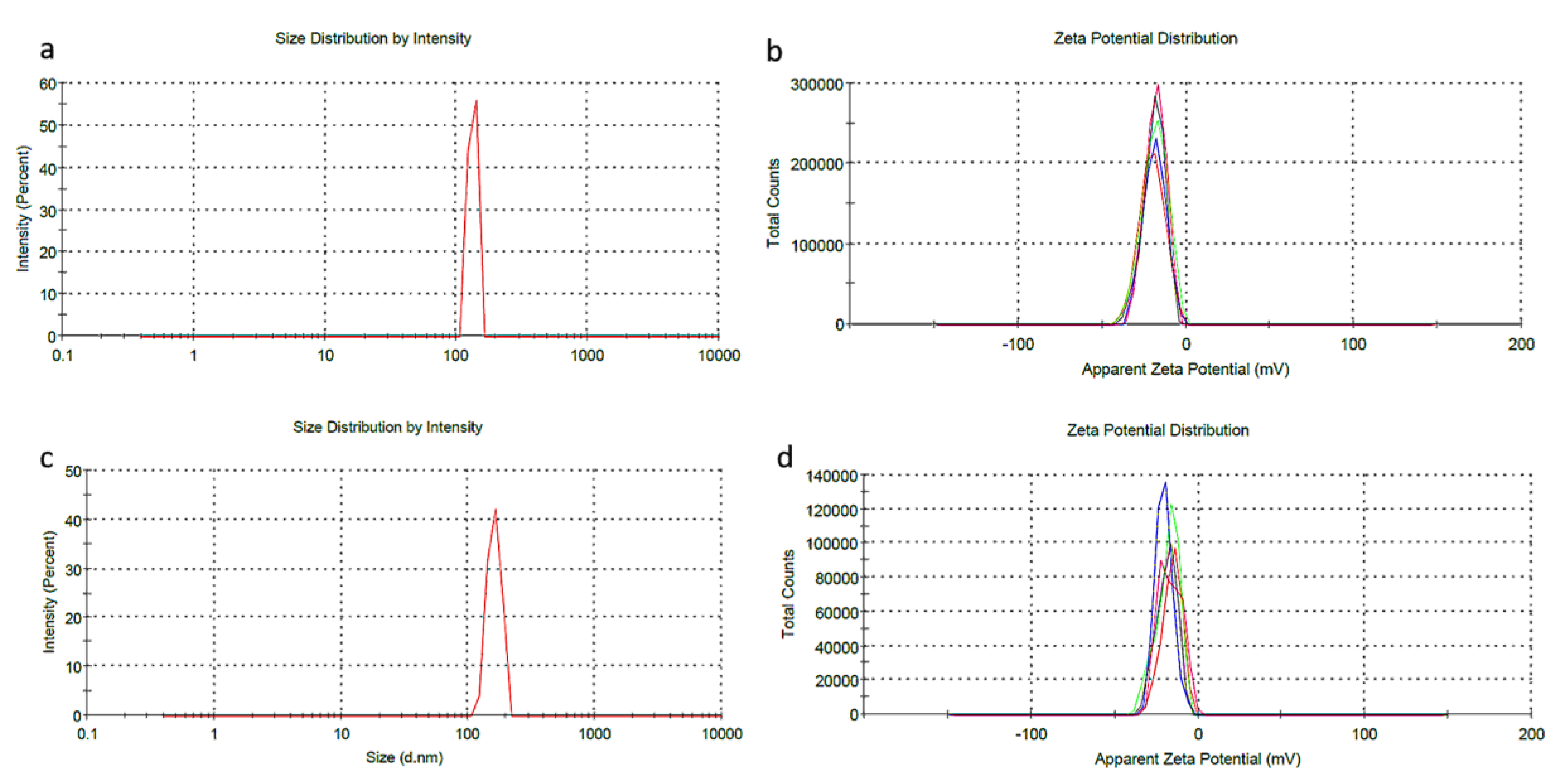
3.4. DLS Analysis
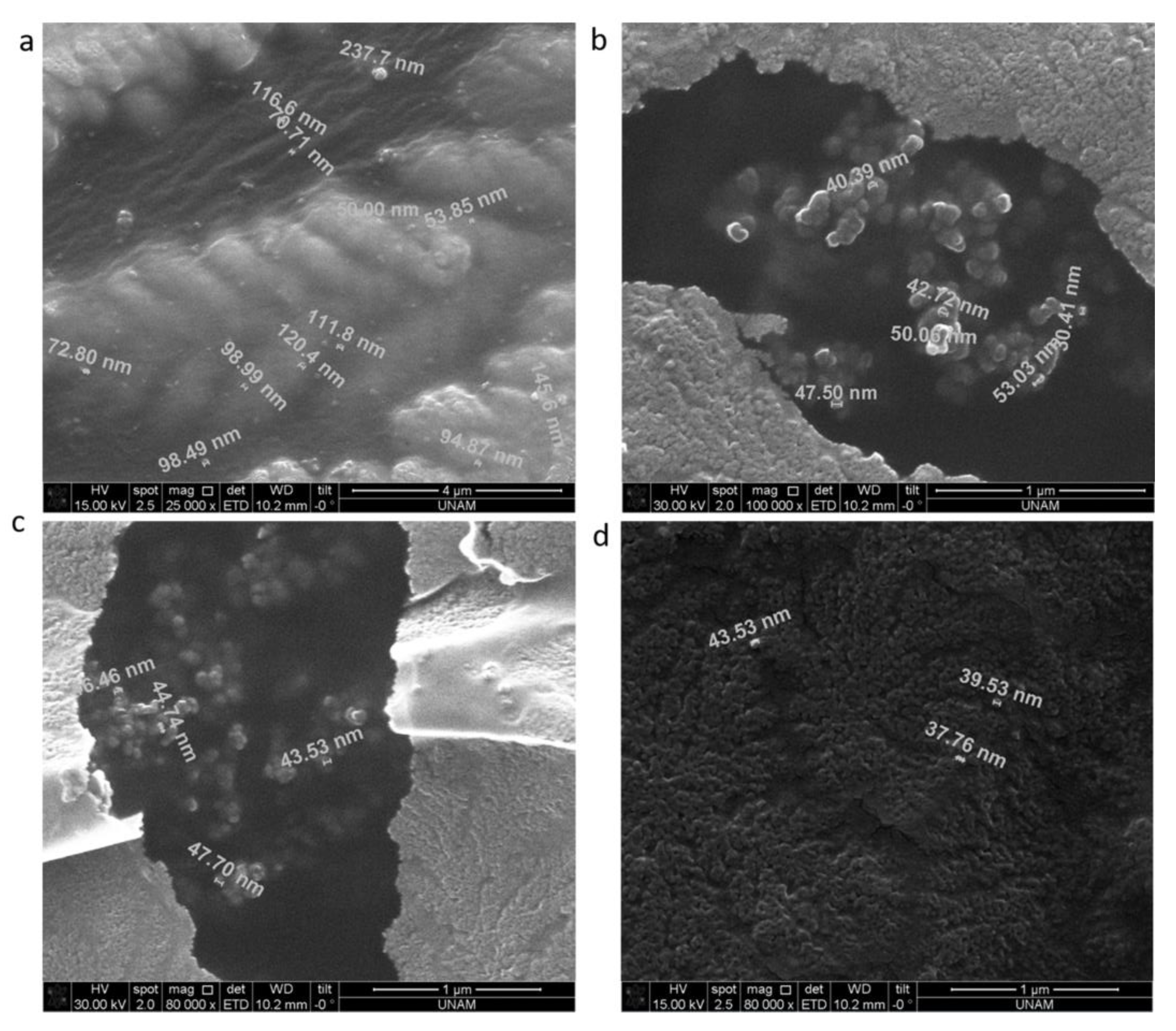
3.5. SEM Analysis
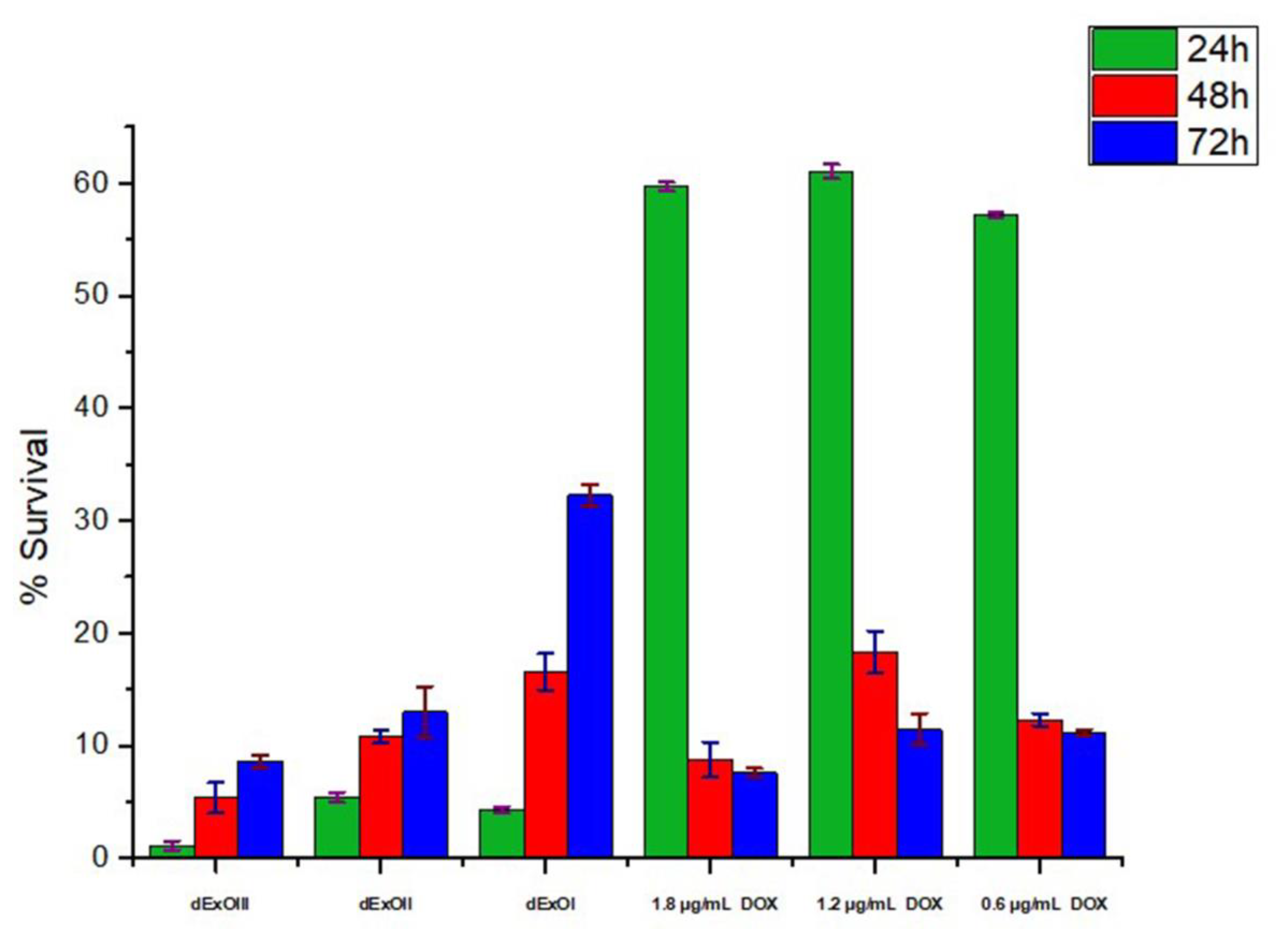
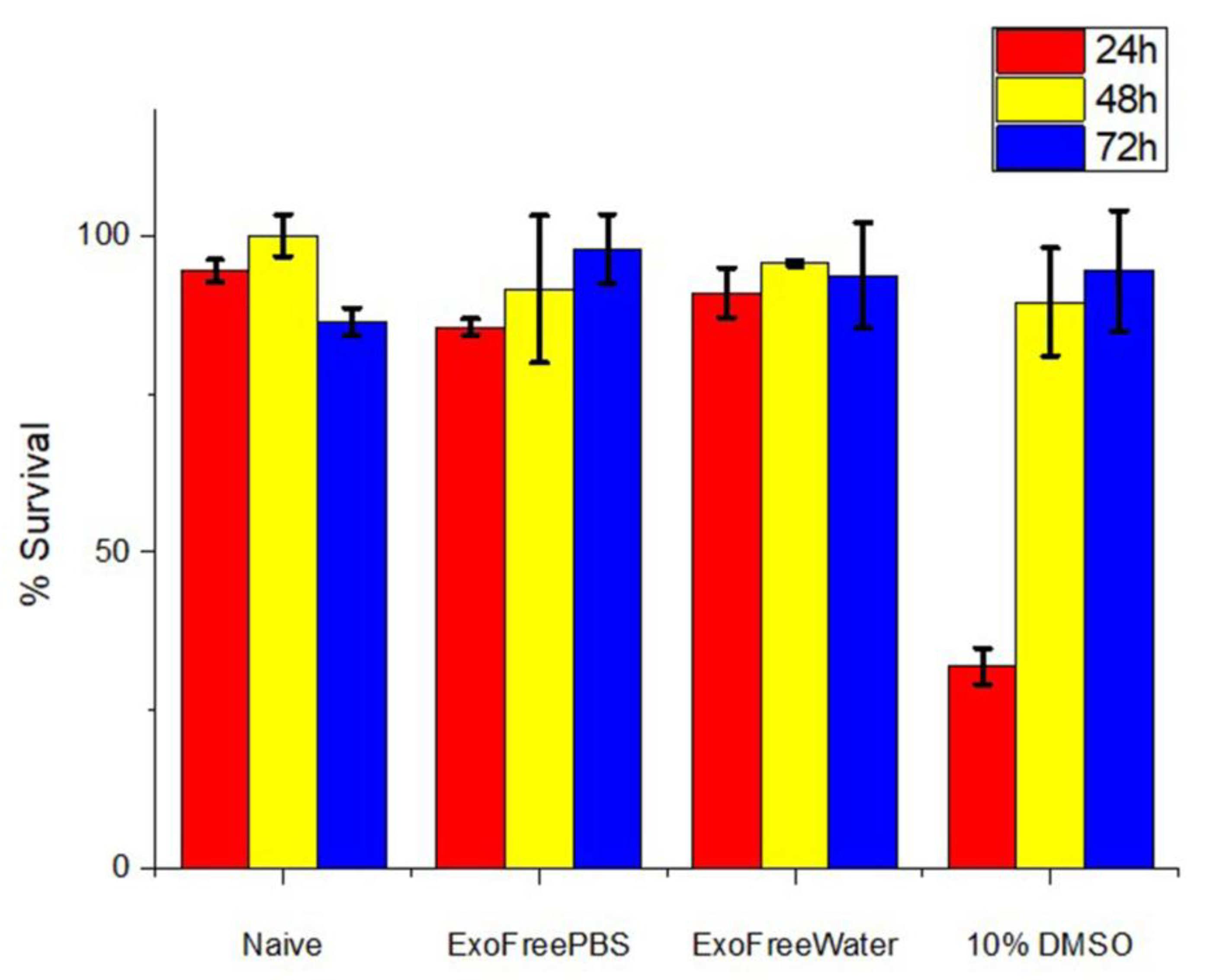
3.6. In Vitro Cytotoxic Assay
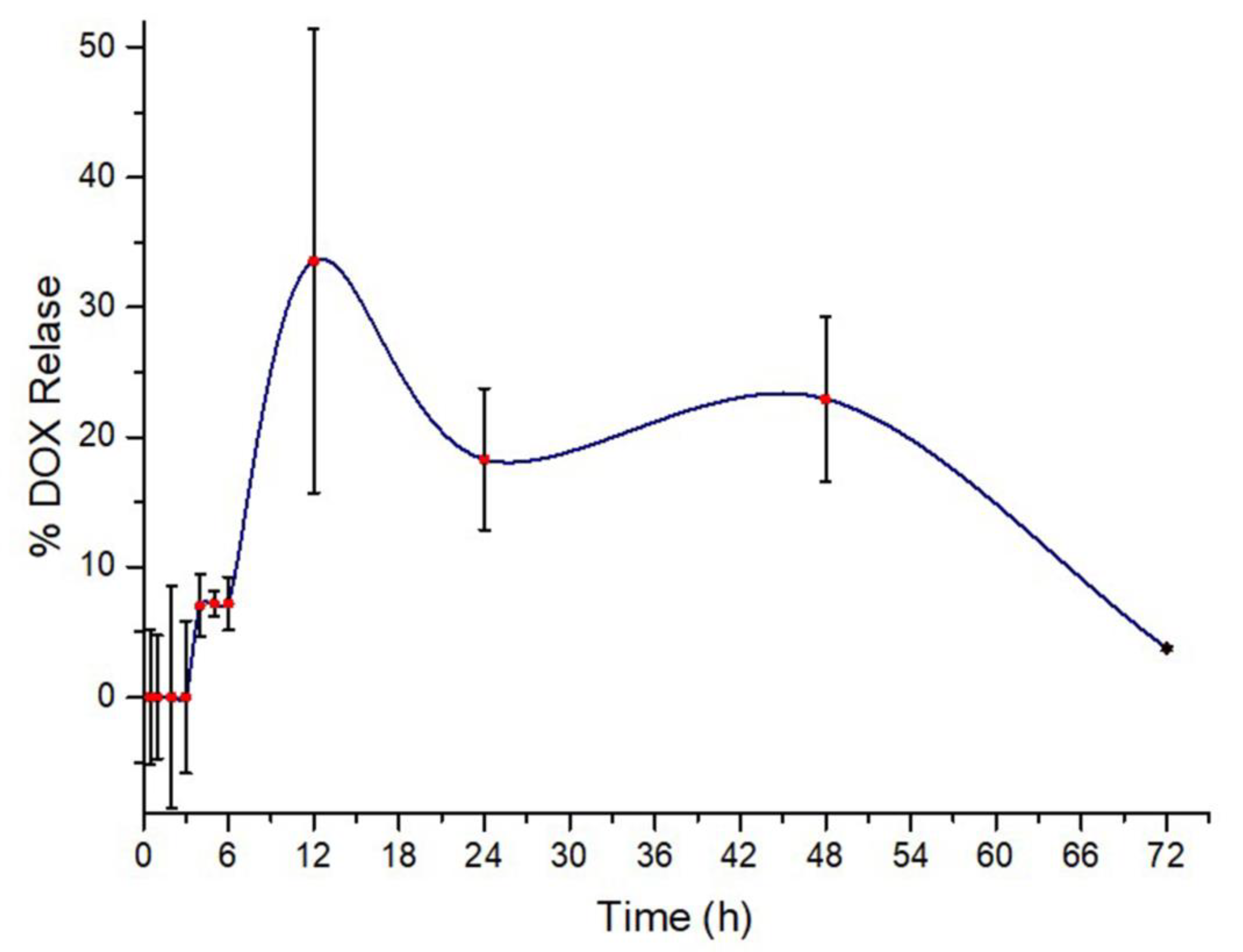
3.7. Drug Release Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nieto-Miguel, T.; Galindo, S.; López-Paniagua, M.; Pérez, I.; Herreras, J.M.; Calonge, M. Cell therapy using extraocular mesenchymal stem cells. In Corneal Regeneration; Springer: Berlin/Heidelberg, Germany, 2019; pp. 231–262. [Google Scholar]
- Hean, J.; Mager, I.; Wood, M.; Andaloussi, S.E.; Wiklander, O.; Nordin, J. Exosomes Comprising Therapeutic Polypeptides. U.S. Patents 2019/0167810A1, 6 June 2019. [Google Scholar]
- Kowal, J.; Tkach, M. Dendritic cell extracellular vesicles. Int. Rev. Cell Mol. Biol. 2019, 349, 213–249. [Google Scholar] [PubMed]
- Lee, Y.; El Andaloussi, S.; Wood, M.J. Exosomes and microvesicles: Extracellular vesicles for genetic information transfer and gene therapy. Hum. Mol. Genet. 2012, 21, R125–R134. [Google Scholar] [CrossRef] [PubMed]
- Willms, E. Development of Targeted Exosomes for Delivery of Biotherapeutics. Ph.D. Thesis, University of Oxford, Oxford, UK, 2018. [Google Scholar]
- Pang, X.-L.; Wang, Z.-G.; Liu, L.; Feng, Y.-H.; Wang, J.-X.; Xie, H.-C.; Yang, X.-L.; Li, J.-F.; Feng, G.-W. Immature dendritic cells derived exosomes promotes immune tolerance by regulating T cell differentiation in renal transplantation. Aging (Albany NY) 2019, 11, 8911–8924. [Google Scholar] [CrossRef] [PubMed]
- Matzinger, P. The danger model: A renewed sense of self. Science 2002, 296, 301–305. [Google Scholar] [CrossRef]
- Lutz, M.B.; Schuler, G. Immature, semi-mature and fully mature dendritic cells: Which signals induce tolerance or immunity? Trends Immunol. 2002, 23, 445–449. [Google Scholar] [CrossRef]
- Mellman, I. Dendritic cells: Master regulators of the immune response. Cancer Immunol. Res. 2013, 1, 145–149. [Google Scholar] [CrossRef]
- Yong, T.; Zhang, X.; Bie, N.; Zhang, H.; Zhang, X.; Li, F.; Hakeem, A.; Hu, J.; Gan, L.; Santos, H.A. Tumor exosome-based nanoparticles are efficient drug carriers for chemotherapy. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Gilligan, K.E.; Dwyer, R.M. Engineering exosomes for cancer therapy. Int. J. Mol. Sci. 2017, 18, 1122. [Google Scholar] [CrossRef]
- Wiklander, O.P.; Brennan, M.Á.; Lötvall, J.; Breakefield, X.O.; Andaloussi, S.E. Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 2019, 11, eaav8521. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- van Dommelen, S.M.; Vader, P.; Lakhal, S.; Kooijmans, S.; van Solinge, W.W.; Wood, M.J.; Schiffelers, R.M. Microvesicles and exosomes: Opportunities for cell-derived membrane vesicles in drug delivery. J. Control. Release 2012, 161, 635–644. [Google Scholar] [CrossRef]
- Vader, P.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Emerging targets for cancer therapy. Trends Mol. Med. 2014, 20, 385–393. [Google Scholar] [CrossRef] [PubMed]
- El-Andaloussi, S.; Lee, Y.; Lakhal-Littleton, S.; Li, J.; Seow, Y.; Gardiner, C.; Alvarez-Erviti, L.; Sargent, I.L.; Wood, M.J. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat. Protoc. 2012, 7, 2112. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Shen, C.; Rey-Ladino, J.; Yu, H.; Brunham, R.C. Characterization of murine dendritic cell line JAWS II and primary bone marrow-derived dendritic cells in Chlamydia muridarum antigen presentation and induction of protective immunity. Infect. Immun. 2008, 76, 2392–2401. [Google Scholar] [CrossRef] [PubMed]
- Wiklander, O.P.; Nordin, J.Z.; Gupta, D.; Seow, Y.; Balusu, S.; Liang, X.-M.; Corso, G.; Felldin, U.; Conceicao, M.; Gustafsson, M. Engineered Extracellular Vesicles as Therapeutic Decoys. J. Extracell. Vesicles 2018, 7, 80–81. [Google Scholar]
- Yang, F.; Teves, S.S.; Kemp, C.J.; Henikoff, S. Doxorubicin, DNA torsion, and chromatin dynamics. Biochim. Biophys. Acta 2014, 1845, 84–89. [Google Scholar] [CrossRef]
- Melguizo, C.; Cabeza, L.; Prados, J.; Ortiz, R.; Caba, O.; Rama, A.R.; Delgado, Á.V.; Arias, J.L. Enhanced antitumoral activity of doxorubicin against lung cancer cells using biodegradable poly (butylcyanoacrylate) nanoparticles. Drug Des. Dev. Ther. 2015, 9, 6433. [Google Scholar]
- Deng, Z.J.; Morton, S.W.; Ben-Akiva, E.; Dreaden, E.C.; Shopsowitz, K.E.; Hammond, P.T. Layer-by-layer nanoparticles for systemic codelivery of an anticancer drug and siRNA for potential triple-negative breast cancer treatment. ACS Nano 2013, 7, 9571–9584. [Google Scholar] [CrossRef]
- Zickler, A.M.; Andaloussi, S.E. Functional extracellular vesicles aplenty. Nat. Biomed. Eng. 2020, 4, 9–11. [Google Scholar] [CrossRef]
- Lässer, C.; Eldh, M.; Lötvall, J. Isolation and characterization of RNA-containing exosomes. J. Vis. Exp. 2012, 59, e3037. [Google Scholar] [CrossRef]
- Mutlu, E.C. Overcoming Challenges Across Production, Isolation and Antineoplastic Drug Loading of Exosomes. In Proceedings of the 5th World Congress on Electrical Engineering and Computer Systems and Sciences (EECSS’19), Lisbon, Portugal, 21–23 August 2018. [Google Scholar]
- Mutlu, E.; Kaya, Ö.; Yildirim, A.B.; Çetinkaya, A. Exosome Production, Isolation and Characterization from A549 Epithelial Carcinoma Cells. Hacet. J. Biol. Chem. 2020, 8, 307. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Bradford, H.F.; Ward, H.K. On glutaminase activity in mammalian synaptosomes. Brain Res. 1976, 110, 115–125. [Google Scholar] [CrossRef]
- Sirelkhatim, A.; Mahmud, S.; Seeni, A.; Kaus, N.H.M.; Ann, L.C.; Bakhori, S.K.M.; Hasan, H.; Mohamad, D. Review on zinc oxide nanoparticles: Antibacterial activity and toxicity mechanism. Nanomicro Lett. 2015, 7, 219–242. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine 2016, 12, 655–664. [Google Scholar] [CrossRef]
- Baek, N.; Seo, O.W.; Kim, M.; Hulme, J.; An, S.S.A. Monitoring the effects of doxorubicin on 3D-spheroid tumor cells in real-time. OncoTargets Ther. 2016, 9, 7207. [Google Scholar] [CrossRef]
- Cole, S. Rapid chemosensitivity testing of human lung tumor cells using the MTT assay. Cancer Chemother. Pharmacol. 1986, 17, 259–263. [Google Scholar] [CrossRef]
- Sari-Chmayssem, N.; Taha, S.; Mawlawi, H.; Guégan, J.-P.; Jeftić, J.; Benvegnu, T. Extracted and depolymerized alginates from brown algae Sargassum vulgare of Lebanese origin: Chemical, rheological, and antioxidant properties. J. Appl. Phycol. 2016, 28, 1915–1929. [Google Scholar] [CrossRef]
- Srivastava, A.; Amreddy, N.; Pareek, V.; Chinnappan, M.; Ahmed, R.; Mehta, M.; Razaq, M.; Munshi, A.; Ramesh, R. Progress in extracellular vesicle biology and their application in cancer medicine. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1621. [Google Scholar] [CrossRef]
- Cvjetkovic, A.; Crescitelli, R.; Lässer, C.; Zabeo, D.; Widlund, P.; Nyström, T.; Höög, J.L.; Lötvall, J. Extracellular vesicles in motion. Matters 2017. [Google Scholar] [CrossRef][Green Version]
- Agrawal, A.K.; Aqil, F.; Jeyabalan, J.; Spencer, W.A.; Beck, J.; Gachuki, B.W.; Alhakeem, S.S.; Oben, K.; Munagala, R.; Bondada, S. Milk-derived exosomes for oral delivery of paclitaxel. Nanomedicine 2017, 13, 1627–1636. [Google Scholar] [CrossRef] [PubMed]
- Im, H.; Yang, K.S.; Lee, H.; Castro, C.M. Nanotechnology Platforms for Cancer Exosome Analyses. In Diagnostic and Therapeutic Applications of Exosomes in Cancer; Elsevier: Amsterdam, The Netherlands, 2018; pp. 119–128. [Google Scholar]
- Li, X.; Corbett, A.L.; Taatizadeh, E.; Tasnim, N.; Little, J.P.; Garnis, C.; Daugaard, M.; Guns, E.; Hoorfar, M.; Li, I.T. Challenges and opportunities in exosome research—Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019, 3, 011503. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xu, X.; Li, B.; Situ, B.; Pan, W.; Hu, Y.; An, T.; Yao, S.; Zheng, L. Single-exosome-counting immunoassays for cancer diagnostics. Nano Lett. 2018, 18, 4226–4232. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-P.; Lin, Z.-X.; Jiang, X.-Y.; Yu, X.-Y. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol. Sin. 2018, 39, 542–551. [Google Scholar] [CrossRef]
- Wiklander, O.P.; Nordin, J.Z.; O’Loughlin, A.; Gustafsson, Y.; Corso, G.; Mäger, I.; Vader, P.; Lee, Y.; Sork, H.; Seow, Y. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles 2015, 4, 26316. [Google Scholar] [CrossRef]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome theranostics: Biology and translational medicine. Theranostics 2018, 8, 237. [Google Scholar] [CrossRef]
- Armstrong, J.P.; Stevens, M.M. Strategic design of extracellular vesicle drug delivery systems. Adv. Drug Deliv. Rev. 2018, 130, 12–16. [Google Scholar] [CrossRef]
- Zlotogorski-Hurvitz, A.; Dekel, B.Z.; Malonek, D.; Yahalom, R.; Vered, M. FTIR-based spectrum of salivary exosomes coupled with computational-aided discriminating analysis in the diagnosis of oral cancer. J. Cancer Res. Clin. Oncol. 2019, 145, 685–694. [Google Scholar] [CrossRef]
- He, F.; Ye, Z.-Y.; Zhao, L.-D.; Yin, B.-C.; Ye, B.-C. Probing exosome internalization pathways through confocal microscopy imaging. Chem. Commun. 2019, 55, 14015–14018. [Google Scholar] [CrossRef]
- Mukherjee, S.; Kotcherlakota, R.; Haque, S.; Bhattacharya, D.; Kumar, J.M.; Chakravarty, S.; Patra, C.R. Improved delivery of doxorubicin using rationally designed PEGylated platinum nanoparticles for the treatment of melanoma. Mater. Sci. Eng. C 2020, 108, 110375. [Google Scholar] [CrossRef]
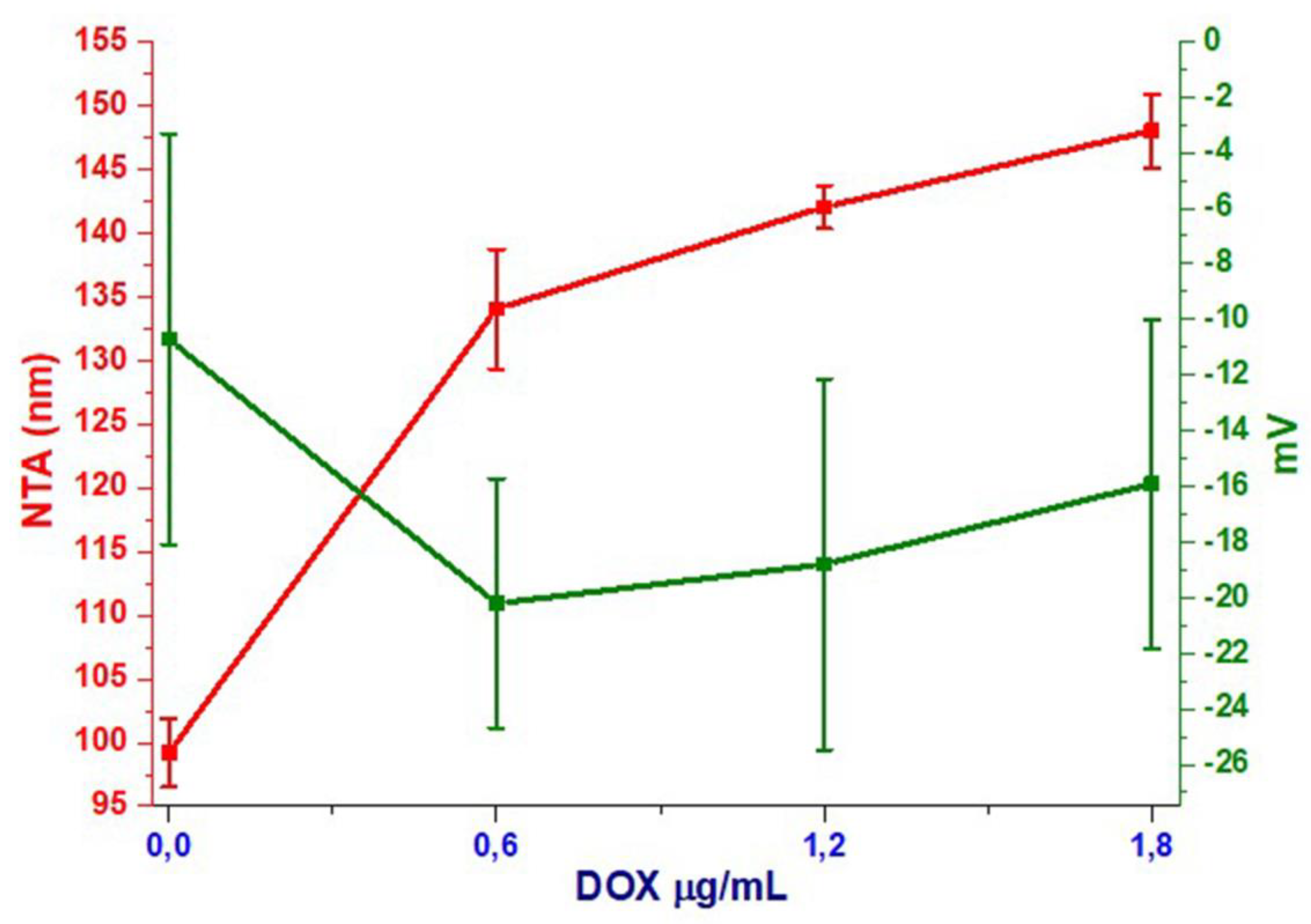
| Sample/Method | Size by NTA (nm) | Z-Intensity (nm) | Zeta Potential (mv) |
|---|---|---|---|
| Naïve | 99.2 ± 2.7 | 209.4 ± 81.7 | −10.7 ± 7.39 |
| dExOIII | 148 ± 2.9 | 161.2 ± 19.26 | −15.9 ± 5.91 |
| dExOII | 142 ± 1.7 | 162.5 ± 17.14 | −18.8 ± 6.67 |
| dExOI | 134 ± 4.7 | 133.2 ± 9.6 | −20.2 ± 4.48 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mutlu, E.C.; Kaya, Ö.; Wood, M.; Mager, I.; Topkara, K.Ç.; Çamsarı, Ç.; Birinci Yildirim, A.; Çetinkaya, A.; Acarel, D.; Odabaşı Bağcı, J. Efficient Doxorubicin Loading to Isolated Dexosomes of Immature JAWSII Cells: Formulated and Characterized as the Bionanomaterial. Materials 2020, 13, 3344. https://doi.org/10.3390/ma13153344
Mutlu EC, Kaya Ö, Wood M, Mager I, Topkara KÇ, Çamsarı Ç, Birinci Yildirim A, Çetinkaya A, Acarel D, Odabaşı Bağcı J. Efficient Doxorubicin Loading to Isolated Dexosomes of Immature JAWSII Cells: Formulated and Characterized as the Bionanomaterial. Materials. 2020; 13(15):3344. https://doi.org/10.3390/ma13153344
Chicago/Turabian StyleMutlu, Esra Cansever, Özge Kaya, Matthew Wood, Imre Mager, Kübra Çelik Topkara, Çağrı Çamsarı, Arzu Birinci Yildirim, Ayhan Çetinkaya, Diğdem Acarel, and Jale Odabaşı Bağcı. 2020. "Efficient Doxorubicin Loading to Isolated Dexosomes of Immature JAWSII Cells: Formulated and Characterized as the Bionanomaterial" Materials 13, no. 15: 3344. https://doi.org/10.3390/ma13153344
APA StyleMutlu, E. C., Kaya, Ö., Wood, M., Mager, I., Topkara, K. Ç., Çamsarı, Ç., Birinci Yildirim, A., Çetinkaya, A., Acarel, D., & Odabaşı Bağcı, J. (2020). Efficient Doxorubicin Loading to Isolated Dexosomes of Immature JAWSII Cells: Formulated and Characterized as the Bionanomaterial. Materials, 13(15), 3344. https://doi.org/10.3390/ma13153344






