Myofibroblast Gene Expression Profile after Tooth Extraction in the Rabbit
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Animal Model and Management
2.3. Surgical Phase
2.4. Terminal Procedure
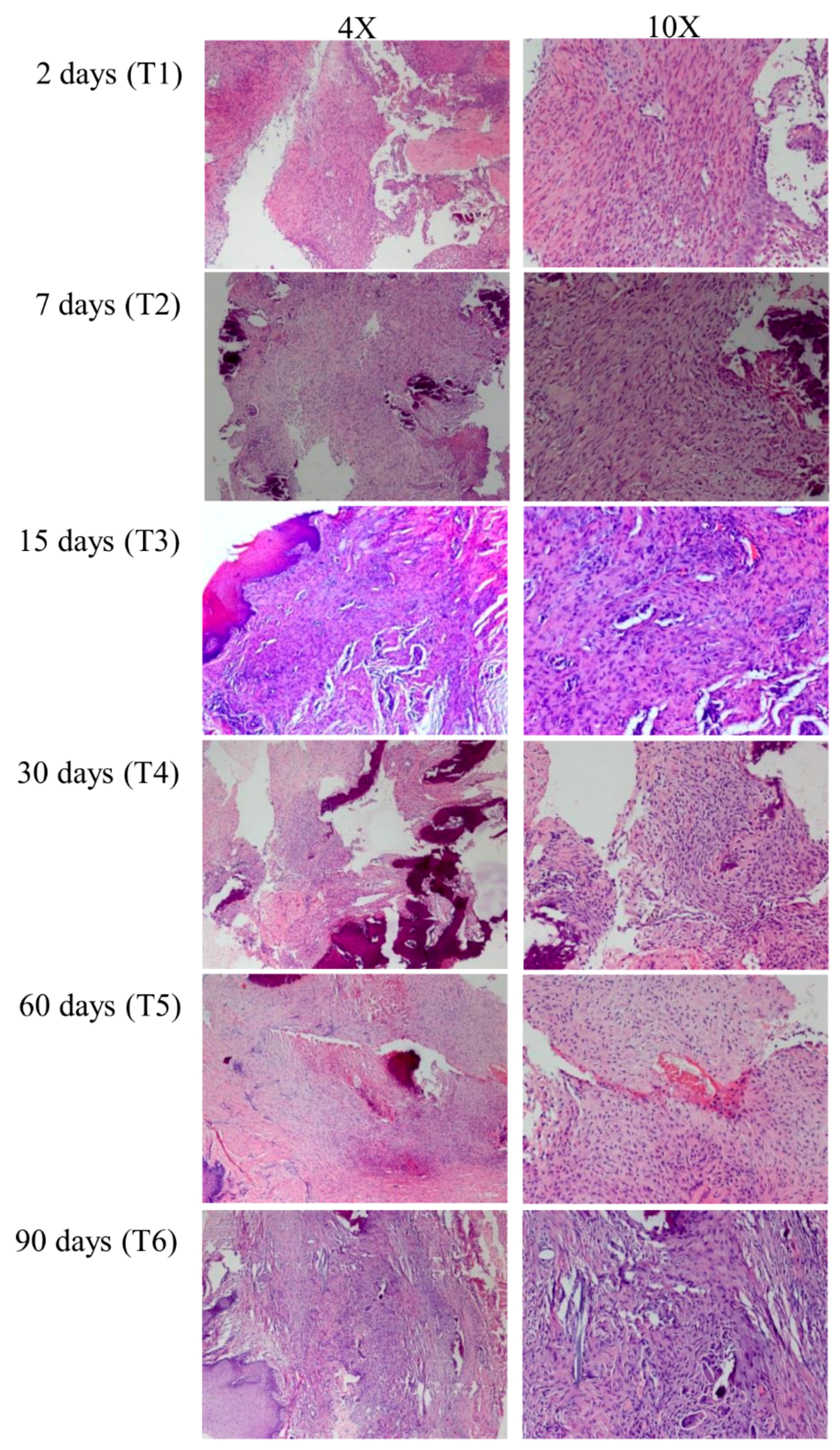
2.5. Histological Preparation
2.6. RNA Purification
2.7. nCounter Nanostring Technology
2.8. nCounter Custom Panel
2.9. Gene Expression Analysis
2.10. Statistical Analysis
3. Results
3.1. Gene Expression Profile of Post-Extractive Sockets
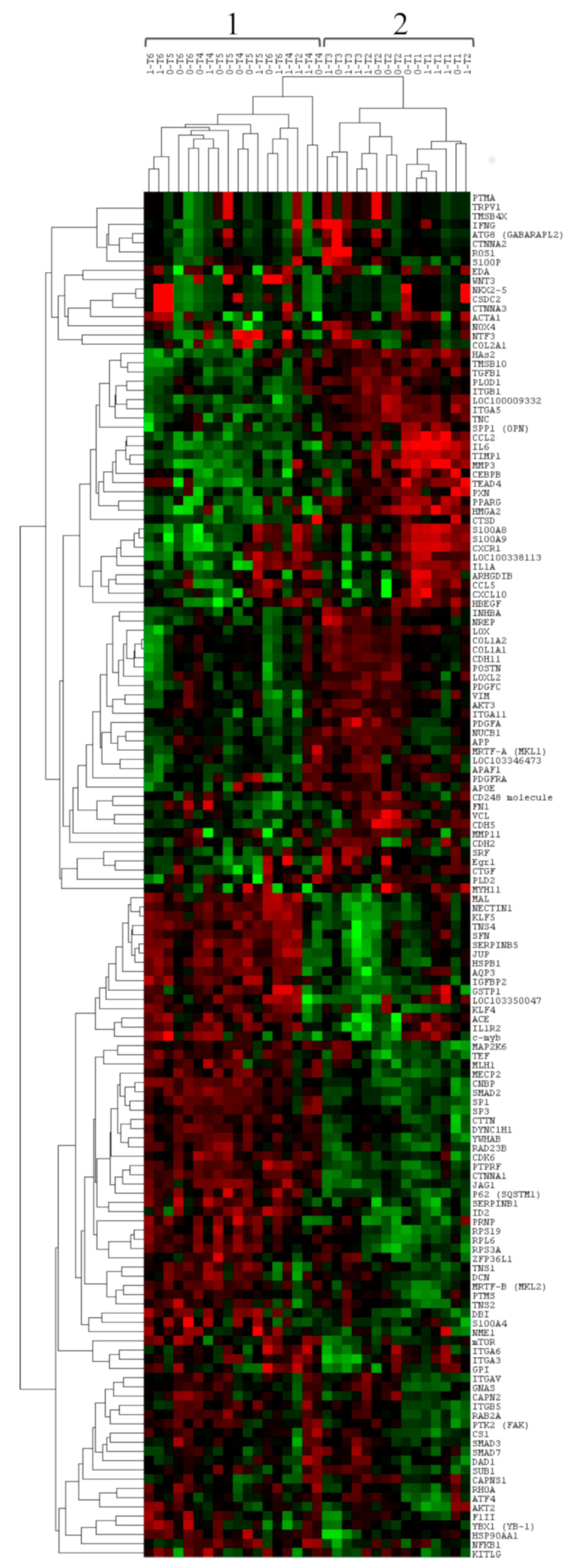
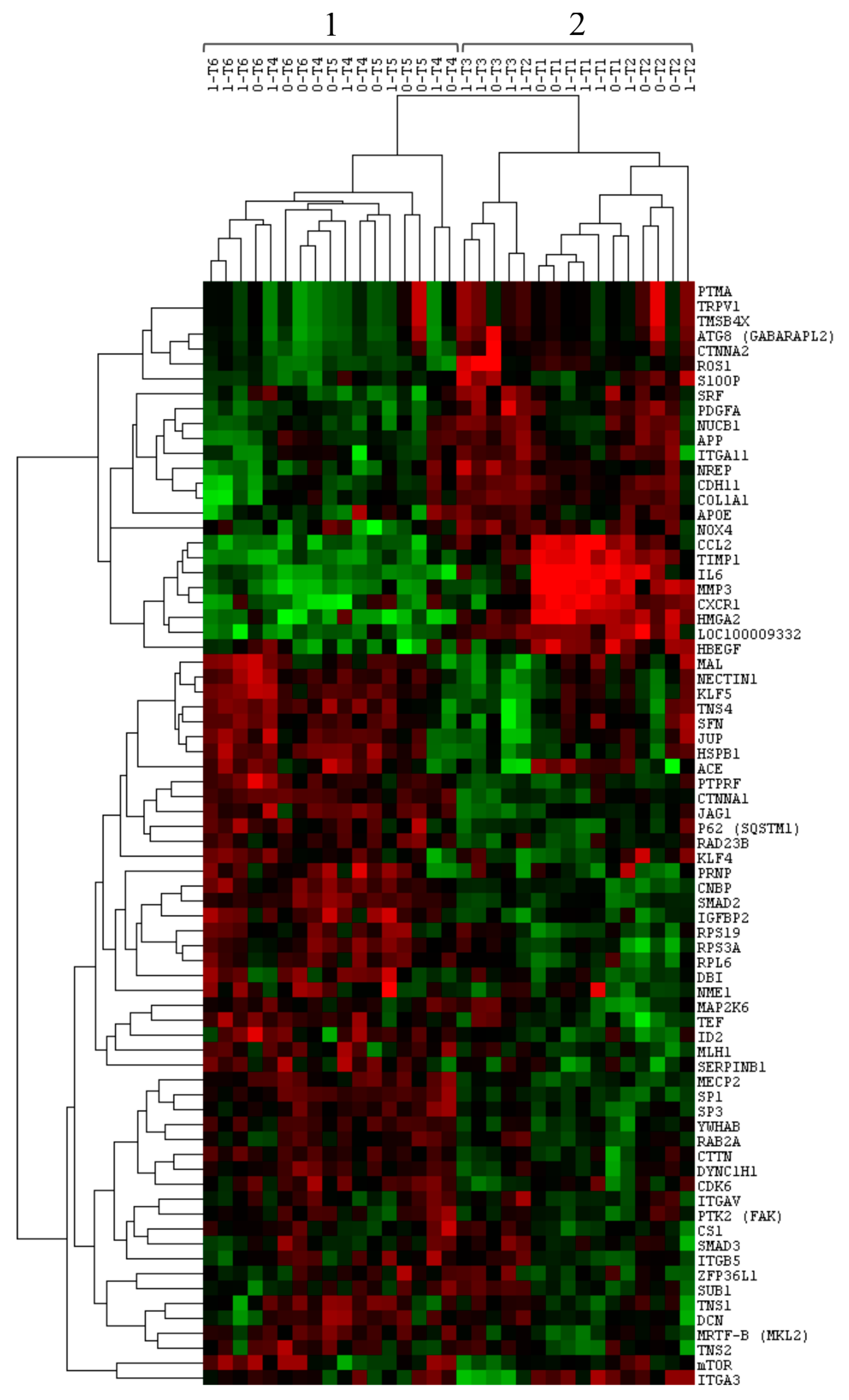
3.2. Comparison of Gene Expression between Group 0 and Group 1
3.3. Comparison of Gene Expression among Time-Related Groups
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Schropp, L.; Wenzel, A.; Kostopoulos, L.; Karring, T. Bone healing and soft tissue contour changes following single-tooth extraction: A clinical and radiographic 12-month prospective study. Int. J. Periodontics Restor. Dent. 2003, 23, 313–323. [Google Scholar]
- Araújo, M.G.; Sukekava, F.; Wennström, J.L.; Lindhe, J. Ridge alterations following implant placement in fresh extraction sockets: An experimental study in the dog. J. Clin. Periodontol. 2005, 32, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.L.; Wong, T.L.T.; Wong, M.C.M.; Lang, N.P. A systematic review of post-extractional alveolar hard and soft tissue dimensional changes in humans. Clin. Oral Implants Res. 2012, 23, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Marconcini, S.; Giammarinaro, E.; Derchi, G.; Alfonsi, F.; Covani, U.; Barone, A. Clinical outcomes of implants placed in ridge-preserved versus nonpreserved sites: A 4-year randomized clinical trial. Clin. Implant Dent. Relat. Res. 2018, 20, 906–914. [Google Scholar] [CrossRef] [PubMed]
- Covani, U.; Ricci, M.; Bozzolo, G.; Mangano, F.; Zini, A.; Barone, A. Analysis of the pattern of the alveolar ridge remodelling following single tooth extraction. Clin. Oral Implants Res. 2011, 22, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Araújo, M.G.; Silva, C.O.; Misawa, M.; Sukekava, F. Alveolar socket healing: What can we learn? Periodontol 2000 2015, 68, 122–134. [Google Scholar] [CrossRef]
- Discepoli, N.; Vignoletti, F.; Laino, L.; de Sanctis, M.; Muñoz, F.; Sanz, M. Early healing of the alveolar process after tooth extraction: An experimental study in the beagle dog. J. Clin. Periodontol. 2013, 40, 638–644. [Google Scholar] [CrossRef]
- Gabbiani, G.; Hirschel, B.J.; Ryan, G.B.; Statkov, P.R.; Majno, G. Granulation tissue as a contractile organ. A study of structure and function. J. Exp. Med. 1972, 135, 719–734. [Google Scholar] [CrossRef]
- Desmoulière, A.; Chaponnier, C.; Gabbiani, G. Tissue repair, contraction, and the myofibroblast. Wound Repair Regen Off. Publ. Wound Heal. Soc. Eur. Tissue Repair Soc. 2005, 13, 7–12. [Google Scholar] [CrossRef]
- Hinz, B.; Mastrangelo, D.; Iselin, C.E.; Chaponnier, C.; Gabbiani, G. Mechanical tension controls granulation tissue contractile activity and myofibroblast differentiation. Am. J. Pathol. 2001, 159, 1009–1020. [Google Scholar] [CrossRef]
- Hinz, B. Matrix mechanics and regulation of the fibroblast phenotype. Periodontol 2000 2013, 63, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Hinz, B.; McCulloch, C.A.; Coelho, N.M. Mechanical regulation of myofibroblast phenoconversion and collagen contraction. Exp. Cell Res. 2019, 379, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Hinz, B. Masters and servants of the force: The role of matrix adhesions in myofibroblast force perception and transmission. Eur. J. Cell Biol. 2006, 85, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Lalani, Z.; Wong, M.; Brey, E.M.; Mikos, A.G.; Duke, P.J. Spatial and temporal localization of transforming growth factor-beta1, bone morphogenetic protein-2, and platelet-derived growth factor-A in healing tooth extraction sockets in a rabbit model. J. Oral Maxillofac Surg. Off J. Am. Assoc. Oral Maxillofac Surg. 2003, 61, 1061–1072. [Google Scholar] [CrossRef]
- Sanders, Y.Y.; Tollefsbol, T.O.; Varisco, B.M.; Hagood, J.S. Epigenetic regulation of thy-1 by histone deacetylase inhibitor in rat lung fibroblasts. Am. J. Respir. Cell Mol. Biol. 2011, 45, 16–23. [Google Scholar] [CrossRef]
- Lugenbiel, P.; Wenz, F.; Govorov, K.; Syren, P.; Katus, H.A.; Thomas, D. Atrial myofibroblast activation and connective tissue formation in a porcine model of atrial fibrillation and reduced left ventricular function. Life Sci. 2017, 181, 1–8. [Google Scholar] [CrossRef]
- Mah, W.; Jiang, G.; Olver, D.; Cheung, G.; Kim, B.; Larjava, H.; Häkkinen, L. Human gingival fibroblasts display a non-fibrotic phenotype distinct from skin fibroblasts in three-dimensional cultures. PLoS ONE 2014, 9. [Google Scholar] [CrossRef][Green Version]
- Tomasek, J.J.; Gabbiani, G.; Hinz, B.; Chaponnier, C.; Brown, R.A. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat. Rev. Mol. Cell Biol. 2002, 3, 349–363. [Google Scholar] [CrossRef]
- Hinz, B.; Gabbiani, G. Cell-matrix and cell-cell contacts of myofibroblasts: Role in connective tissue remodeling. Thromb Haemost. 2003, 90, 993–1002. [Google Scholar] [CrossRef]
- Warburton, G.; Nares, S.; Angelov, N.; Brahim, J.S.; Dionne, R.A.; Wahl, S.M. Transcriptional events in a clinical model of oral mucosal tissue injury and repair. Wound Repair Regen Off Publ. Wound Heal. Soc. Eur. Tissue Repair Soc. 2005, 13, 19–26. [Google Scholar] [CrossRef]
- Vescarelli, E.; Pilloni, A.; Dominici, F.; Angeloni, A.; Polimeni, A.; Ceccarelli, S.; Marchese, C. Autophagy activation is required for myofibroblast differentiation during healing of oral mucosa. J. Clin. Periodontol 2017, 44, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Devlin, H.; Sloan, P. Early bone healing events in the human extraction socket. Int. J. Oral Maxillofac. Surg. 2002, 31, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Vogel, V.; Sheetz, M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006, 7, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Hinz, B. Myofibroblasts. Exp. Eye Res. 2016, 142, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Darby, I.A.; Laverdet, B.; Bonté, F.; Desmoulière, A. Fibroblasts and myofibroblasts in wound healing. Clin. Cosmet. Investig. Dermatol. 2014, 7, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.A.; Byers, P.D. Swelling of cartilage and expansion of the collagen network. Calcif. Tissue Int. 1989, 45, 260–261. [Google Scholar] [CrossRef]
- Karaca, Ç.; Er, N.; Gülşahı, A.; Köseoğlu, O.T. Alveolar ridge preservation with a free gingival graft in the anterior maxilla: Volumetric evaluation in a randomized clinical trial. Int. J. Oral Maxillofac. Surg. 2015, 44, 774–780. [Google Scholar] [CrossRef]
- Shenoy, V.B.; Wang, H.; Wang, X. A chemo-mechanical free-energy-based approach to model durotaxis and extracellular stiffness-dependent contraction and polarization of cells. Interface Focus. 2016, 6. [Google Scholar] [CrossRef]
| Myofibroblast-Activation-Related Genes |
| ACTA1, COL1A1, COL1A2, COL2A1, CDH2, CDH11, S100A4, TAGLN, VIM, FN1, TNS4, TNS2, tensin-3-like, TNS1, MYH11, VCL, PXN |
| Myofibroblast-Inducing Genes |
| SMAD3, SMAD2, MECP2, HMGA2, SRF, TEAD4, SP1, SP3, CEBPB, CSl, c-myb, MRTF-A (MKL1), MRTF-B (MKL2), FlII, TRPV1, TNC, IL6, TEF |
| Autophagy-Related Genes |
| ATG8 (GABARAPL2), P62 (SQSTM1), AKT2, AKT3, mTOR |
| Myofibroblast-Suppressing Genes |
| SMAD7, NFKB1, KLF4, PPARG, NKX2-5, IL1A |
| Growth Factors and Cytokines |
| TGFB1, NREP, WNT3, JAG1, PTK2 (FAK), NOX4, IFNG, CXCL10, PDGFA, PDGFC, HAS2, PDGFRA |
| Genes Coding for ECM Components |
| ROS1, LOX, LOXL2, PLOD1, PLD2, SPP1 (OPN), POSTN, CTGF, EDA, Egr1 |
| Genes Coding for Surface Proteins |
| CD248 molecule, ITGA11, ITGB1, ITGA3, ITGAV, ITGB5, ITGA5 |
| Constitutive Expressed Genes in Normal Oral Mucosa |
| Adhesion molecules: CTNNA1, CTNNA2, CTNNA3, DCN, CDH5, NECTIN1, ITGA6 Chemokine/cytokine/growth factors: S100A9, S100A8, TMSB10, TMSB4X, CCL5, ACE, KITLG, INHBA, PTMS, GPI Receptor: IL1R2, HBEGF, interleukin-2 receptor subunit alpha Cell cycle/apoptosis: RPS19, CDK6, PTMA, RPS3A, DYNC1H1, CAPNS1, APAF1, CAPN2, DAD1 Metabolism: AQP3 Protease/protein turnover: APP, PRNP, SERPINB5, IGFBP2, SERPINB1, CTSD Signal transduction: DBI, RHOA, SFN, JUP, S100P, protein S100-A7-like, CTTN, NTF3, PTPRF, MAP2K6, NME1, ARHGDIB, YWHAB, RAB2A Transcription: MLH1, GNAS, RPL6, CSDC2, ZFP36L1, KLF5, YBX1, ATF4, NUCB1, CNBP, RAD23B, SUB1 Homeostasis & detoxification: HSPB1, GSTP1, HSP90AA1 |
| Deregulated Genes at 48 h Post-Surgery |
| Chemokine/cytokine/growth factors: CCL2 Protease/protein turnover: MMP3, MMP11, TIMP1 Signal Transduction: CXCR1, MAL Metabolism: APOE Transcription: ID2 |
| Housekeeping Genes |
| RPL13A, UCHL-1, GAPDH, TUBA1B, ACTB, RPS9, ACTA2, HMBS, HPRT1, LDHA, TBP, NONO, EEF1E1, PPIH, PPIA |
| Myofibroblast-Activation-Related Genes | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | p-Values | T1 vs. T2 | T1 vs. T3 | T1 vs. T4 | T1 vs. T5 | T1 vs. T6 | T2 vs. T4 | T2 vs. T5 | T2 vs. T6 | T3 vs. T4 | T3 vs. T5 | T3 vs. T6 |
| CDH11 | 0.0183 | 0.042 | 0.006 | 0.089 | 0.221 | 0.811 | 0.743 | 0.420 | 0.023 | 0.172 | 0.079 | 0.003 Up |
| COL1A1 | 0.0093 | 0.052 | 0.002 Up | 0.029 | 0.121 | 1.000 | 0.811 | 0.698 | 0.052 | 0.196 | 0.071 | 0.002 Up |
| TAGLN | 0.0096 | 0.438 | 0.330 | 0.199 | 0.310 | 0.018 | 0.039 | 0.073 | 0.002 Up | 0.043 | 0.071 | 0.004 |
| TNS1 | 0.0240 | 0.095 | 0.021 | 0.002 Up | 0.004 | 0.023 | 0.161 | 0.221 | 0.550 | 0.845 | 0.961 | 0.643 |
| TNS2 | 0.0081 | 0.189 | 0.001 Up | 0.006 | 0.039 | 0.009 | 0.152 | 0.455 | 0.189 | 0.242 | 0.084 | 0.205 |
| TNS4 | 0.0006 | 0.511 | 0.922 | 0.007 | 0.025 | 0.0002 Up | 0.042 | 0.114 | 0.003 Up | 0.022 | 0.054 | 0.002 Up |
| Myofibroblast-Inducing Genes | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | p-Values | T1 vs. T2 | T1 vs. T3 | T1 vs. T4 | T1 vs. T5 | T1 vs. T6 | T2 vs. T4 | T2 vs. T5 | T2 vs. T6 | T3 vs. T4 | T3 vs. T5 | T3 vs. T6 |
| CSl | 0.0032 | 0.101 | 0.001 Up | 0.002 Up | 0.025 | 0.001 Up | 0.128 | 0.550 | 0.114 | 0.450 | 0.131 | 0.480 |
| HMGA2 | 0.0009 | 0.858 | 0.380 | 0.013 | 0.003 Up | 0.001 Up | 0.021 | 0.005 | 0.010 | 0.252 | 0.124 | 0.060 |
| IL6 | 0.0003 | 0.220 | 0.340 | 0.0004 Up | 0.0004 Up | 0.0003 Up | 0.023 | 0.024 | 0.020 | 0.057 | 0.058 | 0.052 |
| MECP2 | 0.0011 | 0.144 | 0.088 | 0.0001 Up | 0.002 Up | 0.001 Up | 0.020 | 0.101 | 0.052 | 0.165 | 0.407 | 0.283 |
| MRTF-B (MKL2) | 0.0096 | 0.107 | 0.034 | 0.001 Up | 0.005 | 0.005 | 0.064 | 0.221 | 0.244 | 0.480 | 0.845 | 0.884 |
| SMAD2 | 0.0008 | 0.339 | 0.283 | 0.0002 Up | 0.003 Up | 0.002 Up | 0.006 | 0.039 | 0.027 | 0.051 | 0.165 | 0.131 |
| SMAD3 | 0.0253 | 0.069 | 0.003 Up | 0.003 Up | 0.022 | 0.037 | 0.244 | 0.633 | 0.788 | 0.609 | 0.283 | 0.214 |
| SP1 | 0.0008 | 0.420 | 0.075 | 0.0001 Up | 0.004 | 0.020 | 0.003 Up | 0.039 | 0.020 | 0.196 | 0.575 | 0.435 |
| SP3 | 0.0051 | 0.083 | 0.068 | 0.0001 Up | 0.005 | 0.004 | 0.042 | 0.282 | 0.244 | 0.214 | 0.643 | 0.592 |
| SRF | 0.0226 | 0.095 | 0.001 Up | 0.064 | 0.765 | 0.199 | 0.858 | 0.170 | 0.698 | 0.093 | 0.003 Up | 0.032 |
| TEF | 0.0007 | 0.633 | 0.048 | 0.002 Up | 0.011 | 0.0003 Up | 0.007 | 0.039 | 0.020 | 0.542 | 0.922 | 0.330 |
| TRPV1 | 0.0293 | 0.009 | 0.001 Up | 0.079 | 0.105 | 0.119 | 0.400 | 0.328 | 0.298 | 0.076 | 0.059 | 0.053 |
| Genes Coding for Surface Proteins | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | p-Values | T1 vs. T2 | T1 vs. T3 | T1 vs. T4 | T1 vs. T5 | T1 vs. T6 | T2 vs. T4 | T2 vs. T5 | T2 vs. T6 | T3 vs. T4 | T3 vs. T5 | T3 vs. T6 |
| ITGA11 | 0.0311 | 0.022 | 0.002 Down | 0.039 | 0.101 | 0.339 | 0.811 | 0.511 | 0.179 | 0.150 | 0.075 | 0.019 |
| ITGA3 | 0.0121 | 0.339 | 0.075 | 0.045 | 0.698 | 0.325 | 0.296 | 0.179 | 0.976 | 0.001 Down | 0.144 | 0.010 |
| ITGAV | 0.0374 | 0.027 | 0.017 | 0.002 Down | 0.029 | 0.009 | 0.403 | 0.976 | 0.698 | 0.922 | 0.542 | 0.789 |
| ITGB5 | 0.0141 | 0.049 | 0.014 | 0.0003 Down | 0.012 | 0.034 | 0.114 | 0.591 | 0.881 | 0.661 | 0.679 | 0.465 |
| KLF4 | 0.0095 | 0.037 | 0.018 | 0.009 | 0.034 | 0.0001 Down | 0.612 | 0.976 | 0.101 | 0.807 | 0.526 | 0.495 |
| Constitutive Expressed Genes in Normal Oral Mucosa | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | p-Values | T1 vs. T2 | T1 vs. T3 | T1 vs. T4 | T1 vs. T5 | T1 vs. T6 | T2 vs. T4 | T2 vs. T5 | T2 vs. T6 | T3 vs. T4 | T3 vs. T5 | T3 vs. T6 |
| S100P | 0.0017 | 0.001 Down | 0.0004 Down | 0.006 | 0.004 | 0.136 | 0.550 | 0.654 | 0.064 | 0.196 | 0.242 | 0.021 |
| ACE | 0.0072 | 0.325 | 0.407 | 0.121 | 0.121 | 0.045 | 0.011 | 0.011 | 0.003 Down | 0.036 | 0.036 | 0.014 |
| APP | 0.0144 | 0.012 | 0.003 Down | 0.009 | 0.069 | 0.438 | 0.929 | 0.492 | 0.083 | 0.394 | 0.137 | 0.019 |
| CCL2 | 0.0021 | 0.095 | 0.845 | 0.014 | 0.003 Down | 0.0003 Up | 0.438 | 0.179 | 0.056 | 0.071 | 0.023 | 0.006 |
| CDK6 | 0.0105 | 0.232 | 0.884 | 0.002 Down | 0.034 | 0.009 | 0.049 | 0.355 | 0.152 | 0.015 | 0.113 | 0.046 |
| CNBP | 0.0004 | 0.531 | 0.144 | 0.001 Down | 0.004 | 0.0002 Down | 0.005 | 0.023 | 0.003 Down | 0.180 | 0.367 | 0.131 |
| CTNNA1 | 0.0010 | 0.199 | 0.733 | 0.001 Down | 0.016 | 0.0004 Down | 0.034 | 0.257 | 0.025 | 0.015 | 0.102 | 0.011 |
| CTNNA2 | 0.0276 | 0.012 | 0.002 Down | 0.091 | 0.016 | 0.132 | 0.403 | 0.903 | 0.308 | 0.080 | 0.243 | 0.057 |
| CTTN | 0.0040 | 0.039 | 0.330 | 0.001 Down | 0.009 | 0.0003 Down | 0.221 | 0.571 | 0.128 | 0.088 | 0.242 | 0.051 |
| DBI | 0.0040 | 0.232 | 0.015 | 0.004 | 0.007 | 0.0004 Down | 0.089 | 0.128 | 0.020 | 0.942 | 0.826 | 0.661 |
| DCN | 0.0126 | 0.078 | 0.012 | 0.001 Down | 0.009 | 0.101 | 0.089 | 0.403 | 0.905 | 0.751 | 0.697 | 0.242 |
| DYNC1H1 | 0.0142 | 0.037 | 0.262 | 0.001 Down | 0.014 | 0.005 | 0.179 | 0.720 | 0.455 | 0.093 | 0.380 | 0.232 |
| HBEGF | 0.0366 | 0.811 | 0.232 | 0.269 | 0.005 | 0.170 | 0.179 | 0.003 Up | 0.107 | 0.770 | 0.283 | 0.942 |
| HSPB1 | 0.0004 | 0.189 | 0.981 | 0.005 | 0.002 Down | 0.0001 Down | 0.128 | 0.083 | 0.013 | 0.022 | 0.014 | 0.020 |
| IGFBP2 | 0.0001 | 0.531 | 0.807 | 0.003 | 0.0001 Down | 0.003 Down | 0.022 | 0.020 | 0.017 | 0.032 | 0.004 | 0.027 |
| JUP | 0.00101 | 0.199 | 0.733 | 0.002 Down | 0.017 | 0.001 Down | 0.078 | 0.269 | 0.039 | 0.005 | 0.022 | 0.002 Down |
| KLF5 | 0.0004 | 0.591 | 0.643 | 0.004 | 0.029 | 0.001 Down | 0.018 | 0.101 | 0.005 | 0.005 | 0.025 | 0.001 Down |
| MAP2K6 | 0.0012 | 0.199 | 0.003 Down | 0.0001 Down | 0.009 | 0.004 | 0.011 | 0.179 | 0.107 | 0.922 | 0.380 | 0.510 |
| MLH1 | 0.0034 | 0.257 | 0.157 | 0.0001 Down | 0.014 | 0.002 Down | 0.012 | 0.189 | 0.052 | 0.119 | 0.559 | 0.273 |
| NECTIN1 | 0.0002 | 0.387 | 0.609 | 0.001 Down | 0.034 | 0.001 Down | 0.018 | 0.210 | 0.009 | 0.002 Down | 0.025 | 0.001 Down |
| NME1 | 0.0328 | 0.511 | 0.542 | 0.025 | 0.073 | 0.003 Down | 0.114 | 0.257 | 0.020 | 0.223 | 0.394 | 0.068 |
| NUCB1 | 0.0198 | 0.018 | 0.001 Down | 0.029 | 0.095 | 0.325 | 0.858 | 0.492 | 0.170 | 0.137 | 0.057 | 0.014 |
| PRNP | 0.0077 | 0.511 | 0.071 | 0.013 | 0.002 Down | 0.005 | 0.069 | 0.014 | 0.029 | 0.826 | 0.465 | 0.609 |
| PTMA | 0.0293 | 0.009 | 0.001 Down | 0.079 | 0.105 | 0.119 | 0.400 | 0.328 | 0.298 | 0.076 | 0.059 | 0.053 |
| PTPRF | 0.0005 | 0.161 | 0.592 | 0.0003 Down | 0.017 | 0.0001 Down | 0.032 | 0.325 | 0.016 | 0.018 | 0.157 | 0.010 |
| RAB2A | 0.0144 | 0.029 | 0.022 | 0.001 Down | 0.005 | 0.008 | 0.210 | 0.531 | 0.633 | 0.609 | 1.000 | 0.903 |
| RAD23B | 0.0121 | 0.069 | 0.318 | 0.001 Down | 0.009 | 0.003 Down | 0.152 | 0.420 | 0.232 | 0.097 | 0.252 | 0.144 |
| RPL6 | 0.0012 | 0.743 | 0.023 | 0.002 Down | 0.005 | 0.002 Down | 0.005 | 0.014 | 0.007 | 0.770 | 1.000 | 0.826 |
| RPS19 | 0.0005 | 0.339 | 0.002 Down | 0.002 Down | 0.001 Down | 0.001 Down | 0.029 | 0.023 | 0.013 | 0.592 | 0.643 | 0.770 |
| RPS3A | 0.0003 | 0.952 | 0.041 | 0.001 Down | 0.002 Down | 0.009 | 0.010 | 0.020 | 0.010 | 0.450 | 0.609 | 0.922 |
| SERPINB1 | 0.0069 | 0.387 | 0.009 | 0.003 | 0.025 | 0.002 Down | 0.039 | 0.170 | 0.029 | 0.826 | 0.435 | 0.903 |
| SFN | 0.0077 | 0.339 | 0.592 | 0.034 | 0.032 | 0.003 Down | 0.244 | 0.232 | 0.042 | 0.023 | 0.022 | 0.030 |
| SUB1 | 0.0041 | 0.049 | 0.0002 Down | 0.003 Down | 0.004 | 0.056 | 0.310 | 0.355 | 0.952 | 0.232 | 0.205 | 0.038 |
| TMSB4X | 0.0293 | 0.009 | 0.001 Down | 0.079 | 0.105 | 0.119 | 0.400 | 0.328 | 0.298 | 0.076 | 0.059 | 0.053 |
| YWHAB | 0.0125 | 0.189 | 0.097 | 0.002 Down | 0.008 | 0.002 Down | 0.078 | 0.179 | 0.073 | 0.394 | 0.609 | 0.380 |
| ZFP36L1 | 0.0056 | 0.455 | 0.001 Down | 0.004 | 0.016 | 0.095 | 0.034 | 0.095 | 0.355 | 0.367 | 0.205 | 0.061 |
| Genes Coding for ECM Components | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | p-Values | T1 vs. T2 | T1 vs. T3 | T1 vs. T4 | T1 vs. T5 | T1 vs. T6 | T2 vs. T4 | T2 vs. T5 | T2 vs. T6 | T3 vs. T4 | T3 vs. T5 | T3 vs. T6 |
| ROS1 | 0.0198 | 0.016 | 0.001 Down | 0.188 | 0.023 | 0.142 | 0.271 | 0.890 | 0.343 | 0.025 | 0.147 | 0.035 |
| Growth Factors and Cytokines | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | p-Values | T1 vs. T2 | T1 vs. T3 | T1 vs. T4 | T1 vs. T5 | T1 vs. T6 | T2 vs. T4 | T2 vs. T5 | T2 vs. T6 | T3 vs. T4 | T3 vs. T5 | T3 vs. T6 |
| JAG1 | 0.0004 | 0.244 | 0.751 | 0.0001 Down | 0.009 | 0.009 | 0.008 | 0.152 | 0.144 | 0.001 Down | 0.015 | 0.014 |
| NOX4 | 0.0407 | 0.083 | 0.002 Down | 0.045 | 0.161 | 0.492 | 0.788 | 0.743 | 0.296 | 0.144 | 0.051 | 0.011 |
| NREP | 0.0128 | 0.128 | 0.008 | 0.078 | 0.144 | 0.531 | 0.811 | 0.952 | 0.032 | 0.223 | 0.144 | 0.002 Up |
| PDGFA | 0.0257 | 0.032 | 0.003 Down | 0.098 | 0.446 | 0.633 | 0.622 | 0.165 | 0.095 | 0.095 | 0.016 | 0.008 |
| PTK2 (FAK) | 0.0169 | 0.018 | 0.043 | 0.001 Down | 0.013 | 0.005 | 0.269 | 0.905 | 0.676 | 0.421 | 1.000 | 0.807 |
| Autophagy-Related Genes | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | p-Values | T1 vs. T2 | T1 vs. T3 | T1 vs. T4 | T1 vs. T5 | T1 vs. T6 | T2 vs. T4 | T2 vs. T5 | T2 vs. T6 | T3 vs. T4 | T3 vs. T5 | T3 vs. T6 |
| ATG8 | 0.0276 | 0.012 | 0.002 Down | 0.091 | 0.016 | 0.132 | 0.403 | 0.903 | 0.308 | 0.080 | 0.243 | 0.057 |
| mTOR | 0.0026 | 0.032 | 0.751 | 0.018 | 0.152 | 0.0001 Down | 0.834 | 0.474 | 0.083 | 0.108 | 0.394 | 0.004 |
| P62 | 0.0008 | 0.095 | 0.330 | 0.001 Down | 0.003 Down | < 0.0001 Down | 0.121 | 0.199 | 0.018 | 0.097 | 0.150 | 0.021 |
| Deregulated Genes at 48 h Post-Surgery | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | p-Values | T1 vs. T2 | T1 vs. T3 | T1 vs. T4 | T1 vs. T5 | T1 vs. T6 | T2 vs. T4 | T2 vs. T5 | T2 vs. T6 | T3 vs. T4 | T3 vs. T5 | T3 vs. T6 |
| APOE | 0.0094 | 0.007 | 0.003 Down | 0.005 | 0.121 | 0.339 | 0.905 | 0.257 | 0.083 | 0.465 | 0.079 | 0.025 |
| CXCR1 | 0.0033 | 0.221 | 0.010 | 0.010 | 0.001 Up | 0.001 Up | 0.174 | 0.049 | 0.028 | 0.652 | 0.961 | 0.817 |
| ID2 | 0.0013 | 0.455 | 0.084 | 0.000 Down | 0.018 | 0.002 Down | 0.003 Down | 0.107 | 0.020 | 0.180 | 0.845 | 0.435 |
| MAL | 0.0021 | 0.310 | 0.626 | 0.083 | 0.025 | 0.000 Down | 0.474 | 0.221 | 0.012 | 0.057 | 0.021 | 0.001 Down |
| MMP3 | 0.0005 | 0.511 | 0.092 | 0.002 Up | 0.0004 Up | 0.001 Up | 0.014 | 0.004 | 0.007 | 0.386 | 0.241 | 0.288 |
| TIMP1 | 0.0010 | 0.511 | 0.380 | 0.006 | 0.003 Up | 0.0001 Up | 0.037 | 0.022 | 0.003 Up | 0.172 | 0.125 | 0.038 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marconcini, S.; Denaro, M.; Cosola, S.; Gabriele, M.; Toti, P.; Mijiritsky, E.; Proietti, A.; Basolo, F.; Giammarinaro, E.; Covani, U. Myofibroblast Gene Expression Profile after Tooth Extraction in the Rabbit. Materials 2019, 12, 3697. https://doi.org/10.3390/ma12223697
Marconcini S, Denaro M, Cosola S, Gabriele M, Toti P, Mijiritsky E, Proietti A, Basolo F, Giammarinaro E, Covani U. Myofibroblast Gene Expression Profile after Tooth Extraction in the Rabbit. Materials. 2019; 12(22):3697. https://doi.org/10.3390/ma12223697
Chicago/Turabian StyleMarconcini, Simone, Maria Denaro, Saverio Cosola, Mario Gabriele, Paolo Toti, Eitan Mijiritsky, Agnese Proietti, Fulvio Basolo, Enrica Giammarinaro, and Ugo Covani. 2019. "Myofibroblast Gene Expression Profile after Tooth Extraction in the Rabbit" Materials 12, no. 22: 3697. https://doi.org/10.3390/ma12223697
APA StyleMarconcini, S., Denaro, M., Cosola, S., Gabriele, M., Toti, P., Mijiritsky, E., Proietti, A., Basolo, F., Giammarinaro, E., & Covani, U. (2019). Myofibroblast Gene Expression Profile after Tooth Extraction in the Rabbit. Materials, 12(22), 3697. https://doi.org/10.3390/ma12223697







