Bacterial Adhesion on Femtosecond Laser-Modified Polyethylene
Abstract
1. Introduction
2. Materials and Methods
2.1. Laser Processing
2.2. Contact Angle Measurement
2.3. Microorganisms, Culture Conditions, and Bacterial Adhesion Assays
2.4. Environmental Scanning Electron Microscopy and Data Analysis
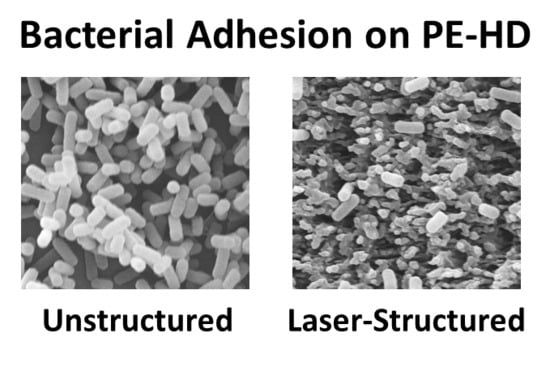
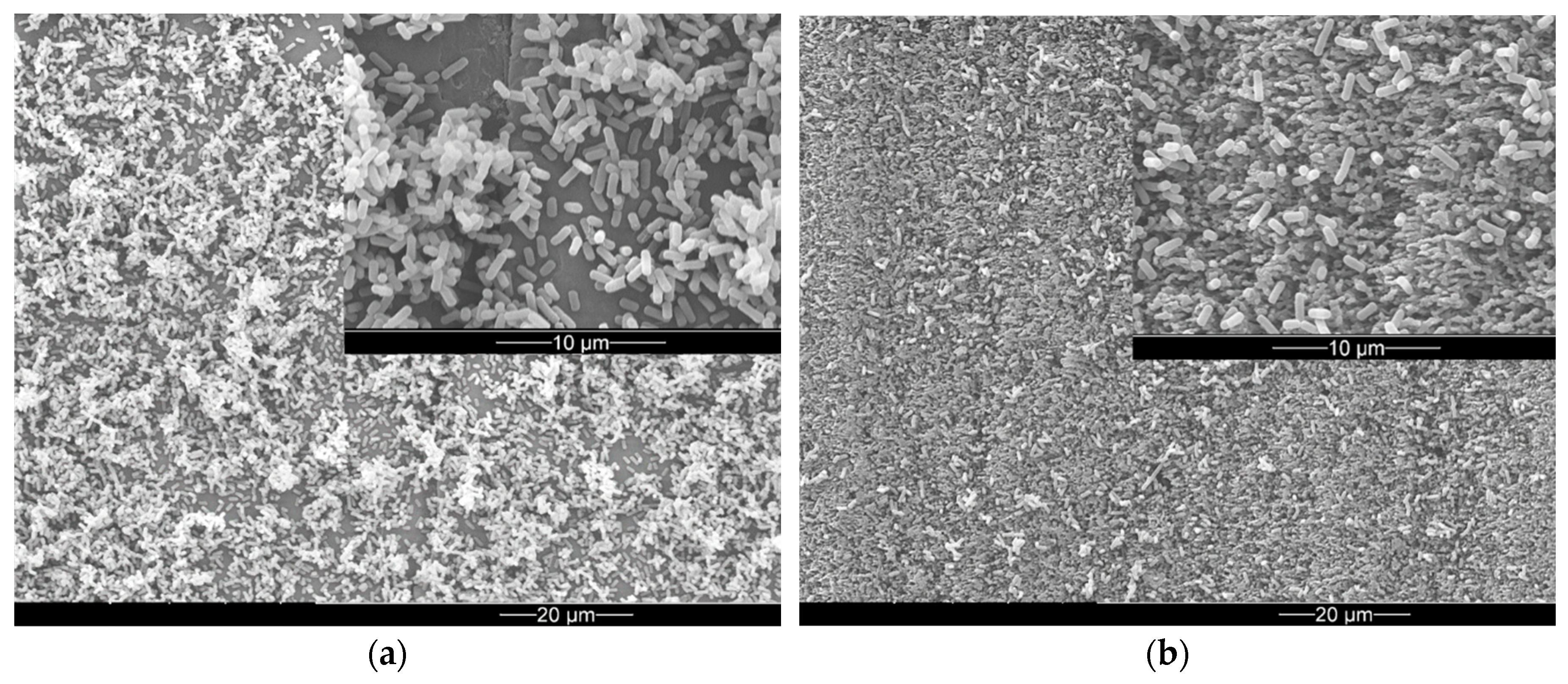
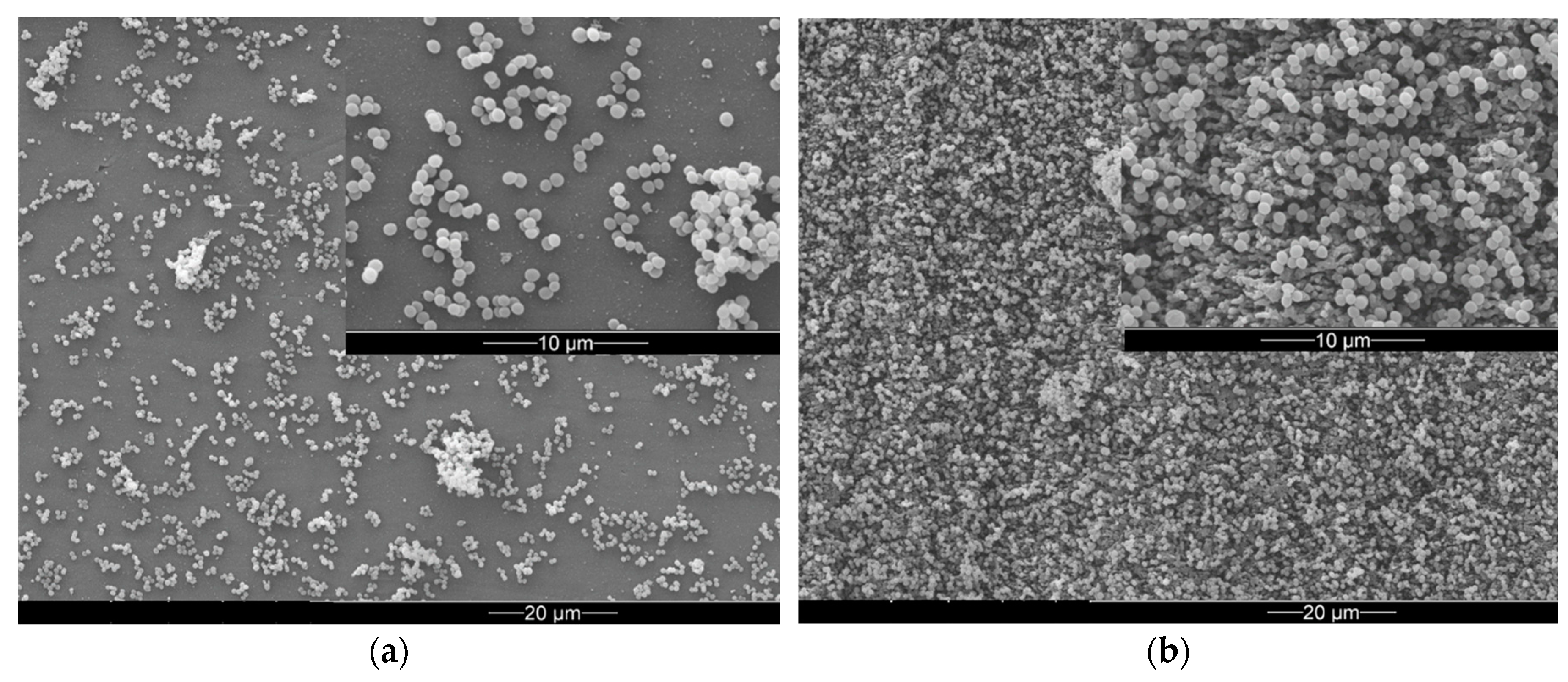
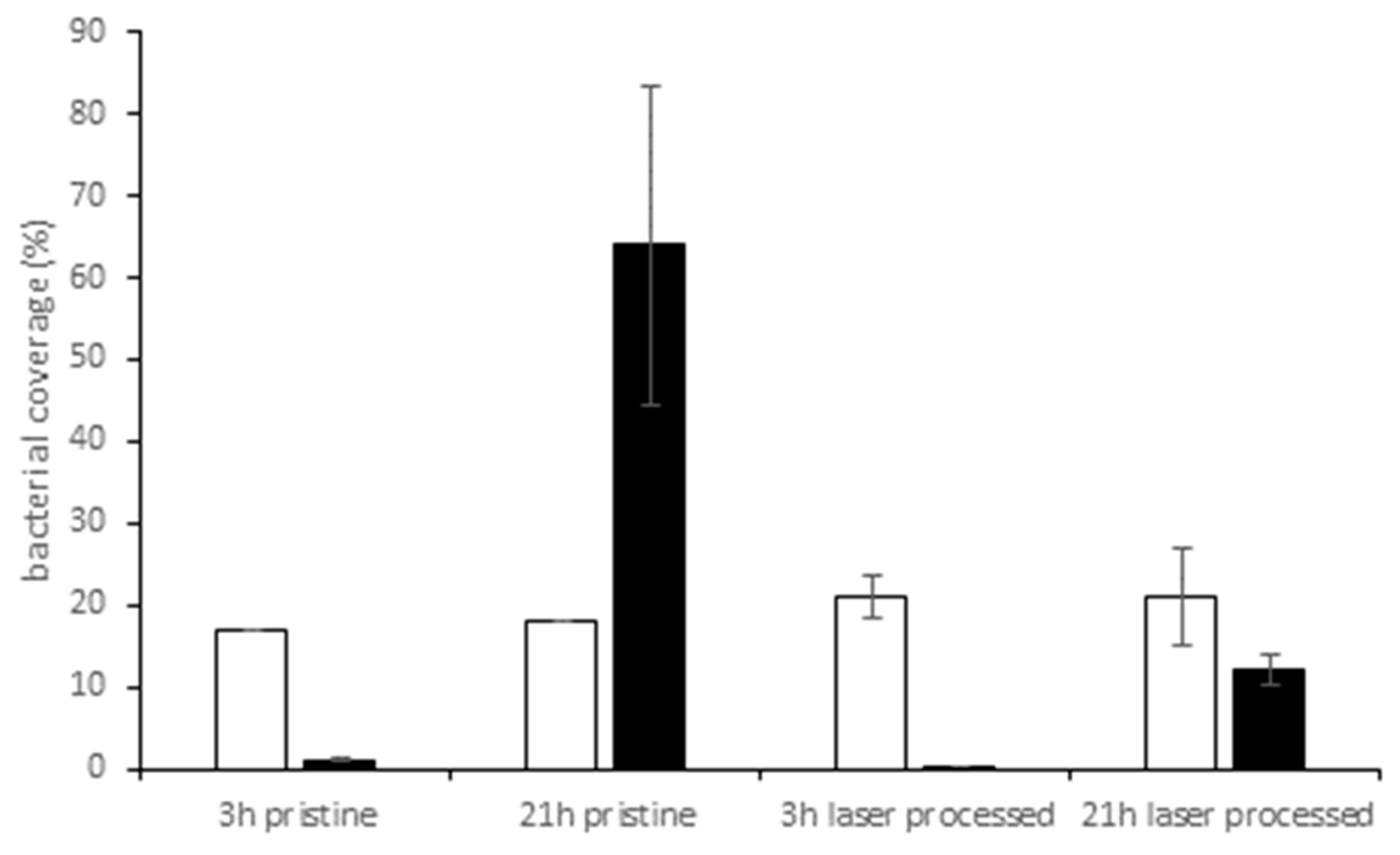
3. Results
3.1. Laser Surface Processing of Polyethylene
3.2. Bacterial Adhesion and Biofilm Formation on PE Surfaces
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zobell, C.E. The Effect of Solid Surfaces upon Bacterial Activity. J. Bacteriol. 1943, 46, 39–56. [Google Scholar] [PubMed]
- Costerton, J. Introduction to biofilm. Int. J. Antimicrob. Agents 1999, 11, 217–221. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial Life on Surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Dufour, D.; Leung, V.; Levesque, C.M. Bacterial biofilm: Structure, function, and antimicrobial resistance. Endod. Top. 2012, 22, 2–16. [Google Scholar] [CrossRef]
- Luppens, S.B.I.; Reij, M.W.; Van Der Heijden, R.W.L.; Rombouts, F.M.; Abee, T. Development of a Standard Test to Assess the Resistance of Staphylococcus aureus Biofilm Cells to Disinfectants. Appl. Environ. Microbiol. 2002, 68, 4194–4200. [Google Scholar] [CrossRef]
- Xu, D.; Jia, R.; Li, Y.; Gu, T. Advances in the treatment of problematic industrial biofilms. World J. Microbiol. Biotechnol. 2017, 33, 97. [Google Scholar] [CrossRef]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the Food Industry: Health Aspects and Control Methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef]
- Chan, S.; Pullerits, K.; Keucken, A.; Persson, K.M.; Paul, C.J.; Rådström, P. Bacterial release from pipe biofilm in a full-scale drinking water distribution system. NPJ Biofilms Microbiomes 2019, 5, 9. [Google Scholar] [CrossRef]
- Bixler, G.D.; Bhushan, B. Biofouling: Lessons from nature. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2012, 370, 2381–2417. [Google Scholar] [CrossRef]
- Di Pippo, F.; Di Gregorio, L.; Congestri, R.; Tandoi, V.; Rossetti, S. Biofilm growth and control in cooling water industrial systems. FEMS Microbiol. Ecol. 2018, 94, fiy044. [Google Scholar] [CrossRef]
- Winn, M.; Foulkes, J.M.; Perni, S.; Simmons, M.J.H.; Overton, T.W.; Goss, R.J.M. Biofilms and their engineered counterparts: A new generation of immobilised biocatalysts. Catal. Sci. Technol. 2012, 2, 1544–1547. [Google Scholar] [CrossRef]
- Perni, S.; Hackett, L.; Goss, R.J.; Simmons, M.J.; Overton, T.W. Optimisation of engineered Escherichia coli biofilms for enzymatic biosynthesis of l-halotryptophans. AMB Express 2013, 3, 66. [Google Scholar] [CrossRef] [PubMed]
- Campoccia, D.; Montanaro, L.; Arciola, C.R. A review of the biomaterials technologies for infection-resistant surfaces. Biomaterials 2013, 34, 8533–8554. [Google Scholar] [CrossRef] [PubMed]
- Sjollema, J.; Zaat, S.A.; Fontaine, V.; Ramstedt, M.; Luginbuehl, R.; Thevissen, K.; Li, J.; Van Der Mei, H.C.; Busscher, H.J. In vitro methods for the evaluation of antimicrobial surface designs. Acta Biomater. 2018, 70, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Tambe, S.M.; Sampath, L.; Modak, S.M. In vitro evaluation of the risk of developing bacterial resistance to antiseptics and antibiotics used in medical devices. J. Antimicrob. Chemother. 2001, 47, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.K.; Schumacher, J.F.; Sampson, E.M.; Burne, R.A.; Antonelli, P.J.; Brennan, A.B. Impact of engineered surface microtopography on biofilm formation of Staphylococcus aureus. Biointerphases 2007, 2, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Z.; Zhang, B.T.; Liu, Y.; Suo, X.K.; Li, H. Influence of surface topography on bacterial adhesion: A review. Biointerphases 2018, 13, 060801. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Cheng, Y.; Wang, S.-Y.; Borca-Tasciuc, D.A.; Worobo, R.W.; Moraru, C.I. Bacterial attachment and biofilm formation on surfaces are reduced by small-diameter nanoscale pores: How small is small enough? NPJ Biofilms Microbiomes 2015, 1, 15022. [Google Scholar] [CrossRef]
- Rizzello, L.; Cingolani, R.; Pompa, P.P. Nanotechnology tools for antibacterial materials. Nanomedicine 2013, 8, 807–821. [Google Scholar] [CrossRef]
- Scardino, A.J.; de Nys, R. Mini review: Biomimetic models and bioinspired surfaces for fouling control. Biofouling 2011, 27, 73–86. [Google Scholar] [CrossRef]
- Hasan, J.; Jain, S.; Padmarajan, R.; Purighalla, S.; Sambandamurthy, V.K.; Chatterjee, K. Multi-scale surface topography to minimize adherence and viability of nosocomial drug-resistant bacteria. Mater. Des. 2018, 140, 332–344. [Google Scholar] [CrossRef] [PubMed]
- Elbourne, A.; Crawford, R.J.; Ivanova, E.P. Nano-structured antimicrobial surfaces: From nature to synthetic analogues. J. Colloid Interface Sci. 2017, 508, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Berton, P.; Moraes, C.; Rogers, R.D.; Tufenkji, N. Nanodarts, nanoblades, and nanospikes: Mechano-bactericidal nanostructures and where to find them. Adv. Colloid Interface Sci. 2018, 252, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Modaresifar, K.; Azizian, S.; Ganjian, M.; Fratila-Apachitei, L.E.; Zadpoor, A.A. Bactericidal effects of nanopatterns: A systematic review. Acta Biomater. 2019, 83, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.M.; Zuber, F.; Maniura-Weber, K.; Brugger, J.; Ren, Q. Nanostructured surface topographies have an effect on bactericidal activity. J. Nanobiotechnol. 2018, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Valle, J.; Burgui, S.; Langheinrich, D.; Gil, C.; Solano, C.; Helbig, R.; Lasagni, A.; Lasa, I.; Toledo-Arana, A.; Toledo-Arana, A. Evaluation of Surface Microtopography Engineered by Direct Laser Interference for Bacterial Anti-Biofouling. Macromol. Biosci. 2015, 15, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Günther, D.; Friedrichs, J.; Rößler, F.; Helbig, R.; Lasagni, A.; Werner, C. The impact of structure dimensions on initial bacterial adhesion. Biomater. Sci. 2016, 4, 1074–1078. [Google Scholar]
- Rosenkranz, A.; Hans, M.; Gachot, C.; Thome, A.; Bonk, S.; Mücklich, F. Direct Laser Interference Patterning: Tailoring of Contact Area for Frictional and Antibacterial Properties. Lubricants 2016, 4, 2. [Google Scholar] [CrossRef]
- Gillett, A.; Waugh, D.; Lawrence, J.; Swainson, M.; Dixon, R. Laser surface modification for the prevention of biofouling by infection causing Escherichia Coli. J. Laser Appl. 2016, 28, 022503. [Google Scholar] [CrossRef]
- Bonse, J.; Krüger, J.; Höhm, S.; Rosenfeld, A. Femtosecond laser-induced periodic surface structures. J. Laser Appl. 2012, 24, 042006. [Google Scholar] [CrossRef]
- Vorobyev, A.Y.; Guo, C.L. Direct femtosecond laser surface nano/microstructuring and its applications. Laser Photonics Rev. 2013, 7, 385–407. [Google Scholar] [CrossRef]
- Heitz, J.; Arenholz, E.; Sauerbrey, R.; Phillips, H.M. Femtosecond excimer-laser-induced structure formation on polymers. Appl. Phys. A 1994, 59, 289–293. [Google Scholar] [CrossRef]
- Mezera, M.; van Drongelen, M.; Römer, G.R.B.E. Laser-Induced Periodic Surface Structures (LIPSS) on Polymers Processed with Picosecond Laser Pulses. J. Laser Micro Nano. 2018, 13, 105–116. [Google Scholar] [CrossRef]
- Rebollar, E.; Ezquerra, T.A.; Castillejo, M. Laser Nanostructuring of Polymers. In Pulsed Laser Ablation: Advances and Applications in Nanoparticles and Nanostructuring Thin Films, 1st ed.; Mihailescu, I.N., Caricato, A.P., Eds.; Pan Stanford Publishing PTE Ltd: Singapore, 2018; pp. 471–497. [Google Scholar]
- Danso, D.; Chow, J.; Streit, W.R. Plastics: Microbial Degradation, Environmental and Biotechnological Perspectives. Appl. Environ. Microbiol. 2019. [Google Scholar] [CrossRef]
- Pruitt, L.; Furmanski, J. Polymeric biomaterials for load-bearing medical devices. JOM 2009, 61, 14–20. [Google Scholar] [CrossRef]
- Maitz, M.F. Applications of synthetic polymers in clinical medicine. Biosurf. Biotribol. 2015, 1, 161–176. [Google Scholar] [CrossRef]
- Rossetti, F.F.; Siegmann, K.; Köser, J.; Wegner, I.; Keskin, I.; Schlotterbeck, G.; Winkler, M. Antimicrobial Polyethylene through Melt Compounding with Quaternary Ammonium Salts. Int. J. Polym. Sci. 2017, 2017, 2823604. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, Y.; Ji, J.; Yan, Q.; Huang, A.; Chu, P.K. Antimicrobial polyethylene with controlled copper release. J. Biomed. Mater. Res. Part A 2007, 83, 838–844. [Google Scholar] [CrossRef]
- Seyfriedsberger, G.; Rametsteiner, K.; Kern, W. Polyethylene compounds with antimicrobial surface properties. Eur. Polym. J. 2006, 42, 3383–3389. [Google Scholar] [CrossRef]
- Erdmann, M.; Böhning, M.; Niebergall, U. Physical and chemical effects of biodiesel storage on high-density polyethylene: Evidence of co-oxidation. Polym. Degrad. Stab. 2019, 161, 139–149. [Google Scholar] [CrossRef]
- Rédei, G.P. Encyclopedia of Genetics, Genomics, Proteomics and Informatics; Springer: Dordrecht, The Netherlands, 2008; p. 1087. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Zhang, X.X.; Wang, L.; Levanen, E. Superhydrophobic surfaces for the reduction of bacterial adhesion. RSC Adv. 2013, 3, 12003–12020. [Google Scholar] [CrossRef]
- Hasan, J.; Raj, S.; Yadav, L.; Chatterjee, K. Engineering a nanostructured “super surface” with superhydrophobic and superkilling properties. RSC Adv. 2015, 5, 44953–44959. [Google Scholar] [CrossRef]
- Yuan, Y.; Hays, M.P.; Hardwidge, P.R.; Kim, J. Surface characteristics influencing bacterial adhesion to polymeric substrates. RSC Adv. 2017, 7, 14254–14261. [Google Scholar] [CrossRef]
- Falde, E.J.; Yohe, S.T.; Colson, Y.L.; Grinstaff, M.W. Superhydrophobic materials for biomedical applications. Biomaterials 2016, 104, 87–103. [Google Scholar] [CrossRef]
- Hwang, G.B.; Page, K.; Patir, A.; Nair, S.P.; Allan, E.; Parkin, I.P. The Anti-Biofouling Properties of Superhydrophobic Surfaces are Short-Lived. ACS Nano 2018, 12, 6050–6058. [Google Scholar] [CrossRef]
- Lamb, R.; Zhang, H.; Lewis, J. Engineering nanoscale roughness on hydrophobic surface—Preliminary assessment of fouling behaviour. Sci. Technol. Adv. Mater. 2005, 6, 236–239. [Google Scholar]
- Lutey, A.H.A.; Gemini, L.; Romoli, L.; Lazzini, G.; Fuso, F.; Faucon, M.; Kling, R. Towards Laser-Textured Antibacterial Surfaces. Sci. Rep. 2018, 8, 10112. [Google Scholar] [CrossRef]
- Hsu, L.C.; Fang, J.; Borca-Tasciuc, D.A.; Worobo, R.W.; Moraru, C.I. Effect of Micro- and Nanoscale Topography on the Adhesion of Bacterial Cells to Solid Surfaces. Appl. Environ. Microbiol. 2013, 79, 2703–2712. [Google Scholar] [CrossRef]
- Fadeeva, E.; Truong, V.K.; Stiesch, M.; Chichkov, B.N.; Crawford, R.J.; Wang, J.; Ivanova, E.P. Bacterial Retention on Superhydrophobic Titanium Surfaces Fabricated by Femtosecond Laser Ablation. Langmuir 2011, 27, 3012–3019. [Google Scholar] [CrossRef]
- Epperlein, N.; Menzel, F.; Schwibbert, K.; Koter, R.; Bonse, J.; Sameith, J.; Krüger, J.; Toepel, J. Influence of femtosecond laser produced nanostructures on biofilm growth on steel. Appl. Surf. Sci. 2017, 418, 420–424. [Google Scholar] [CrossRef]
- Hou, S.; Gu, H.; Smith, C.; Ren, D. Microtopographic Patterns Affect Escherichia coli Biofilm Formation on Poly(dimethylsiloxane) Surfaces. Langmuir 2011, 27, 2686–2691. [Google Scholar] [CrossRef]
- Gu, H.; Chen, A.; Song, X.; Brasch, M.E.; Henderson, J.H.; Ren, D. How Escherichia coli lands and forms cell clusters on a surface: a new role of surface topography. Sci. Rep. 2016, 6, 29516. [Google Scholar] [CrossRef]
- Epstein, A.K.; Hochbaum, A.I.; Kim, P.; Aizenberg, J. Control of bacterial biofilm growth on surfaces by nanostructural mechanics and geometry. Nanotechnology 2011, 22, 494007. [Google Scholar] [CrossRef]
- Friedlander, R.S.; Vlamakis, H.; Kim, P.; Khan, M.; Kolter, R.; Aizenberg, J. Bacterial flagella explore microscale hummocks and hollows to increase adhesion. Proc. Natl. Acad. Sci. USA 2013, 110, 5624–5629. [Google Scholar] [CrossRef]


| Surface | Contact Angle |
|---|---|
| Pristine polyethylene | 64.5° |
| Femtosecond laser-processed polyethylene | 120.5° |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwibbert, K.; Menzel, F.; Epperlein, N.; Bonse, J.; Krüger, J. Bacterial Adhesion on Femtosecond Laser-Modified Polyethylene. Materials 2019, 12, 3107. https://doi.org/10.3390/ma12193107
Schwibbert K, Menzel F, Epperlein N, Bonse J, Krüger J. Bacterial Adhesion on Femtosecond Laser-Modified Polyethylene. Materials. 2019; 12(19):3107. https://doi.org/10.3390/ma12193107
Chicago/Turabian StyleSchwibbert, Karin, Friederike Menzel, Nadja Epperlein, Jörn Bonse, and Jörg Krüger. 2019. "Bacterial Adhesion on Femtosecond Laser-Modified Polyethylene" Materials 12, no. 19: 3107. https://doi.org/10.3390/ma12193107
APA StyleSchwibbert, K., Menzel, F., Epperlein, N., Bonse, J., & Krüger, J. (2019). Bacterial Adhesion on Femtosecond Laser-Modified Polyethylene. Materials, 12(19), 3107. https://doi.org/10.3390/ma12193107