Tomographic Collection of Block-Based Sparse STEM Images: Practical Implementation and Impact on the Quality of the 3D Reconstructed Volume
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Samples
2.2. Experimental Setup of the Electron Microscope
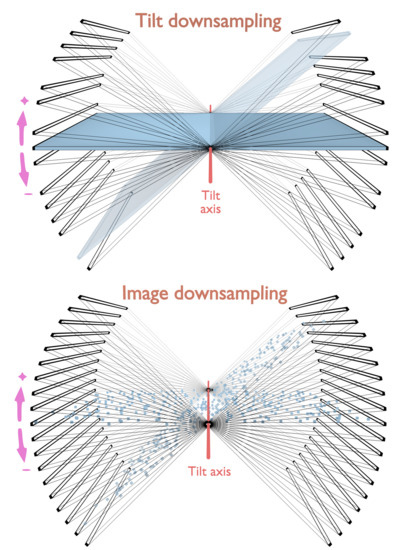
2.3. Tilt-Series Collection, Alignment and Reconstruction
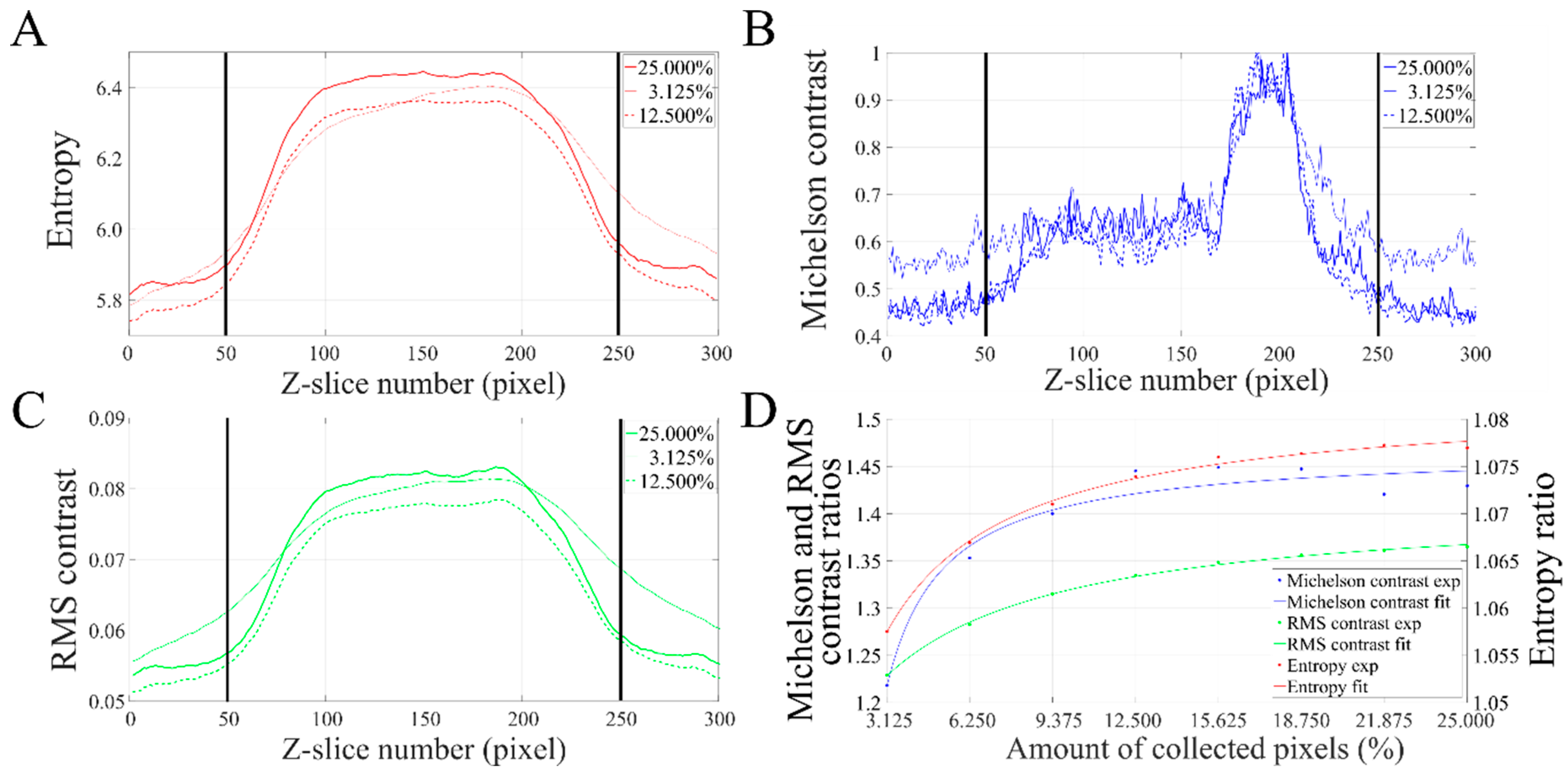
2.4. Reconstruction Quality Assessment
2.5. Data Presentation
3. Results
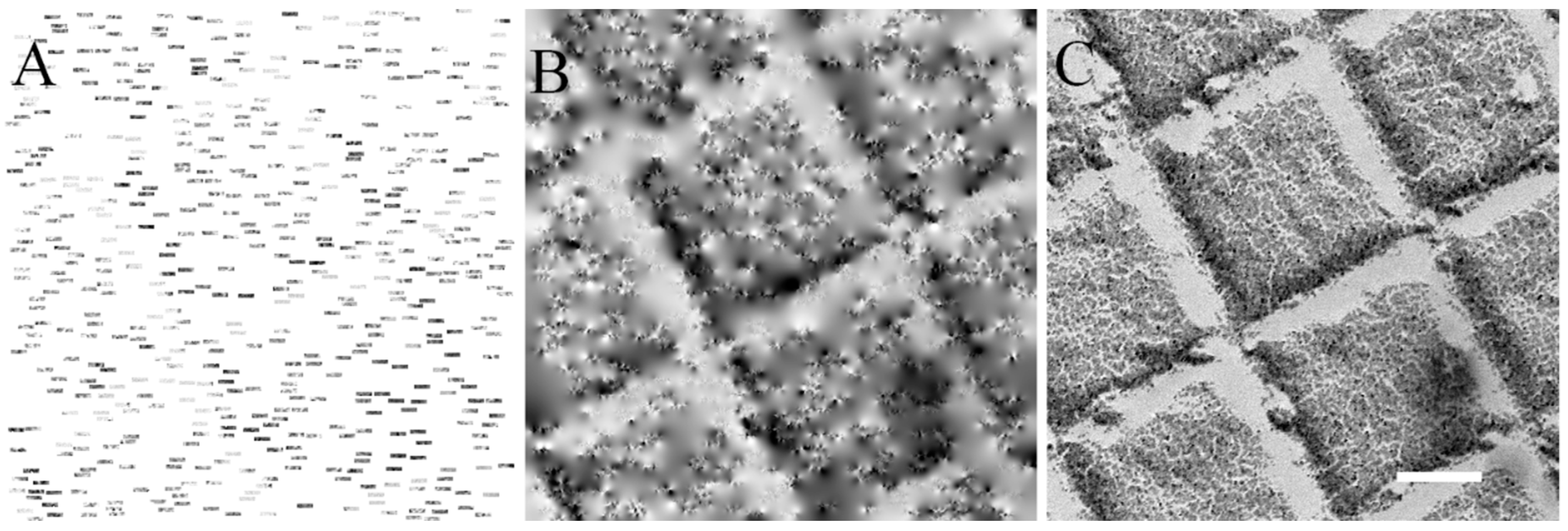
3.1. Practical Implementation in 2D
3.1.1. Collection of Sparse STEM Images
3.1.2. Accuracy of the Electron Beam Positioning during Sparse Imaging
3.2. Practical Implementation in 3D
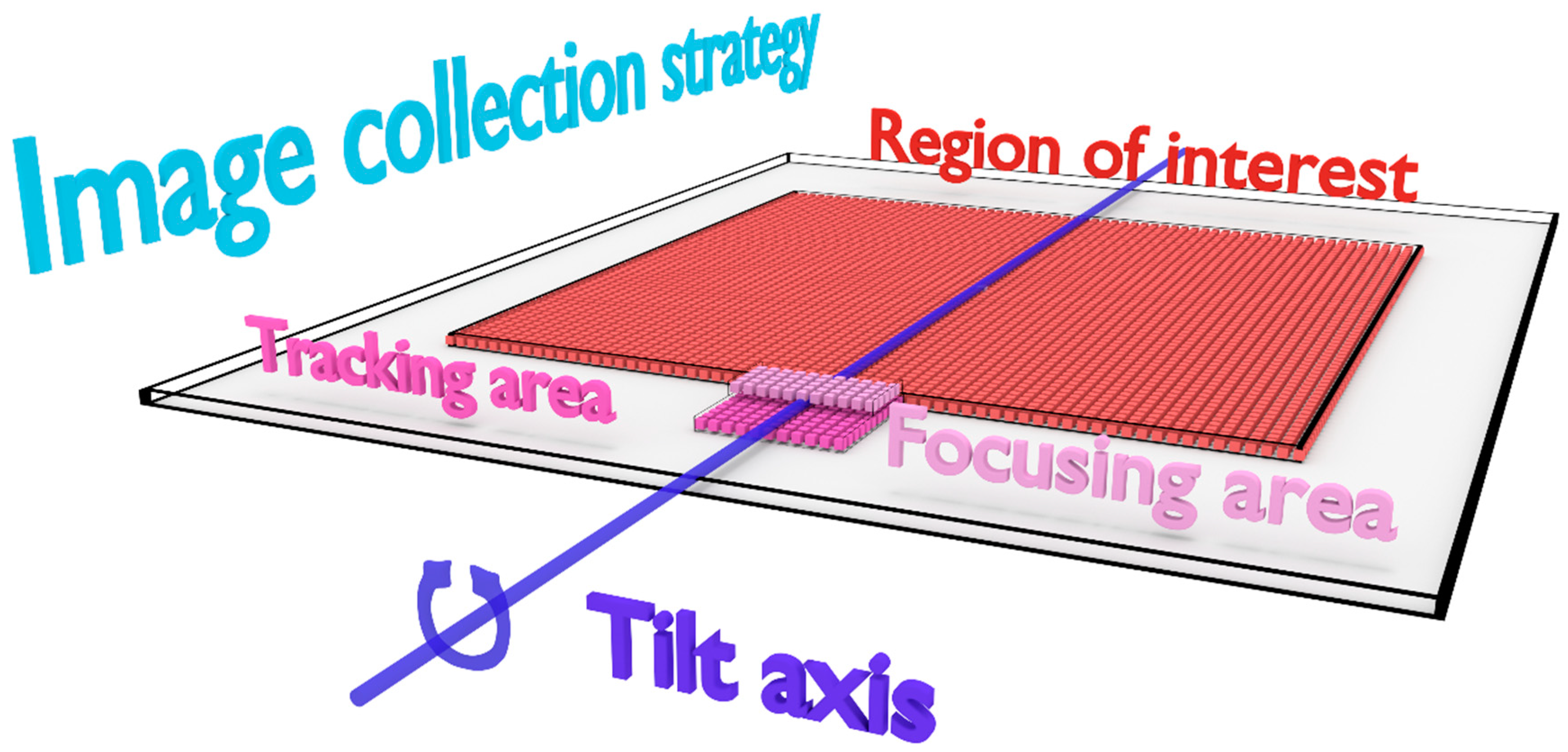
3.2.1. Collection of Sparse STEM Tilt-Series
- Step 1—Focusing: determination of the 0 nm focus value by successive acquisitions of fully-collected images on the focusing area in a focus range.
- Step 2—Tracking: acquisition of a fully-collected image on the tracking area and determination of the drift that occurred compared to the prior tilt angle by cross-correlation.
- Step 3—Sparse data collection: acquisition of the block-based sparse image on the region of interest.
- Step 4—End of current tilt: rotation to the next tilt angle and back to step 1.
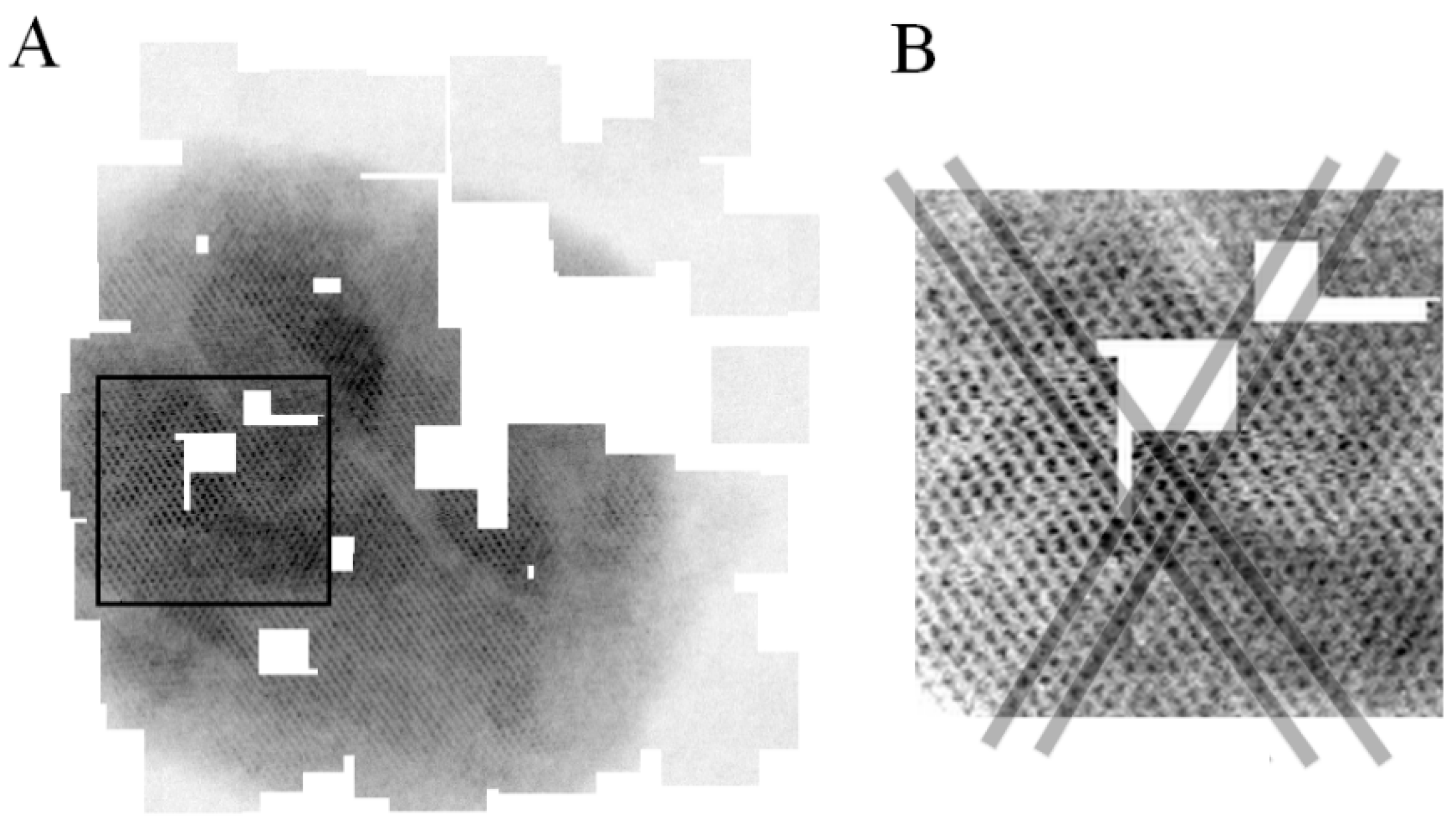
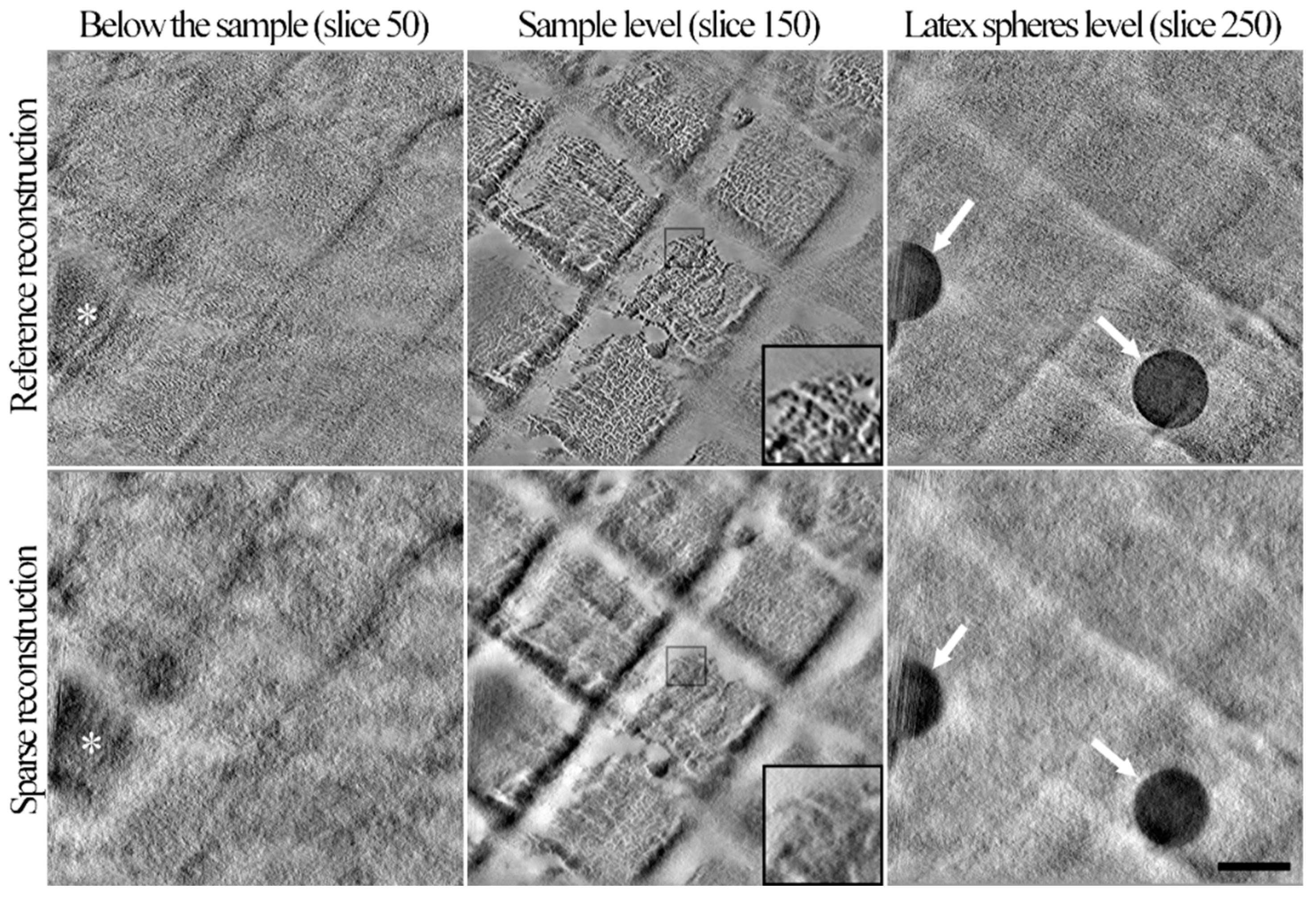
3.2.2. Validation of the Sparse STEM Tomography Workflow
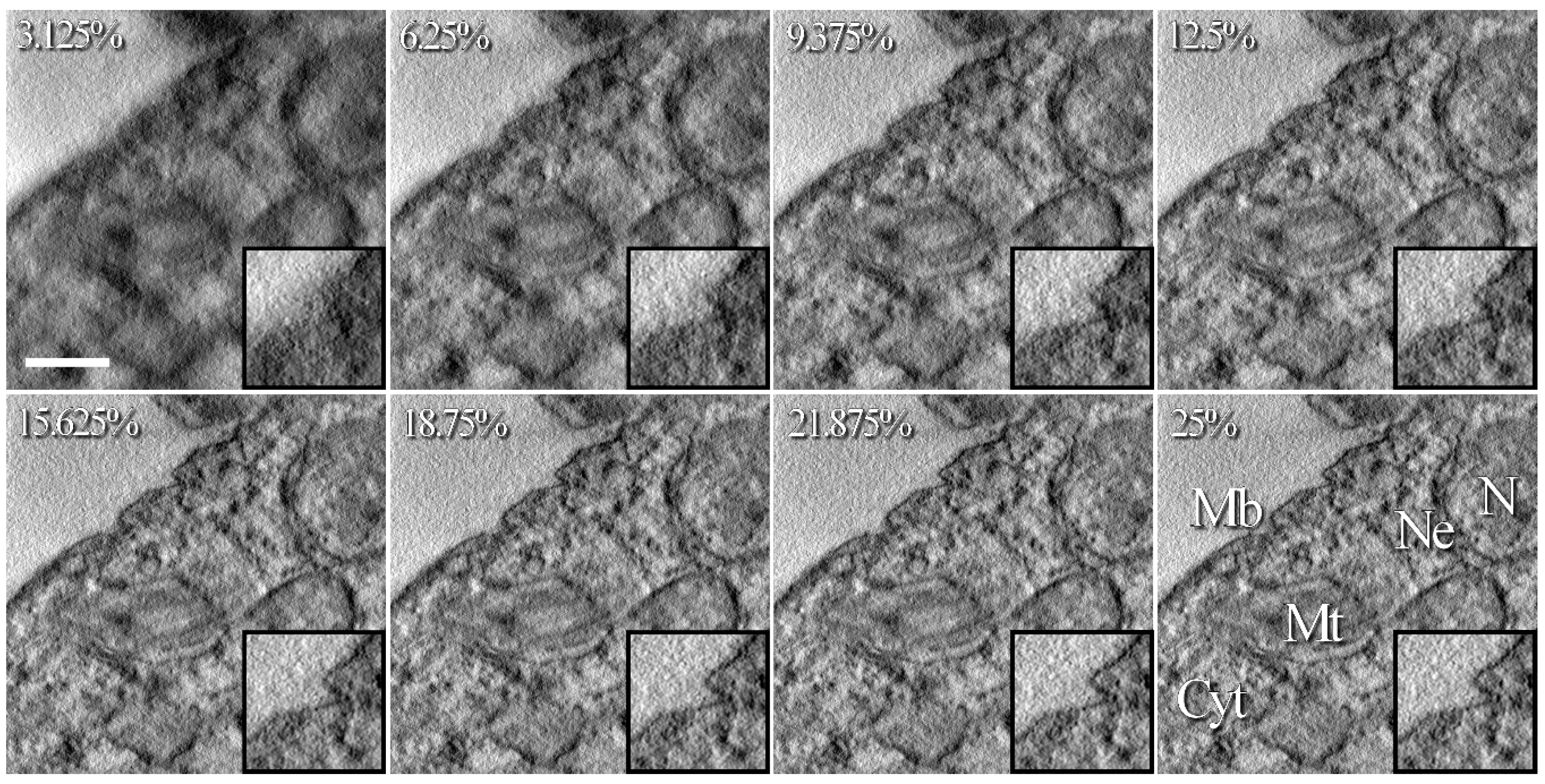
3.3. Application on a Biological Sample
4. Discussion
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Lacomble, S.; Vaughan, S.; Gadelha, C.; Morphew, M.K.; Shaw, M.K.; McIntosh, J.R.; Gull, K. Basal body movements orchestrate membrane organelle division and cell morphogenesis in Trypanosoma brucei. J. Cell Sci. 2010, 123, 2884–2891. [Google Scholar] [CrossRef] [PubMed]
- Lucic, V.; Rigort, A.; Baumeister, W. Cryo-electron tomography: The challenge of doing structural biology in situ. J. Cell Biol. 2013, 202, 407–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, L.; Jensen, G.J. Electron tomography of cells. Q. Rev. Biophys. 2012, 45, 27–56. [Google Scholar] [CrossRef] [PubMed]
- Oikonomou, C.M.; Jensen, G.J. A new view into prokaryotic cell biology from electron cryotomography. Nat. Rev. Microbiol. 2017, 15, 128. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, M.; Engel, B.D.; Laugks, T.; Mahamid, J.; Plitzko, J.M.; Baumeister, W. Cryo-focused Ion Beam Sample Preparation for Imaging Vitreous Cells by Cryo-electron Tomography. Bio-Protocol 2015, 5, 1575. [Google Scholar] [CrossRef]
- Mahamid, J.; Pfeffer, S.; Schaffer, M.; Villa, E.; Danev, R.; Cuellar, L.K.; Forster, F.; Hyman, A.A.; Plitzko, J.M.; Baumeister, W. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 2016, 351, 969–972. [Google Scholar] [CrossRef]
- Bertiaux, E.; Mallet, A.; Fort, C.; Blisnick, T.; Bonnefoy, S.; Jung, J.; Lemos, M.; Marco, S.; Vaughan, S.; Trepout, S.; et al. Bidirectional intraflagellar transport is restricted to two sets of microtubule doublets in the trypanosome flagellum. J. Cell Biol. 2018, 217, 4284–4297. [Google Scholar] [CrossRef] [Green Version]
- Soto, G.E.; Young, S.J.; Martone, M.E.; Deerinck, T.J.; Lamont, S.; Carragher, B.O.; Hama, K.; Ellisman, M.H. Serial section electron tomography: A method for three-dimensional reconstruction of large structures. NeuroImage 1994, 1, 230–243. [Google Scholar] [CrossRef]
- Midgley, P.A.; Weyland, M. 3D electron microscopy in the physical sciences: The development of Z-contrast and EFTEM tomography. Ultramicroscopy 2003, 96, 413–431. [Google Scholar] [CrossRef]
- Pennycook, S.J.; Nellist, P.D. Scanning Transmission Electron Microscopy; Springer: New York, NY, USA, 2011. [Google Scholar]
- Hohmann-Marriott, M.F.; Sousa, A.A.; Azari, A.A.; Glushakova, S.; Zhang, G.; Zimmerberg, J.; Leapman, R.D. Nanoscale 3D cellular imaging by axial scanning transmission electron tomography. Nat. Methods 2009, 6, 729–731. [Google Scholar] [CrossRef] [Green Version]
- Aoyama, K.; Takagi, T.; Hirase, A.; Miyazawa, A. STEM tomography for thick biological specimens. Ultramicroscopy 2008, 109, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.A.; Hohmann-Marriott, M.F.; Zhang, G.; Leapman, R.D. Monte Carlo electron-trajectory simulations in bright-field and dark-field STEM: Implications for tomography of thick biological sections. Ultramicroscopy 2009, 109, 213–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sousa, A.A.; Azari, A.A.; Zhang, G.; Leapman, R.D. Dual-axis electron tomography of biological specimens: Extending the limits of specimen thickness with bright-field STEM imaging. J. Struct. Biol. 2011, 174, 107–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, S.G.; Houben, L.; Elbaum, M. Cryo-scanning transmission electron tomography of vitrified cells. Nat. Methods 2014, 11, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Rez, P.; Larsen, T.; Elbaum, M. Exploring the theoretical basis and limitations of cryo-STEM tomography for thick biological specimens. J. Struct. Biol. 2016, 196, 466–478. [Google Scholar] [CrossRef] [PubMed]
- Batenburg, K.J.; Bals, S.; Sijbers, J.; Kubel, C.; Midgley, P.A.; Hernandez, J.C.; Kaiser, U.; Encina, E.R.; Coronado, E.A.; Van Tendeloo, G. 3D imaging of nanomaterials by discrete tomography. Ultramicroscopy 2009, 109, 730–740. [Google Scholar] [CrossRef] [PubMed]
- Goris, B.; Van den Broek, W.; Batenburg, K.J.; Heidari Mezerji, H.; Bals, S. Electron tomography based on a total variation minimization reconstruction technique. Ultramicroscopy 2012, 113, 120–130. [Google Scholar] [CrossRef]
- Saghi, Z.; Holland, D.J.; Leary, R.; Falqui, A.; Bertoni, G.; Sederman, A.J.; Gladden, L.F.; Midgley, P.A. Three-dimensional morphology of iron oxide nanoparticles with reactive concave surfaces. A compressed sensing-electron tomography (CS-ET) approach. Nano Lett. 2011, 11, 4666–4673. [Google Scholar] [CrossRef] [PubMed]
- Saghi, Z.; Divitini, G.; Winter, B.; Leary, R.; Spiecker, E.; Ducati, C.; Midgley, P.A. Compressed sensing electron tomography of needle-shaped biological specimens--Potential for improved reconstruction fidelity with reduced dose. Ultramicroscopy 2016, 160, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Donati, L.; Nilchian, M.; Trepout, S.; Messaoudi, C.; Marco, S.; Unser, M. Compressed sensing for STEM tomography. Ultramicroscopy 2017, 179, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Padgett, E.; Hovden, R.; Muller, D.A. Sampling limits for electron tomography with sparsity-exploiting reconstructions. Ultramicroscopy 2018, 186, 94–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monier, E.; Oberlin, T.; Brun, N.; Tencé, M.; de Frutos, M.; Dobigeon, N. Reconstruction of partially sampled multi-band images—Application to STEM-EELS imaging. IEEE Trans. Comput. Imaging 2018, 4, 585–598. [Google Scholar] [CrossRef]
- Stevens, A.; Yang, H.; Carin, L.; Arslan, I.; Browning, N.D. The potential for Bayesian compressive sensing to significantly reduce electron dose in high-resolution STEM images. Microscopy 2014, 63, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Saghi, Z.; Benning, M.; Leary, R.; Macias-Montero, M.; Borras, A.; Midgley, P.A. Reduced-dose and high-speed acquisition strategies for multi-dimensional electron microscopy. Adv. Struct. Chem. Imaging 2015, 1, 7. [Google Scholar] [CrossRef] [Green Version]
- Béché, A.; Goris, B.; Freitag, B.; Verbeeck, J. Development of a fast electromagnetic shutter for compressive sensing imaging in scanning transmission electron microscopy. Appl. Phys. Lett. 2016, 108, 093103. [Google Scholar] [CrossRef]
- Anderson, H.S.; Ilic-Helms, J.; Rohrer, B.; Wheeler, J.; Larson, K. Sparse Imaging for Fast Electron Microscopy. In Proceedings of the Computational Imaging XI. International Society for Optics and Photonics, Burlingame, CA, USA, 14 February 2013; Volume 8657, p. 86570C. [Google Scholar] [CrossRef]
- Li, X.; Dyck, O.; Kalinin, S.V.; Jesse, S. Compressed Sensing of Scanning Transmission Electron Microscopy (STEM) With Nonrectangular Scans. Microsc. Microanal. 2018, 24, 623–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanrompay, H.; Béché, A.; Verbeeck, J.; Bals, S. Experimental Evaluation of Undersampling Schemes for Electron Tomography of Nanoparticles. Part. Part. Syst. Charact. 2019, 1900096. [Google Scholar] [CrossRef]
- Ferroni, M.; Signoroni, A.; Sanzogni, A.; Masini, L.; Migliori, A.; Ortolani, L.; Pezza, A.; Morandi, V. Biological application of Compressed Sensing Tomography in the Scanning Electron Microscope. Sci. Rep. 2016, 6, 33354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guay, M.D.; Czaja, W.; Aronova, M.A.; Leapman, R.D. Compressed Sensing Electron Tomography for Determining Biological Structure. Sci. Rep. 2016, 6, 27614. [Google Scholar] [CrossRef] [Green Version]
- Pichon, B.P.; Gerber, O.; Lefevre, C.; Florea, I.; Fleutot, I.; Baaziz, W.; Pauly, M.; Ohlmann, M.; Ulhaq, C.; Ersen, O.; et al. Microstructural and Magnetic Investigations of Wüstite-Spinel Core-Shell Cubic-Shaped Nanoparticles. Chem. Mater. 2011, 23, 2886–2900. [Google Scholar] [CrossRef]
- Brun, R.; Schonenberger. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 1979, 36, 289–292. [Google Scholar] [PubMed]
- Trépout, S.; Tassin, A.M.; Marco, S.; Bastin, P. STEM tomography analysis of the trypanosome transition zone. J. Struct. Biol. 2018, 202, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D. Robust smoothing of gridded data in one and higher dimensions with missing values. Comput. Stat. Data Anal. 2010, 54, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Garcia, D.; Liu, Y.; de Jeu, R.; Dolman, A.J. A three-dimensional gap filling method for large geophysical datasets: Application to global satellite soil moisture observations. Environ. Model. Softw. 2012, 30, 139–142. [Google Scholar] [CrossRef]
- Kremer, J.R.; Mastronarde, D.N.; McIntosh, J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996, 116, 71–76. [Google Scholar] [CrossRef]
- Mastronarde, D.N.; Held, S.R. Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 2017, 197, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Peli, E. Contrast in complex images. J. Opt. Soc. Am. A Opt. Image Sci. 1990, 7, 2032–2040. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Sorzano, C.O.; Messaoudi, C.; Eibauer, M.; Bilbao-Castro, J.R.; Hegerl, R.; Nickell, S.; Marco, S.; Carazo, J.M. Marker-free image registration of electron tomography tilt-series. BMC Bioinform. 2009, 10, 124. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Snapp, R.R.; Ruiz, T.; Radermacher, M. Projection-based volume alignment. J. Struct. Biol. 2013, 182, 93–105. [Google Scholar] [CrossRef] [Green Version]
- Tomonaga, S.; Baba, M.; Baba, N. Alternative automatic alignment method for specimen tilt-series images based on back-projected volume data cross-correlations. Microscopy 2014, 63, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Kvashnin, D.G.; Bourgeois, L.; Fernando, J.F.S.; Firestein, K.; Sorokin, P.B.; Fukata, N.; Golberg, D. Mechanical, Electrical, and Crystallographic Property Dynamics of Bent and Strained Ge/Si Core–Shell Nanowires As Revealed by in situ Transmission Electron Microscopy. Nano Lett. 2018, 18, 7238–7246. [Google Scholar] [CrossRef] [PubMed]
- Karakulina, O.M.; Demortière, A.; Dachraoui, W.; Abakumov, A.M.; Hadermann, J. In Situ Electron Diffraction Tomography Using a Liquid-Electrochemical Transmission Electron Microscopy Cell for Crystal Structure Determination of Cathode Materials for Li-Ion batteries. Nano Lett. 2018, 18, 6286–6291. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trépout, S. Tomographic Collection of Block-Based Sparse STEM Images: Practical Implementation and Impact on the Quality of the 3D Reconstructed Volume. Materials 2019, 12, 2281. https://doi.org/10.3390/ma12142281
Trépout S. Tomographic Collection of Block-Based Sparse STEM Images: Practical Implementation and Impact on the Quality of the 3D Reconstructed Volume. Materials. 2019; 12(14):2281. https://doi.org/10.3390/ma12142281
Chicago/Turabian StyleTrépout, Sylvain. 2019. "Tomographic Collection of Block-Based Sparse STEM Images: Practical Implementation and Impact on the Quality of the 3D Reconstructed Volume" Materials 12, no. 14: 2281. https://doi.org/10.3390/ma12142281
APA StyleTrépout, S. (2019). Tomographic Collection of Block-Based Sparse STEM Images: Practical Implementation and Impact on the Quality of the 3D Reconstructed Volume. Materials, 12(14), 2281. https://doi.org/10.3390/ma12142281