Optical Biosensors Based on Photonic Crystals Supporting Bound States in the Continuum
Abstract
1. Introduction
2. Materials and Methods
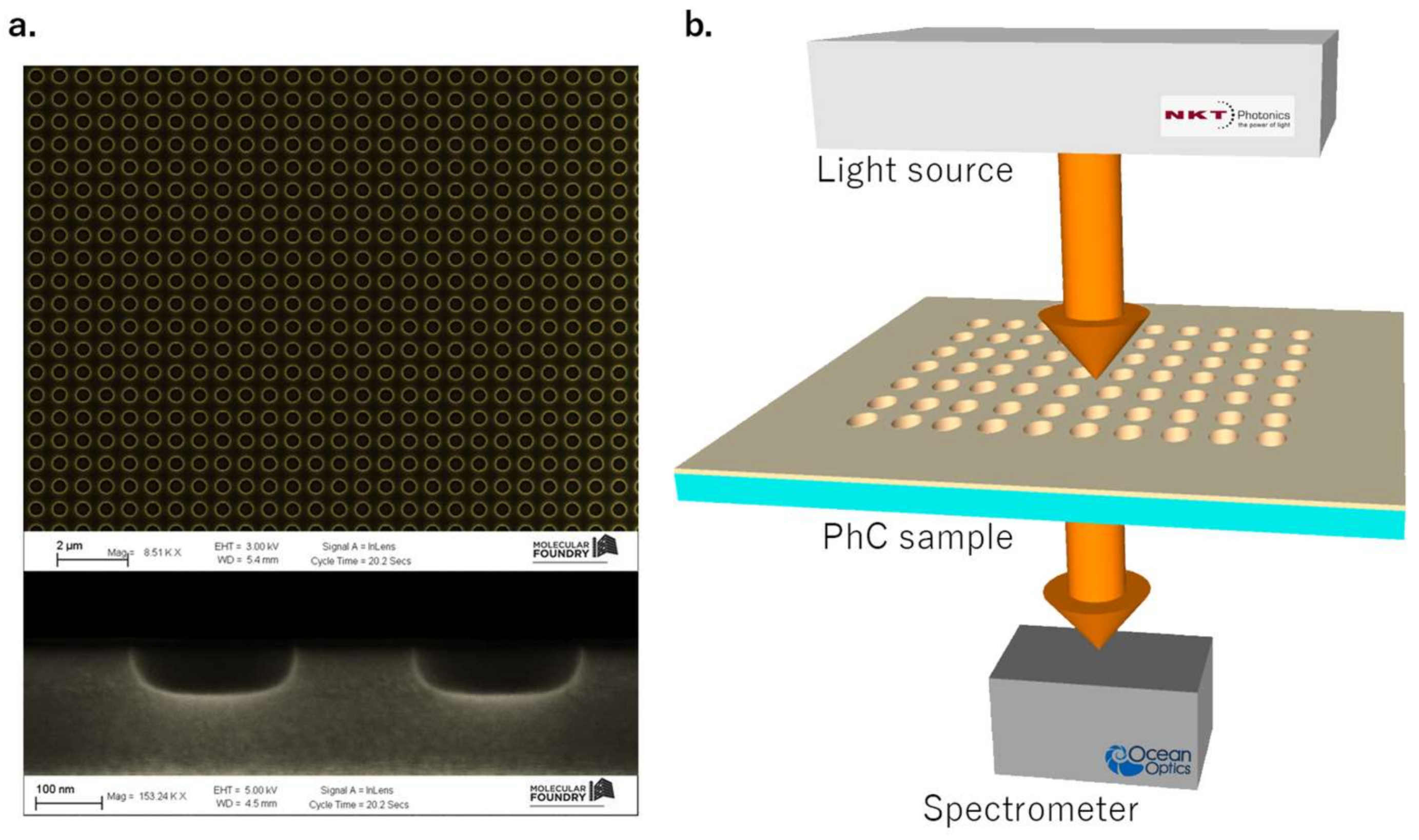
2.1. Design, Fabrication, and Optical Characterization
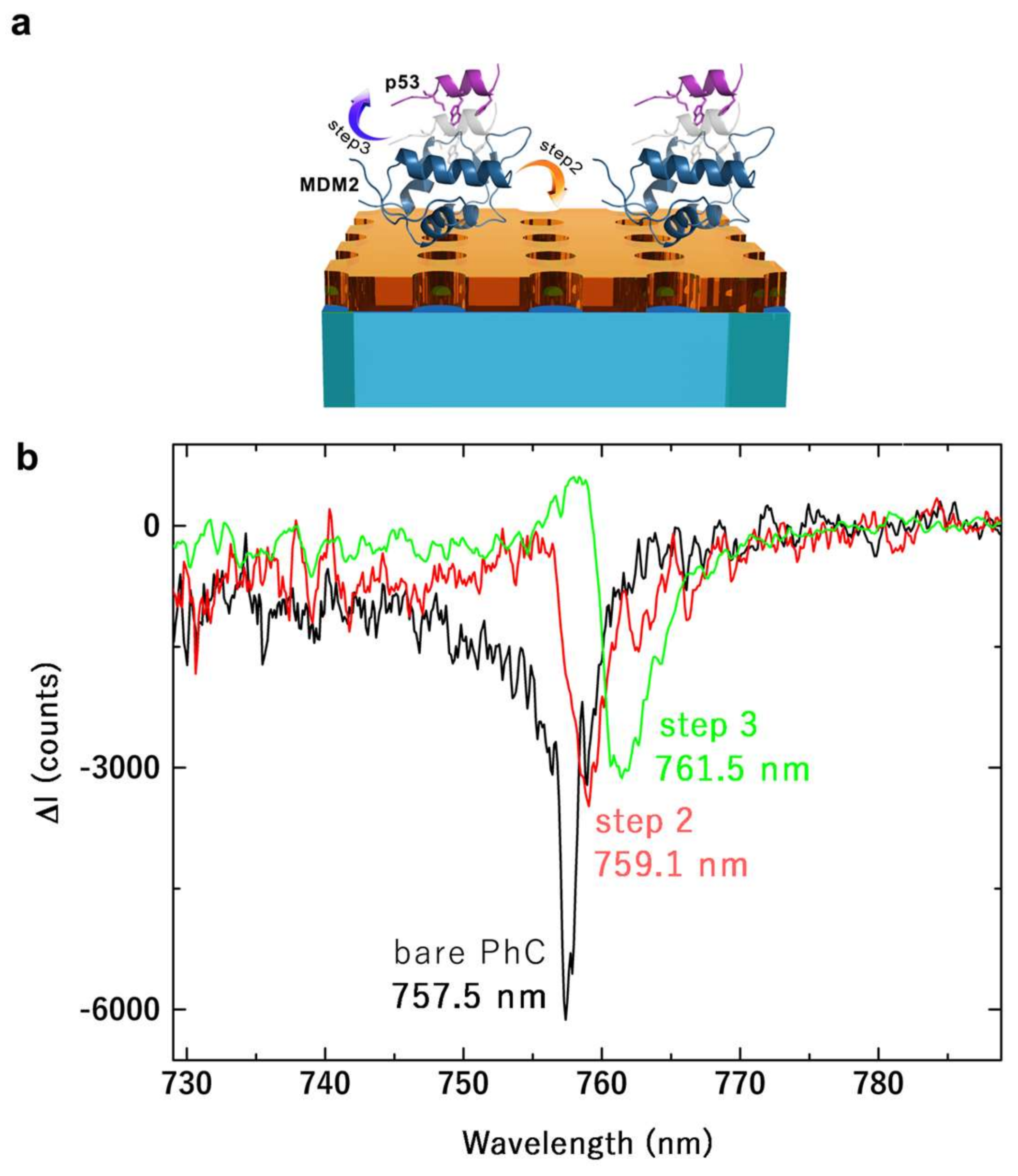
2.2. Surface Modification
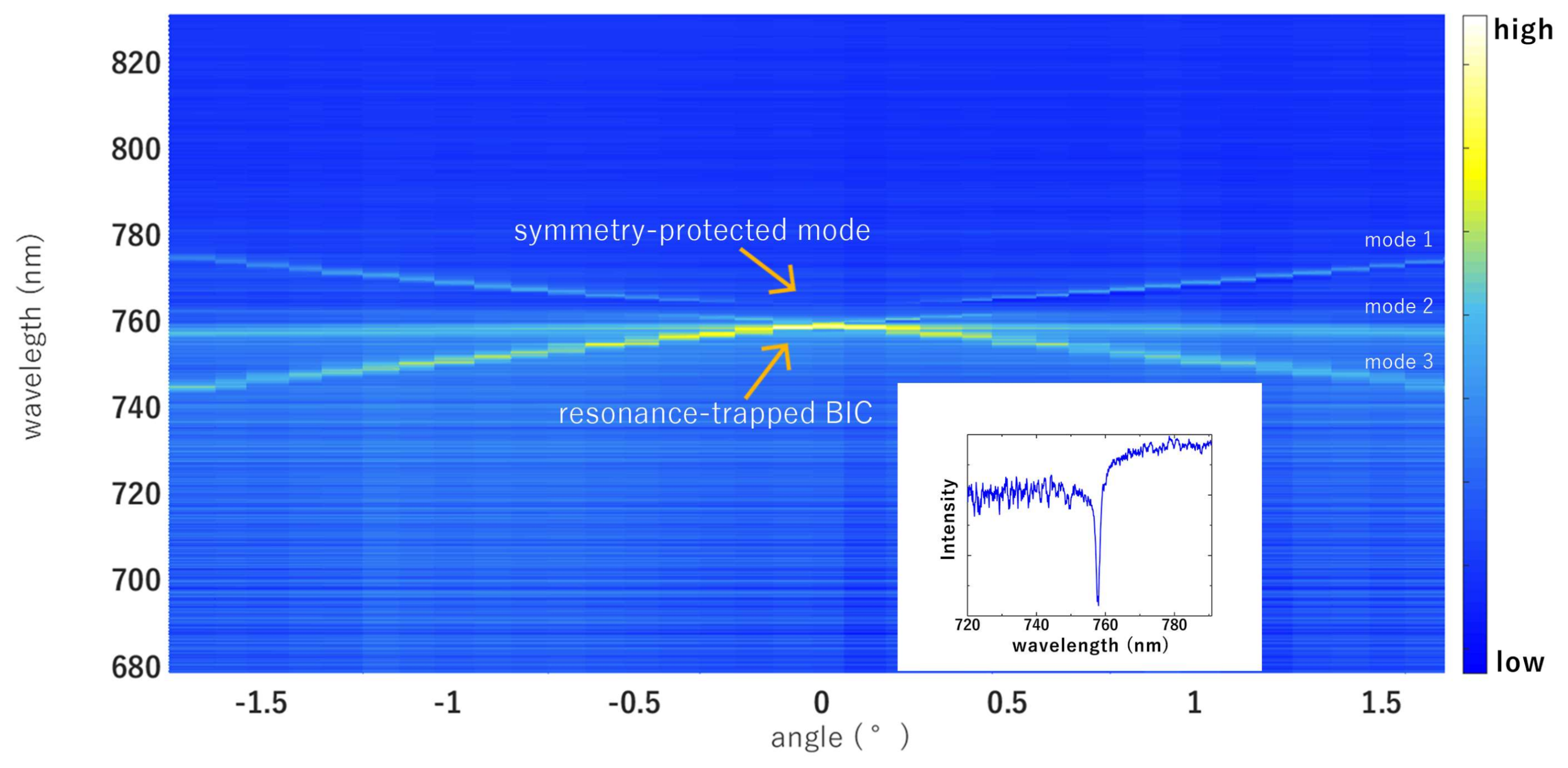
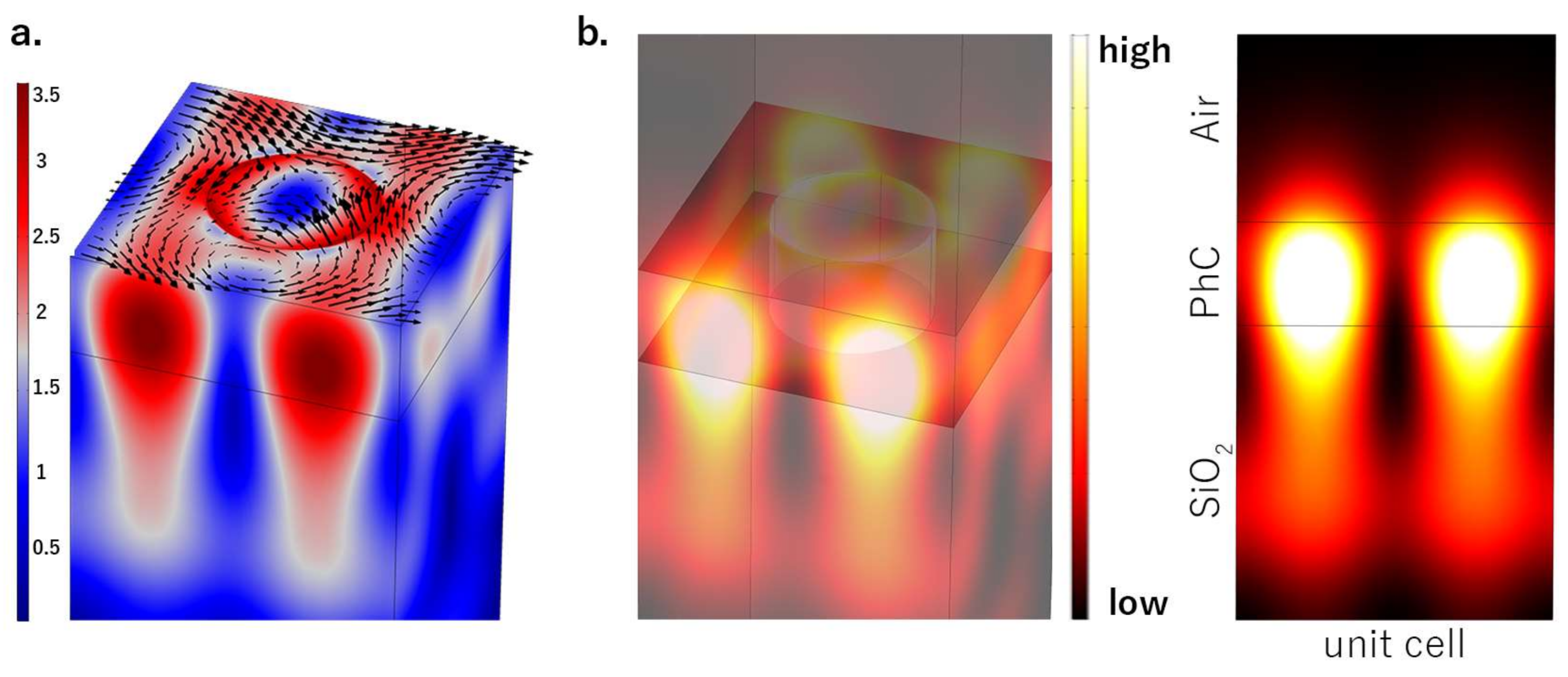
3. Results and Discussion
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
Chemicals and Reagents
References
- Borisov, S.M.; Wolfbeis, O.S. Optical biosensors. Chem. Rev. 2008, 108, 423–461. [Google Scholar] [CrossRef] [PubMed]
- Ligler, F.S.; Ligler, F.S. Perspective on Optical Biosensors and Integrated Sensor Systems. Anal. Chem. 2009, 81, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Wijaya, E.; Lenaerts, C.; Maricot, S.; Hastanin, J.; Habraken, S.; Vilcot, J.P.; Boukherroub, R.; Szunerits, S. Surface plasmon resonance-based biosensors: From the development of different SPR structures to novel surface functionalization strategies. Curr. Opin. Solid State Mater. Sci. 2011, 15, 208–224. [Google Scholar] [CrossRef]
- Shankaran, D.R.; Gobi, K.V.; Miura, N. Recent advancements in surface plasmon resonance immunosensors for detection of small molecules of biomedical, food and environmental interest. Sens. Actuators B Chem. 2007, 121, 158–177. [Google Scholar] [CrossRef]
- Homola, J.; Yee, S.S.; Gauglitz, G. Surface plasmon resonance sensors: Review. Sens. Actuators B Chem. 1999, 54, 3–15. [Google Scholar] [CrossRef]
- Green, R.J.; Frazier, R.A.; Shakesheff, K.M.; Davies, M.C.; Roberts, C.J.; Tendler, S.J.B. Surface plasmon resonance analysis of dynamic biological interactions with biomaterials. Biomaterials 2000, 21, 1823–1835. [Google Scholar] [CrossRef]
- Fan, X.; White, I.M.; Shopova, S.I.; Zhu, H.; Suter, J.D.; Sun, Y. Sensitive optical biosensors for unlabeled targets: A review. Anal. Chim. Acta 2008, 620, 8–26. [Google Scholar] [CrossRef] [PubMed]
- Linnert, T.; Mulvaney, P.; Henglein, A. Surface chemistry of colloidal silver: Surface plasmon damping by chemisorbed iodide, hydrosulfide (SH-), and phenylthiolate. J. Phys. Chem. 1993, 97, 679–682. [Google Scholar] [CrossRef]
- De Tommasi, E.; Chiara De Luca, A.; Cabrini, S.; Rendina, I.; Romano, S.; Mocella, V. Plasmon-like surface states in negative refractive index photonic crystals. Appl. Phys. Lett. 2013, 102, 081113. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Y.; Lv, R. A review for optical sensors based on photonic crystal cavities. Sens. Actuators A Phys. 2015, 233, 374–389. [Google Scholar] [CrossRef]
- Zou, Y.; Chakravarty, S.; Kwong, D.N.; Lai, W.C.; Xu, X.; Lin, X.; Hosseini, A.; Chen, R.T. Cavity-Waveguide Coupling Engineered High Sensitivity Silicon Photonic Crystal Microcavity Biosensors With High Yield. IEEE J. Sel. Top. Quantum Electron. 2014, 20, 171–180. [Google Scholar] [CrossRef]
- Hagino, H.; Takahashi, Y.; Tanaka, Y.; Asano, T.; Noda, S. Effects of fluctuation in air hole radii and positions on optical characteristics in photonic crystal heterostructure nanocavities. Phys. Rev. B Condens. Matter Mater. Phys. 2009, 79, 085112. [Google Scholar] [CrossRef]
- Pergande, D.; Geppert, T.M.; Von Rhein, A.; Schweizer, S.L.; Wehrspohn, R.B.; Moretton, S.; Lambrecht, A. Miniature infrared gas sensors using photonic crystals. J. Appl. Phys. 2011, 109. [Google Scholar] [CrossRef]
- Von Neumann, J.; Wigner, E.P. Über merkwürdige diskrete Eigenwerte. Phys. Z. 1929, 30, 465–467. [Google Scholar]
- Plotnik, Y.; Peleg, O.; Dreisow, F.; Heinrich, M.; Nolte, S.; Szameit, A.; Segev, M. Experimental Observation of Optical Bound States in the Continuum. Phys. Rev. Lett. 2011, 107, 183901. [Google Scholar] [CrossRef] [PubMed]
- Marinica, D.; Borisov, A.; Shabanov, S. Bound States in the Continuum in Photonics. Phys. Rev. Lett. 2008, 100, 183902. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.I.; Miroshnichenko, A.E.; Kivshar, Y.S. Surface bound states in the continuum. Phys. Rev. Lett. 2012, 108, 704011. [Google Scholar] [CrossRef] [PubMed]
- Bulgakov, E.N.; Sadreev, A.F. Bound states in the continuum in photonic waveguides inspired by defects. Phys. Rev. B Condens. Matter Mater. Phys. 2008, 78, 751051. [Google Scholar] [CrossRef]
- Porter, R.; Evans, D.V. Embedded Rayleigh-Bloch surface waves along periodic rectangular arrays. Wave Motion 2005, 43, 29–50. [Google Scholar] [CrossRef]
- Yoon, J.W.; Song, S.H.; Magnusson, R. Critical field enhancement of asymptotic optical bound states in the continuum. Sci. Rep. 2015, 5, 18301. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Song, J.; Dong, L.; Lu, M. Optical bound states in slotted high-contrast gratings. J. Opt. Soc. Am. B 2016, 33, 2472. [Google Scholar] [CrossRef]
- Hsu, C.W.; Zhen, B.; Stone, A.D.; Joannopoulos, J.D.; Soljacic, M. Bound states in the continuum. Nat. Rev. Mater. 2016, 1, 16048. [Google Scholar] [CrossRef]
- Mocella, V.; Romano, S. Giant field enhancement in photonic resonant lattices. Phys. Rev. B Condens. Matter Mater. Phys. 2015, 92, 155117. [Google Scholar] [CrossRef]
- Kodigala, A.; Lepetit, T.; Gu, Q.; Bahari, B.; Fainman, Y.; Kanté, B. Lasing action from photonic bound states in continuum. Nature 2017, 541, 196–199. [Google Scholar] [CrossRef] [PubMed]
- Zhen, B.; Chua, S.-L.; Lee, J.; Rodriguez, A.W.; Liang, X.; Johnson, S.G.; Joannopoulos, J.D.; Soljacic, M.; Shapira, O. Enabling enhanced emission and low-threshold lasing of organic molecules using special Fano resonances of macroscopic photonic crystals. Proc. Natl. Acad. Sci. USA 2013, 110, 13711–13716. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ali, M.A.; Chow, E.K.C.; Dong, L.; Lu, M. An optofluidic metasurface for lateral flow-through detection of breast cancer biomarker. Biosens. Bioelectron. 2018, 107, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, W.; Sun, Y. Optical refractive index sensing based on high-Q bound states in the continuum in free-space coupled photonic crystal slabs. Sensors 2017, 17, 1861. [Google Scholar] [CrossRef]
- Sun, T.; Kan, S.; Marriott, G.; Chang-Hasnain, C. High-contrast grating resonators for label-free detection of disease biomarkers. Sci. Rep. 2016, 6, 27482. [Google Scholar] [CrossRef] [PubMed]
- Lahav, G.; Rosenfeld, N.; Sigal, A.; Geva-Zatorsky, N.; Levine, A.J.; Elowitz, M.B.; Alon, U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 2004, 36, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Michael, D.; Oren, M. The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 2003, 13, 49–58. [Google Scholar] [CrossRef]
- Kubbutat, M.H.; Jones, S.N.; Vousden, K.H. Regulation of p53 stability by Mdm2. Nature 1997, 387, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, L.T.; Vu, B.T.; Craves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In Vivo Activation of the p53 Pathway by Small-MoleculeAntagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef] [PubMed]
- Daniele, S.; Taliani, S.; Da Pozzo, E.; Giacomelli, C.; Costa, B.; Trincavelli, M.L.; Rossi, L.; La Pietra, V.; Barresi, E.; Carotenuto, A.; et al. Apoptosis therapy in cancer: The first single-molecule co-activating p53 and the translocator protein in Glioblastoma. Sci. Rep. 2014, 4, 4749. [Google Scholar] [CrossRef] [PubMed]
- D’Silva, L.; Ozdowy, P.; Krajewski, M.; Rothweiler, U.; Singh, M.; Holak, T.A. Monitoring the effects of antagonists on protein-protein interactions with NMR spectroscopy. J. Am. Chem. Soc. 2005, 127, 13220–13226. [Google Scholar] [CrossRef] [PubMed]
- Reed, D.; Shen, Y.; Shelat, A.A.; Arnold, L.A.; Ferreira, A.M.; Zhu, F.; Mills, N.; Smithson, D.C.; Regni, C.A.; Bashford, D.; et al. Identification and characterization of the first small molecule inhibitor of MDMX. J. Biol. Chem. 2010, 285, 10786–10796. [Google Scholar] [CrossRef] [PubMed]
- Lamberti, A.; Sgammato, R.; Desiderio, D.; Punzo, C.; Raimo, G.; Novellino, E.; Carotenuto, A.; Masullo, M. Native PAGE to study the interaction between the oncosuppressor p53 and its protein ligands. Electrophoresis 2015, 36, 552–555. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, T.; Oishi, S.; Honda, K.; Kondoh, Y.; Saito, T.; Kubo, T.; Kaneda, M.; Ohno, H.; Osada, H.; Fujii, N. Affinity-based screening of MDM2/MDMX-p53 interaction inhibitors by chemical array: Identification of novel peptidic inhibitors. Bioorg. Med. Chem. Lett. 2013, 23, 3802–3805. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Romano, S.; Cabrini, S.; Rendina, I.; Mocella, V. Guided resonance in negative index photonic crystals: A new approach. Light Sci. Appl. 2014, 3, e120. [Google Scholar] [CrossRef]
- Fan, S.; Joannopoulos, J. Analysis of guided resonances in photonic crystal slabs. Phys. Rev. B 2002, 65, 235112. [Google Scholar] [CrossRef]
- Zito, G.; Rusciano, G.; Sasso, A. Enhancement factor statistics of surface enhanced Raman scattering in multiscale heterostructures of nanoparticles. J. Chem. Phys. 2016, 145, 54708. [Google Scholar] [CrossRef] [PubMed]
- Zito, G.; Rusciano, G.; Sasso, A. Dark spots along slowly scaling chains of plasmonic nanoparticles. Opt. Exp. 2016, 24, 13584–13589. [Google Scholar] [CrossRef] [PubMed]
- Caballero, D.; Samitier, J.; Bausells, J.; Errachid, A. Direct patterning of anti-human serum albumin antibodies on aldehyde-terminated silicon nitride surfaces for HSA protein detection. Small 2009, 5, 1531–1534. [Google Scholar] [CrossRef] [PubMed]
- Martucci, N.M.; Rea, I.; Ruggiero, I.; Terracciano, M.; De Stefano, L.; Migliaccio, N.; Palmieri, C.; Scala, G.; Arcari, P.; Rendina, I.; et al. A new strategy for label-free detection of lymphoma cancer cells. Biomed. Opt. Express 2015, 6, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Sgammato, R.; Desiderio, D.; Lamberti, A.; Raimo, G.; Novellino, E.; Carotenuto, A.; Masullo, M. Nondenaturing polyacrylamide gel electrophoresis to study the dissociation of the p53·MDM2/X complex by potentially anticancer compounds. Electrophoresis 2015, 36, 3101–3104. [Google Scholar] [CrossRef] [PubMed]



| θ = −2° | θ = −1° | θ = 0° | θ = 1° | θ = 2° |
|---|---|---|---|---|
| 69 nM | 66 nM | 66 nM | 57 nM | 69 nM |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romano, S.; Lamberti, A.; Masullo, M.; Penzo, E.; Cabrini, S.; Rendina, I.; Mocella, V. Optical Biosensors Based on Photonic Crystals Supporting Bound States in the Continuum. Materials 2018, 11, 526. https://doi.org/10.3390/ma11040526
Romano S, Lamberti A, Masullo M, Penzo E, Cabrini S, Rendina I, Mocella V. Optical Biosensors Based on Photonic Crystals Supporting Bound States in the Continuum. Materials. 2018; 11(4):526. https://doi.org/10.3390/ma11040526
Chicago/Turabian StyleRomano, Silvia, Annalisa Lamberti, Mariorosario Masullo, Erika Penzo, Stefano Cabrini, Ivo Rendina, and Vito Mocella. 2018. "Optical Biosensors Based on Photonic Crystals Supporting Bound States in the Continuum" Materials 11, no. 4: 526. https://doi.org/10.3390/ma11040526
APA StyleRomano, S., Lamberti, A., Masullo, M., Penzo, E., Cabrini, S., Rendina, I., & Mocella, V. (2018). Optical Biosensors Based on Photonic Crystals Supporting Bound States in the Continuum. Materials, 11(4), 526. https://doi.org/10.3390/ma11040526







