Abstract
Antimicrobial surfaces can be applied to break transmission pathways in hospitals. Polyaniline (PANI) and poly(3-aminobenzoic acid) (P3ABA) are novel antimicrobial agents with potential as non-leaching additives to provide contamination resistant surfaces. The activity of PANI and P3ABA were investigated in suspension and as part of absorbent and non-absorbent surfaces. The effect of inoculum size and the presence of organic matter on surface activity was determined. PANI and P3ABA both demonstrated bactericidal activity against Escherichia coli and Staphylococcus aureus in suspension and as part of an absorbent surface. Only P3ABA showed antimicrobial activity in non-absorbent films. The results that are presented in this work support the use of P3ABA to create contamination resistant surfaces.
1. Introduction
Microbial resistance to antimicrobial agents is increasing worldwide and it represents a major threat to the successful treatment of infectious diseases [1]. Development of antimicrobial resistance is an inescapable consequence of natural selection and is associated with exposure to antimicrobial agents [1]. Efforts need to be made to decrease the unnecessary exposure of bacteria to antibiotics to reduce the selective pressure driving the development of resistance so that existing antibiotics retain their efficacy for as long as possible [1]. In part, this can be achieved by controlling the spread of pathogenic bacteria and therefore reducing the number of infections that require antibiotic treatment [2].
Healthcare-associated infections are a major contributor to patient morbidity and mortality, and occur in part due to bacterial contamination of hospital surfaces [3]. Surfaces in hospitals that come into contact with hands are regularly contaminated with nosocomial pathogens [4,5]. Infected patients shed pathogenic bacteria, including methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus spp., into their immediate environment [5,6,7,8,9,10]. Surfaces near shedding patients, such as walls, door handles, bed frames, and light switches, tend to be touched frequently and therefore are more likely to be contaminated [3,10,11]. Once a surface is contaminated, a single hand contact event is sufficient to transmit bacteria from the surface to a person [7,8,12].
Bacteria that have been transferred to a surface can persist for a period of time or actively colonise to form a biofilm. Bacterial persistence on a surface is influenced by dynamic environmental conditions, including organic soiling, humidity, and temperature [4,13,14]. Biofilms are highly recalcitrant to antimicrobial treatments and facilitate the persistence of bacteria on surfaces resulting in surface associated pathogen reservoirs, which increase the risk of transmission [15,16,17]. The level of bacterial transfer that occurs between a contaminated surface and a hand following contact has been demonstrated to occur at a comparable level to direct contact with an infectious patient, which is a well-established transmission route [4,18,19]. Hand washing can help to control the spread of infection in hospitals; however, without the decontamination of surfaces, the reservoirs of pathogens will seed further spread. Nosocomial pathogens isolated from hospital surfaces are typically in the range of 100–10,000 colony forming units (CFU)/cm2 [5,10]. For a microbial burden exceeding 250 CFU/100 cm2, transmission from the surfaces to health care workers and/or patients increases [10,20,21]. Therefore, despite the relatively low inocula present, any contamination of a hospital surface by a pathogen should be considered to be a transmission risk [10,20,21].
The involvement of contaminated surfaces in pathogen transmission pathways in hospitals necessitates the improved control of surface microbiology. Reduction of microbial contamination on hospital surfaces could disrupt transmission pathways and potentially reduce infectious disease incidence rates and the associated antibiotic usage [22]. Utilisation of antibacterial surfaces is a promising means of reducing microbial surface load as well as preventing formation of biofilms and surface associated pathogen reservoirs [2]. An ideal antimicrobial surface would be active against relevant bacteria at appropriate bacterial loads and active in environmental conditions relating to potential applications in terms of temperature, relative humidity, pH, exposure to cleaning products, and contaminating organic matter [23,24,25]. The time that is required for decontamination would need to be sufficiently short to be effective in breaking transmission pathways. Activity needs to be retained for sufficiently long periods of time, and after repeated bacterial challenges, to be cost effective [26,27]. Activity overtime is informed by whether the antimicrobial agent is immobilised on the surface or if it has to be released to elicit an effect [28,29]. The release of an antimicrobial agent over time means that the surface concentration of the agent will fall below the threshold needed to exert antimicrobial activity [27]. An ideal antimicrobial surface would also need to be cheap and easy to make, suitable for large-scale production, and have regulatory approval for the intended use [26,30].
To create an antimicrobial surface, we can take one of two basic approaches. First, a coating may be applied to a material or a modification of the surface chemistry of the material made to provide an antimicrobial surface [31]. Alternatively, the material may be fabricated by incorporating an antimicrobial into the material, which can be challenging as manufacturing procedures can involve extreme environmental conditions, including high temperatures and shear forces, which can negatively impact on bactericidal activity [26,28,32]. Covalent attachment of an antimicrobial agent to a surface may cause side reactions that result in conformational changes in the agent, ultimately causing a loss of activity [33]. Therefore, the method of antimicrobial surface production may affect the resulting surface activity.
The activity of an antimicrobial surface is also influenced by the nature of the surface. Surfaces can be absorbent allowing water droplets to move into the surface or they can be non-absorbent, in which water droplets sit on top of the surface [34,35]. These surface properties may affect the antimicrobial activity as a bacterium in a water droplet would have more contact with the antimicrobial agent if it has absorbed into the surface. Non-absorbent surfaces in hospitals are frequently contaminated with pathogens, and include walls, door handles, and bed frames. Much of the focus of development of antimicrobial surfaces in the published literature is on model non-absorbent surfaces, such as metal coupons and plastic films [36,37,38]. Many absorbent surfaces in hospitals are fabric-based, such as apparel worn by healthcare workers and patient privacy curtains [36,39,40]. Privacy curtains are high-touch areas that are contacted by the hands of the healthcare worker before, during, and after patient care, and are infrequently changed [40,41]. It has been demonstrated that more than 90% of privacy curtains can become contaminated within a week of use [41]. Contaminated absorbent surfaces in hospitals may be involved in pathogen transmission [36,39]. Absorbent surfaces are harder to clean or disinfect than non-absorbent surfaces, while the latter facilitates a greater transfer of bacteria [8,41,42]. Therefore, the development of both absorbent and non-absorbent antimicrobial surfaces would help to curtail the spread of infection in hospitals [1].
Antimicrobial polymers are good candidates for immobilised biocides. These polymers can be either polymeric biocides (the repeating unit is a biocide) or biocidal polymers (the active principle is embodied by the whole macromolecule) [28,29]. In this article, we investigate the antimicrobial activity of polyaniline (PANI) and a functionalised derivative (fPANI), homopolymer poly(3-aminobenzoic acid) (P3ABA), as surface-immobilised biocidal polymers. Utilisation of PANI for potential applications is restricted because of its insolubility in common solvents, which renders it difficult to process [43,44]. fPANIs are easily and inexpensively synthesised using substituted aniline monomers, which improves the solubility, and thus the processability, of the resulting polymer [43,44]. PANI and P3ABA are good candidates for incorporation into surfaces because they have thermal stability up to 300 °C, environmental stability in the conducting form, simple and inexpensive synthetic procedures [45,46,47,48], and have been demonstrated to be biocompatible with mammalian cells [49,50,51,52], all of which increases their commercial viability. Surfaces containing PANI and P3ABA are non-leaching [45,53,54], which promotes activity over a longer period of time and reduces both personal and environmental safety concerns [27].
In this study, we investigated the potential of PANI and P3ABA as surface antimicrobial agents. Initial testing involved the challenge of target organisms in suspension, mirroring standard antimicrobial susceptibility testing methods [55,56]. The target organisms selected were the antimicrobial susceptibility testing strains, Escherichia coli 25922 and S. aureus 6538, which reflect bacteria that are commonly isolated from surfaces in hospitals [10,57]. Susceptibility to antimicrobial activity can be influenced by media composition through its effects on bacterial cell physiology [58]. Therefore, E. coli was challenged in Lennox Broth (LB)—a rich media on which cells grow at high rates—and in minimal A salts with 0.4% succinate as the carbon source, which only contains nutrients that are essential for growth [59,60,61]. The slow growth of bacteria in a minimal media environment is similar to what may occur on surfaces in nature [62].
Following confirmation of activity in suspension, PANI and P3ABA were incorporated into absorbent and non-absorbent surfaces. The effect of incorporation on antimicrobial activity was determined in 96 well plate based assays, which allowed for the the testing of many concentrations and treatment times against one inoculum [27]. Absorbent surfaces were modelled using agar mixed with varying amounts of PANI or P3ABA [63]. Drops of liquid containing bacteria absorbed into the solidified agar test surfaces [64]. Non-absorbent surfaces were established in the form of compression moulded Styrene Ethylene Butylene Styrene (SEBS) films [63]. The activity of non-absorbent surfaces containing PANI and P3ABA were then characterised in relevant environmental conditions, including challenging with a range of inocula and in the presence of organic matter [62]. The experimental strategy is summarised in Figure 1, and, taken together, the results presented demonstrate the activity of PANI and P3ABA in suspension and in surfaces, in application relevant settings. The efficacious activity of P3ABA supports the utilisation of this polymer to create contamination resistant surfaces.
Figure 1.
Experimental strategy. Polyaniline (PANI) and functionalised derivative (fPANI) are tested according to the scheme presented. 1 Activity is measured as reduction in the number of viable cells recovered from surfaces. 2 The Gram positive bacterium S. aureus, and the Gram-negative bacterium E. coli were tested as model species.
2. Results
2.1. Activity of PANI and P3ABA Suspensions against E. coli and S. aureus
To examine the activity of PANI and P3ABA, cell viability assays were performed on E. coli 25922 lux and S. aureus 6538 challenged with 0.5% (w/v) suspensions. Activity was determined in rich media, LB broth, for E. coli and S. aureus, as well as a minimal media, minimal A salts with succinate as the carbon source, for E. coli. At 0.5 h, 1 h, 2 h, and 4 h time points the treated cells were enumerated using the drop count method [64].
E. coli and S. aureus treated with PANI suspension were present at similar cell numbers at the earlier, 0.5 h and 1 h, time points (Figure 2A). For the later, 2 h and 4 h, time points E. coli was knocked down by 1 to 2 logs (measured as the difference between inoculum and the median number of viable cells remaining), while S. aureus cell numbers decreased by only ~0.5 log (Figure 2A). The overall difference in sensitivity between E. coli and S. aureus to 0.5% PANI suspension was statistically significant (linear regression analysis, intercepts are different, p value: less than 0.05).
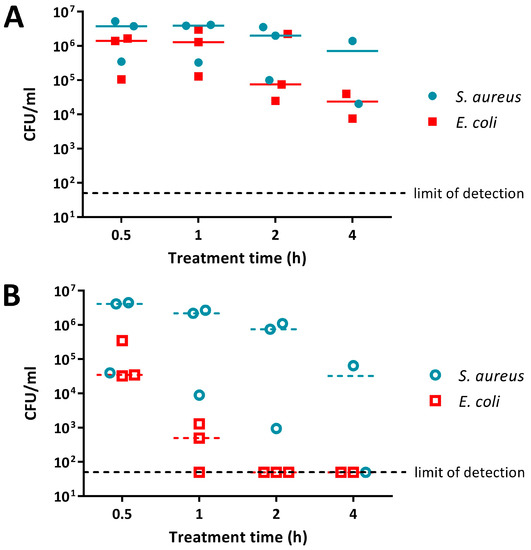
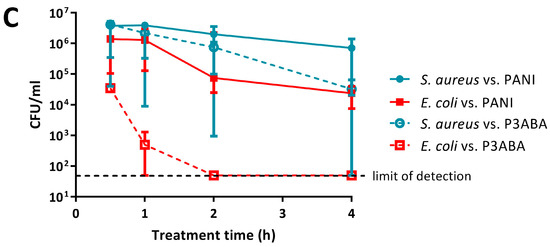
Figure 2.
Sensitivity of E. coli 25922 lux and S. aureus 6538 to PANI and poly(3-aminobenzoic acid) (P3ABA) suspensions. Cell viability assays of ~106 CFU/mL E. coli and S. aureus treated with 0.5% PANI suspension (A) and 0.5% P3ABA suspension (B) in Lennox broth (LB), with each symbol representing the median of three technical replicates and the bar representing the median of each biological replicate. The data in A and B is replotted together in (C) where each point represents the median of biological replicates and the error bars are the range Viable cell counts (colony forming units (CFU)/mL) were obtained for each strain at 0.5 h, 1 h, 2 h, and 4 h time points. The limit of detection is 50 CFU/mL.
Both E. coli and S. aureus were more susceptible to P3ABA suspension when compared to PANI suspension (Figure 2). P3ABA suspension reduced E. coli viable cell numbers to below the limit of detection within 2 h, while S. aureus was knocked down by ~2 log following a 4 h exposure (Figure 2B; i.e., the difference between inoculum and the median number of viable cells remaining was about 2 logs, or 100-fold). As observed for PANI suspension, P3ABA suspension was more active against E. coli as compared to S. aureus (Figure 2B,C). The difference in susceptibility between E. coli and S. aureus to 0.5% P3ABA suspension was statistically significant (linear regression analysis, intercepts are different, p value: less than 0.05). These results confirm that PANI and P3ABA in suspension are active against the model Gram-negative and Gram-positive organisms tested and support investigation of their surface activity.
The antimicrobial activity of 0.5% PANI and P3ABA suspensions in rich and minimal media was determined against E. coli 25922 lux. PANI suspension mediated a greater reduction in the viable cell count in minimal media when compared to rich media (Figure 3A). E. coli in minimal media reached the limit of detection (~4 log reduction) after a 2 h challenge, while a decrease of ~1–2 log was observed in rich media after 4 h (Figure 3A). The greater activity of PANI suspensions against E. coli in minimal media when compared to LB broth was statistically significant (linear regression analysis; intercepts are different, p value: less than 0.05).
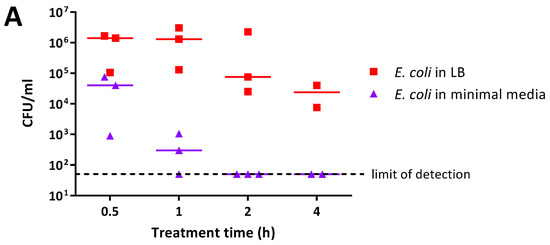
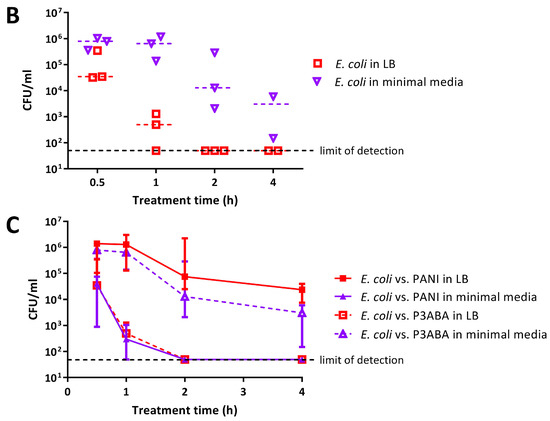
Figure 3.
Sensitivity of E. coli 25922 lux to PANI and P3ABA suspensions in rich and minimal media. Cell viability assays of ~106 CFU/mL E. coli treated with 0.5% PANI suspension (A) and 0.5% P3ABA suspension (B) in Lennox broth (LB) and minimal A salts, with each symbol representing the median of three technical replicates and the bar representing the median of each biological replicate. The data in A and B is replotted together in (C) where each point represents the median of biological replicates and the error bars are the range. Viable cell counts (CFU/mL) were obtained for experimental sample at 0.5 h, 1 h, 2 h, and 4 h time points. The limit of detection is 50 CFU/mL.
P3ABA suspension was more active against E. coli in rich media as compared to minimal media (Figure 3B). E. coli in LB broth was knocked down ~3 log after 1 h and reached the limit of detection by 2 h (Figure 3B). In comparison, the levels of viable E. coli in minimal media were stable at the 0.5 h and 1 h time points (Figure 3B). After 4 h of treatment, P3ABA knocked down E. coli in minimal media by ~2 log (Figure 3B). The difference in activity of P3ABA against E. coli in LB broth and E. coli in minimal media was statistically significant (linear regression analysis, slopes are different, p value: less than 0.05). These results confirm that PANI and P3ABA in suspension are active against E. coli 25922 in rich and minimal media, and support the investigation of their surface activity.
2.2. Activity of Absorbent Surfaces Containing PANI and P3ABA against E. coli and S. aureus
Following the demonstration of activity for PANI and P3ABA against E. coli and S. aureus, we investigated the antibacterial activity of absorbent surfaces containing PANI and P3ABA to simulate surfaces that absorb water droplets, such as fabrics [65]. LB agar as used as a model of an absorbent surface. Agar containing 1% and 2% concentrations of PANI and P3ABA in agar were tested along with a higher concentration of PANI (8% PANI), because PANI was less active than P3ABA in suspension (Figure 2).
To investigate antimicrobial activity, E. coli 25922 lux or S. aureus 6538 were inoculated onto the experimental agar, with cells being rescued at various time points in fresh media. Survival of the PANI or P3ABA in agar challenge was based on growth of rescued cells. A lux-tagged version of E. coli 25922 was used in this work as bioluminescence is a practical alternative to enumeration by plate counts [66,67,68] (Appendix A, Figure A1 and Figure A2). Growth of E. coli 25922 lux was determined by measuring bioluminescence using the VICTOR X Multilabel Plate Reader. Growth of S. aureus 6538 was determined by examining optical density at 600 nm (OD600) using the µQuant™ Microplate Spectrophotometer as a lux-tagged S. aureus 6538 strain was not available for testing.
Agar containing 8% PANI mediated a decrease in bioluminescence levels of E. coli 25922 lux to background levels after 1 h of treatment, while 2% PANI agar required 2 h to reduce surface bacterial load (Figure 4A). The limit of detection for this assay is ~20 CFU (Figure A3). Agar containing 1% PANI did not exhibit antimicrobial activity against E. coli (Figure 4A). P3ABA agar had greater surface antimicrobial activity than PANI agar (Figure 4). Agar containing 1% P3ABA and 2% P3ABA reduced bioluminescence levels from E. coli 25922 lux to background levels within 1 h and 15 min, respectively (Figure 4B).
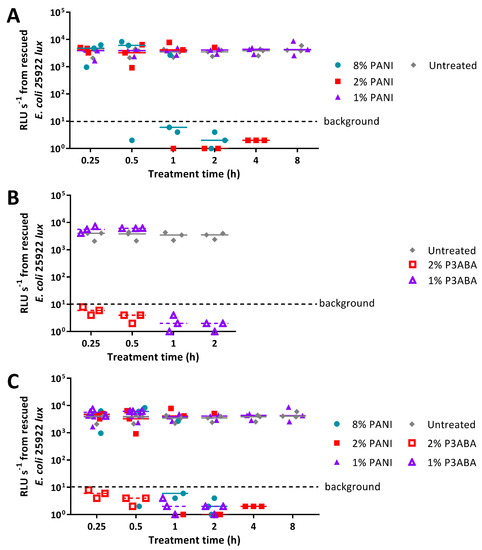
Figure 4.
Sensitivity of E. coli 25922 lux to PANI and P3ABA in agar. (A) ~104 CFU of E. coli was exposed to 8% PANI, 2% PANI, and 1% PANI incorporated into LB agar for 0.25 h, 0.5 h, 1 h, 2 h, 4 h, and 8 h. (B) ~104 CFU of E. coli was exposed to 2% P3ABA and 1% P3ABA incorporated into LB agar for 0.25 h, 0.5 h, 1 h, and 2 h. Following treatment, the cells were rescued by washing the agar surface with LB broth and transferred to a 96 well plate. Each point represents the median of three technical replicates and each bar represents the median of each biological replicate. The data from A and B is combined for comparison in (C). The rescued cells were incubated at 37 °C for 16 h and light release was measured. The vertical axis shows the bioluminescence measurements (relative light units per second, RLU s−1) from the recovered cells with each data point representing an independent experiment and the line representing the median. Background luminescence readings are ~10 RLU s−1.
PANI in agar was less active against S. aureus 6538 than E. coli 25922 lux, reflecting the trend observed with suspension testing (Figure 2A, Figure 4A, and Figure 5A). Agar containing 2% and 8% PANI reduced S. aureus surface load to background levels within 8 h and 4 h, respectively (Figure 5A). The limit of detection for this assay is ~20 CFU (Figure A4). As observed for E. coli, 1% PANI in agar was inactive against S. aureus within the time constraints (8 h) of the experiment (Figure 4A and Figure 5A). The viability of S. aureus cells following treatment with 8% PANI in agar for 2 h, 4 h, and 8 h, and following treatment with 2% PANI in agar for 8 h was significantly different from that of untreated cells (Friedman test, p value: less than 0.05, Dunn’s multiple comparison test).
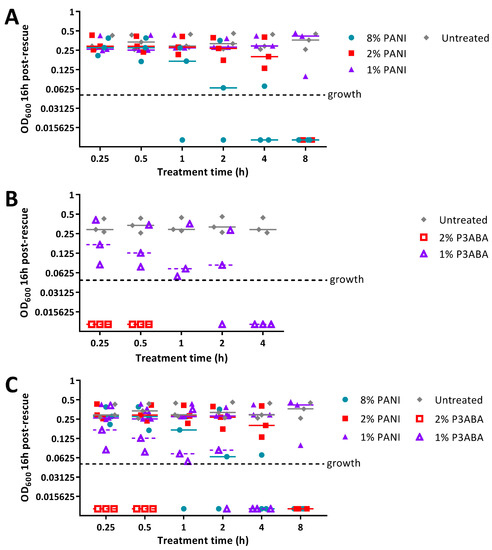
Figure 5.
Sensitivity of S. aureus 6538 to PANI in agar. (A) ~104 CFU of S. aureus was exposed to 8% PANI, 2% PANI, and 1% PANI incorporated into LB agar for 0.25 h, 0.5 h, 1 h, 2 h, 4 h, and 8 h. (B) ~104 CFU of S. aureus was exposed to 2% P3ABA and 1% P3ABA incorporated into LB agar for 0.25 h, 0.5 h, 1 h, 2 h, and 4 h. Following treatment, the cells were rescued by washing the agar surface with LB broth and transferred to a 96 well plate. Each point represents the median of three technical replicates and each bar represents the median of each biological replicate. The data from A and B is combined for comparison in (C). The rescued cells were incubated at 37 °C for 16 h and optical density at 600 nm (OD600) was measured. The vertical axis shows the OD600 measurements from the recovered cells with each data point representing an independent experiment and the line representing the median. OD600 readings above 0.05 are considered as growth.
Agar containing 2% P3ABA effectively decontaminated S. aureus on a surface following a 15 min treatment (Figure 5B), which was consistent with the activity against E. coli (Figure 5A). 1% P3ABA in agar was less active than the higher concentration, with the former requiring a 4 h exposure to reduce bacterial load (Figure 5B). The activity of 2% P3ABA in agar over 15 min and 1% P3ABA in agar over 4 h was statistically significant (Friedman test, p value: less than 0.05, Dunn’s multiple comparison test).
2.3. Activity of Non-Absorbent Surfaces Containing PANI and P3ABA against E. coli and S. aureus
The activity of PANI and P3ABA as surface antimicrobials at non-absorbent surfaces to simulate surfaces that do not absorb water, such as walls and door handles, was investigated using SEBS films containing 5% PANI or 3% P3ABA [69]. The concentrations of the additive in these films are within the range typically used for incorporation into surfaces (0.1–5%) [69] and reflect the greater activity of P3ABA against E. coli and S. aureus compared to PANI as demonstrated in suspension (Figure 2) and in agar (Figure 4 and Figure 5). The activity of PANI and P3ABA films was examined in a ‘micro-surface testing assay’ (MSTA), in which 10 µL of inoculum in LB broth is sandwiched between two pieces of film and recovered at particular time points in fresh LB broth in a 96 well plate [28,70]. Cell viability was determined by measuring the bioluminescence for E. coli 25922 lux and OD600 for S. aureus 6538.
The MSTA for testing 5% PANI and 3% P3ABA films against bacteria was optimised using E. coli 25922 lux. Following either 2 h or 24 h challenges on the films, bacteria were rescued by washing with LB broth and incubated in a 96-well plate for 16 h, after which bioluminescence was measured. The bacteria present in the remaining recovery broth were enumerated using plate counts to verify the ability of bioluminescence levels to infer cell number.
PANI and P3ABA films gave no reduction in bacterial viability for E. coli 25922 lux for after a 2 h challenge, which was indicated by both plate counts and bioluminescence readings (Figure 6A,B). Films containing P3ABA were more active than their PANI counterparts after 24 h exposure, with the former reducing the plate counts and bioluminescence levels by ~2 log relative to the untreated cells (Figure 6A,B). The activity of 3% P3ABA films against E. coli after 24 h treatment was statistically significant (2-way RM ANOVA; CFU/mL p value: less than 0.05; RLU s−1 p value: less than 0.05). The similarity in trends seen between the plate counts and bioluminescence measurements from E. coli 25922 lux that was treated with PANI and P3ABA films confirmed that the bioluminescence-based experimental approach to determining the activity of a non-absorbent surface was appropriate to use for further testing. The results presented show that non-absorbent surfaces containing P3ABA can reduce bacterial load after a 24 h exposure.
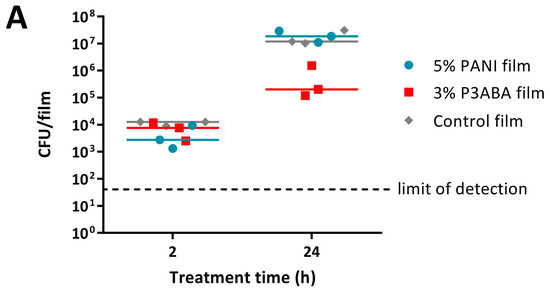
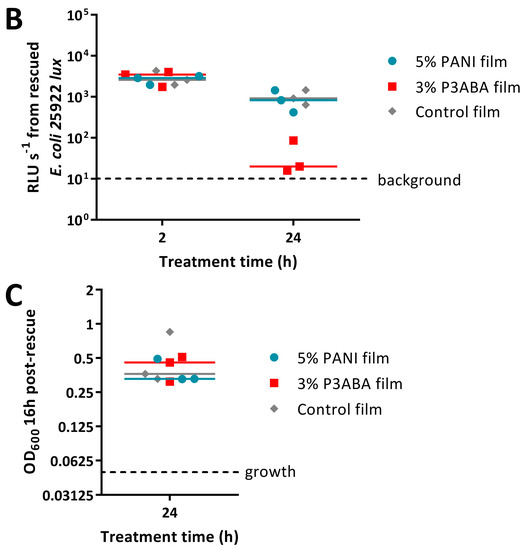
Figure 6.
Sensitivity of E. coli 25922 lux and S. aureus 6538 to PANI and P3ABA films. ~104 CFU of E. coli (A,B) or S. aureus (C) in 10 μL LB broth was sandwiched between two pieces of PANI film, P3ABA film, or control film for 2 h (A,B) and 24 h (A–C). The cells were rescued by washing the film samples with LB broth and transferred to a 96 well plate (B,C). Each point represents the median of three technical replicates and each bar represents the median of each biological replicate. The rescued E. coli cells were also enumerated with plate counts (A). The cells in the 96 well plate were incubated at 37 °C for 16 h and light release (B) or OD600 (C) was measured. The vertical axes show the viable cell counts (A) and bioluminescence measurements (B) from the recovered E. coli cells, and OD600 measurements (C) from the recovered S. aureus cells, with each data point representing an independent experiment and the line representing the median. The limit of detection for the plate counts is 50 CFU/mL. Background luminescence readings are 10 RLU s−1. OD600 readings above 0.05 are considered as growth.
Following from this, the optimised protocol for determining the activity of 5% PANI and 3% P3ABA films was used against S. aureus 6538 with an increase in OD600 above 0.05, indicating the presence of viable cells. S. aureus 6538 was treated for only 24 h as PANI and P3ABA films were not active against E. coli 25922 lux following 2 h treatments (Figure 6A,B) and S. aureus was less sensitive than E. coli to PANI and P3ABA in suspension (Figure 2). Both 5% PANI in films and 3% P3ABA in films displayed no activity against S. aureus 6538 inoculated in LB broth (Figure 6C).
2.4. Characterisation of the Action of PANI and P3ABA Films against E. coli and S. aureus
The activity of 5% PANI and 3% P3ABA films against E. coli and S. aureus, was poorer than expected. We hypothesised that this might be due to inoculating large numbers of bacteria in rich media. To test this hypothesis, films containing PANI and P3ABA were challenged with a range of concentrations of E. coli 25922 lux and S. aureus 6538. The test organisms were washed in saline to simulate a low nutrient environment. A 2 h exposure was used, as this contact time is more effective in disrupting transmission pathways. The influence of the presence of organic matter was examined by challenging PANI and P3ABA films with E. coli 25922 lux washed in LB broth or in 0.85% saline for 2 h.
Bioluminescence from all of the doses of E. coli exposed to films containing 5% PANI was reduced to below background levels (Figure 7A). Films containing 3% P3ABA exhibited similar levels of antimicrobial activity (Figure 7A). The antimicrobial activity of PANI and 3PABA films against each inoculum level tested was significantly different from the control film (2-way RM ANOVA, interaction of film type and CFU dose p value: less than 0.05). These results indicate that PANI and P3ABA films are active against E. coli in low nutrient conditions.
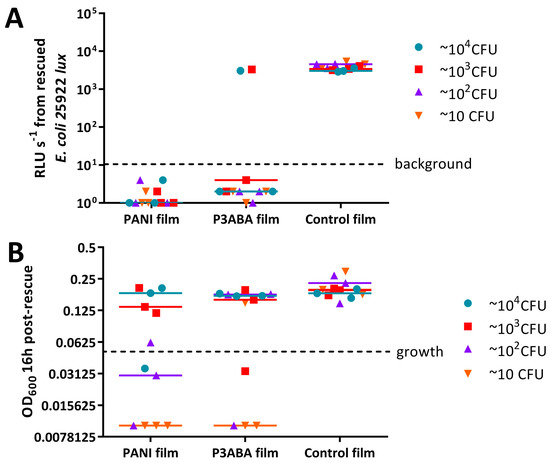
Figure 7.
Activity of PANI and P3ABA films against a range of CFU doses of E. coli 25922 lux and S. aureus 6538. ~10 CFU–~104 CFU of E. coli (A) or S. aureus (B) in 10 μL 0.85% saline was sandwiched between two pieces of PANI film, P3ABA film, or control film for 2 h. The cells were rescued by washing the film samples with LB broth and transferred to a 96 well plate. The rescued cells were incubated at 37 °C for 16 h and light release (A) or OD600 (B) was measured. The vertical axes show the bioluminescence measurements (A) or OD600 (B) from the recovered cells with each data point representing an independent experiment and the line representing the median. Background luminescence readings are 10 RLU s−1. OD600 readings above 0.05 are considered as growth.
OD600 values from ~104 CFU and ~103 CFU doses of S. aureus 6538 treated with 5% PANI and 3% P3ABA films were above OD600 of 0.05, the threshold for growth, indicating that the films were not active against these higher CFU doses (Figure 7B). The OD600 from lower doses of S. aureus that was exposed to PANI and P3ABA films did not increase above the threshold for growth implying killing of the inoculated cells occurred (Figure 7B). The activity of PANI films against ~102 and ~10 CFU, and P3ABA films against ~10 CFU was statistically significant (2-way RM ANOVA, film type p value: less than 0.05, CFU dose p value: less than 0.05). It can be concluded that PANI and P3ABA films are active against low inocula of S. aureus in saline.
The effect of the presence of organic matter on the surface activity of films containing PANI or P3ABA was determined by challenging E. coli 25922 lux in LB broth and 0.85% saline. Bioluminescence levels from E. coli 25922 lux recovered from 5% PANI films and 3% P3ABA films when inoculated in 0.85% saline were below background levels, whereas E. coli 25922 lux inoculated in LB broth released the same amount of light as bacteria that were recovered from control films (Figure 8). This indicates that E. coli in saline was much more sensitive to PANI and P3ABA films than E. coli in LB broth. It is possible that the constituents of LB broth interfere with the contact killing of E. coli on films containing PANI and P3ABA.
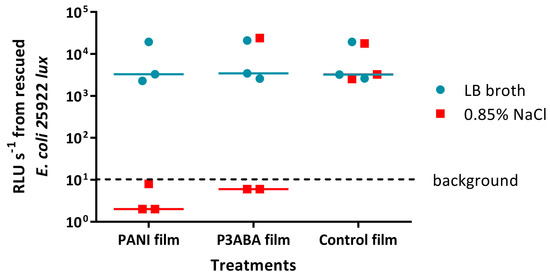
Figure 8.
Activity of PANI and P3ABA films against E. coli 25922 lux in the presence and absence of organic matter. ~104 CFU of E. coli in 10 μL LB broth or 10 μL 0.85% saline was sandwiched between two pieces of PANI film, P3ABA film, or control film for 2 h. The cells were rescued by washing the film samples with LB broth and transferred to a 96 well plate. The rescued cells were incubated at 37 °C for 16 h and light release was measured. The vertical axis shows the bioluminescence measurements (RLU s−1) from the recovered cells with each data point representing an independent experiment and the line representing the median. Background luminescence readings are ~10 RLU s−1.
3. Discussion
PANI and P3ABA are promising additives to materials to create contamination resistance surfaces. Factors that may influence the antibacterial efficacy of the surface were explored over short treatment times (up to 4 h). Disrupting transmission pathways through surface decontamination can be best achieved with an antimicrobial agent that kills over a short period of time [71]. The longer a bacterium persists on a surface, the greater the opportunity to be spread [4]. Therefore, rapid decontamination times will decrease the chance that bacteria may be transferred to a new surface before sterilisation is achieved, and will decrease the likelihood of resistance developing [72].
The activity of PANI and P3ABA was determined against E. coli and S. aureus, representing important pathogens that are found in settings requiring antimicrobial surfaces, such as hospitals and food processing plants. Overall, E. coli had greater susceptibility to both PANI and P3ABA in suspension when compared to S. aureus (Figure 2). A similar trend was observed for PANI or P3ABA in agar (Figure 4 and Figure 5) and in films (Figure 6). These results demonstrate that PANI and P3ABA are active against the model Gram-negative and Gram-positive bacteria, E. coli and S. aureus, respectively, in suspension and in different types of surfaces. The differing levels of activity tha were observed against E. coli and S. aureus highlight how a broad spectrum antimicrobial agent may be more or less effective against a range of bacteria and demonstrates why testing should be done against all the potential target organisms.
The effect of the presence of complex nutrients on the susceptibility of E. coli to PANI and P3ABA in suspension was examined. E. coli was more susceptible to the antimicrobial action of PANI in suspension when incubated in minimal media when compared to LB broth (Figure 3A). The more efficacious activity of PANI in a low nutrient environment supports the incorporation of this antimicrobial agent in surfaces for applications that are associated with only minor contamination with organic matter. In contrast to this, P3ABA was more active against E. coli in rich media relative to minimal media (Figure 3B). The metabolic state of the cell may influence how it responds to bactericidal treatment [73]. The bactericidal action of antimicrobial agents is associated with increased respiration, while bacteriostatic action is characterised by suppressed cellular respiration [73]. The bacteriostatic effect reduces ATP demand and is often the dominant effect blocking bactericidal action [73]. Following from this, if cellular energy output is readily inhibited, such as in cells growing in energy poor conditions, antimicrobial action may result in the inhibition of growth rather than bactericidal killing [73]. Bacterial cells that are highly active may therefore be more susceptible to antimicrobial exposure because of accelerated respiration. The reduced sensitivity of E. coli cells to P3ABA in low nutrient conditions could be reflective of a predisposition to the bacteriostatic effect. The greater activity of P3ABA in the presence of nutrients that facilitate bacterial cell growth supports the use of P3ABA in surfaces in settings that are associated with contamination of organic matter, such as surfaces in the vicinity of patients with gastrointestinal infections, which are commonly contaminated with faecal matter containing the bacteria.
The feasibility of using PANI and P3ABA as additives to create antimicrobial surfaces was examined by determining the activity in suspension. Following confirmation of activity against E. coli and S. aureus (Figure 2), the activity of PANI and P3ABA as agents that are added to absorbent and non-absorbent surfaces was investigated. Overall, both PANI and P3ABA are most active in suspension, followed by in agar and then in films. E. coli treated with 0.5% PANI in suspension for 4 h was reduced in numbers by 2 log (Figure 3A), while 1% PANI in agar did not reduce the viable cell count, even after 8 h of treatment (Figure 4A). For surface incorporated PANI to achieve comparable activity to PANI in suspension, a higher concentration is required. This is demonstrated by total knockdown of E. coli after a 4 h exposure to 2% PANI in agar (Figure 4A); a result that was achieved by a concentration of 0.5% in suspension (Figure 3A). The reduction in activity of surface incorporated PANI and P3ABA is reflective of how immobilisation in a surface can affect bactericidal activity and how different surface matrixes may influence this in different ways [28].
In this study, the antibacterial activity of PANI and an fPANI were determined by the quantification of the viable cells remaining after a period of challenge, using either classical culture-based techniques, or measuring bioluminescence of genetically modified bacteria as a surrogate measure of viability. Future studies may be enhanced by coupling this type of analysis with scanning electron microscopy (SEM) of bacteria on surfaces and fluorescence microscopy after live/dead staining. SEM has previously allowed for visualisation of bacterial killing by fPANIs to the conclusion that the antimicrobial mode of action eventually leads to a loss of cell integrity [74,75]. Fluorescence microscopy of live/dead stained biofilms has allowed for the activity of another fPANI, polysulfanilic acid, to be followed, with the killing of bacteria being established in biofilms and the release of biomass from the surface, imaged [54]. In the study of bacterial attachment to surfaces real time imaging, e.g., using differential interference contrast microscopy [76] may allow for a better understanding of the interaction of bacteria with surfaces and the factors that influence resistance to colonisation.
It is believed that PANI and P3ABA exert antimicrobial action following contact with a bacterial cell [45,77]. Thus, the reduced contact that occurs between a bacterial cell and surface incorporated PANI and P3ABA (relative to in suspension) would mediate the decrease in antimicrobial efficacy. The least amount of contact between the antimicrobial agent and a bacterial cell would occur for non-absorbent surfaces, which mirrors the decreased activity that was observed for PANI and P3ABA in films. 2% PANI and 2% P3ABA in agar (Figure 5) were able to mediate knockdown of S. aureus in 8 h and 15 min, respectively, while 5% PANI and 3% P3ABA in films were unable to reduce bacterial cell numbers after a 24 h treatment (Figure 6C). In this example, higher concentration and treatment time did not ameliorate the reduction of activity for polymers that were incorporated into a non-absorbent surface. The results of this work demonstrate why it is important to test antimicrobial agents, first in suspension (associated with quick and reproducible results) before testing as part of a surface, which should reflect the final application [71].
In real world settings, antimicrobial surfaces may be challenged with a range of inocula. It is well known that the size of the inoculum that is used can influence the magnitude of antimicrobial activity in susceptibility testing [56]. In general, higher inocula need a higher concentration of antimicrobial agent and/or a longer treatment time to achieve knockdown [78]. The surface activity of PANI in film and P3ABA in film was affected by S. aureus inoculum size with activity demonstrated only for lower inocula (Figure 7B). The decreased surface activity in the presence of high numbers of bacteria may be mediated by the piling of bacterial cells on top of each other, thereby reducing direct contact with the antimicrobial agent for a portion of the population [79]. The results that are presented demonstrate the necessity to perform antimicrobial surface testing with appropriate inocula to simulate the potential challenges that would occur in the real world application. Surfaces in hospitals are considered to be contaminated when aerobic colony counts exceed 2.5 CFU/cm2; however, sampling of objects in patient hospital rooms has demonstrated contamination with a range of bacterial loads (up to 104 CFU/m2, equivalent to 102 CFU/cm2), including 103 CFU/m2 (equivalent to 10 CFU/cm2) of MRSA on door handles [80,81,82,83]. Therefore, antimicrobial surfaces in hospitals would need to be active against up to 104 CFU/m2 of contaminants to prevent bacterial spread.
Organic soiling of antimicrobial surfaces is a known cause of loss of activity and thus was investigated for surfaces containing PANI and P3ABA [25,71,84]. Surface activity of both PANI in film and P3ABA in film was decreased in the presence of organic matter (Figure 8). Organic matter can interfere with contact between the bacterial cell and the antimicrobial agent—particularly for charged proteins and polysaccharides that can disrupt charge based interactions—thus providing protection from antimicrobial action [28,85,86]. Additionally, contaminating organic matter may inactivate antimicrobial agents [86]. Typical organic contaminants on hospital surfaces include blood and faecal matter [25,71]. It is important that antimicrobial surfaces are tested in conditions, including contamination with organic matter, relevant to the application to verify that the surfaces will be sufficiently active in these settings [25].
While it is not ideal that a reduction in surface activity was observed, the loss of activity upon soiling is common and the effect of organic soiling can be reduced by regular cleaning. Therefore, antimicrobial surfaces need to be able to withstand any adverse environmental conditions that are associated with cleaning [27]. PANI and P3ABA have thermal stability up to 300 °C and environmental stability in the conducting form [45,46,47,48]. An fPANI containing surface was demonstrated to retain activity against E. coli and S. aureus after 10 repeated challenges if hydrogen peroxide, but not bleach, was the cleaning agent [87]. Future work will include examining the influence of current cleaning procedures on the activity of surface incorporated PANI and P3ABA.
P3ABA containing surfaces demonstrated potential as contamination resistant surfaces for applications. P3ABA as part of a non-absorbent surface reduced E. coli by 2 log after a 24 h incubation (Figure 6B), while an absorbent surface containing 2% P3ABA cleared the bacterial load after 15 min (Figure 4B). The P3ABA containing surfaces in this work indicate a superior performance than has been reported for triclosan, a popular additive claiming antimicrobial activity, which had no effect on the viable cell count of E. coli following a 24 h exposure [88]. Similarly, triclosan-incorporated plastic only inhibited E. coli O157:H7 after a 24 h incubation [89] and triclosan melt-mixed with 4.5% polystyrene inhibited E. coli Y 1090 for 5 h, after which the viable cell number increased [90]. Materials containing P3ABA may therefore have a future as a cost-effective antimicrobial surface to prevent or at least reduce the undesirable spread of micro-organisms.
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
E. coli ATCC 25922 (referred to as E. coli 25922) and S. aureus subsp. aureus ATCC 6538 (referred to as S. aureus 6538) were used in this work because they are routinely used as control organisms to verify that antibiotic susceptibility results are accurate [56,91]. E. coli 25922 was tagged with an integrating plasmid (p16Slux) containing the bacterial luciferase (lux) operon (designated E. coli 25922 lux) [92,93]. E. coli 25922 lux was used for testing of surfaces containing PANI and P3ABA [28]. All strains were grown at 37 °C, with 200 rpm agitation where appropriate. The University of Auckland Institutional Biological Safety Committee approved the construction and use of genetically modified Enterobacteriaceae (GMO04-UA0027).
4.2. Media and Chemicals
PANI and P3ABA were synthesised via chemical oxidation of aniline and 3-aminobenzoic acid monomers, respectively [45]. Cell biology reagents were purchased from Sigma-Aldrich (New South Wales, Australia). Bacteria were cultured in LB broth (BD) or in minimal media. Minimal A medium was used to support growth in a minimal environment, providing only essential nutrients. A 5× minimal A solution was made according to the following: 5 g (NH4)2SO4, 22.5 g KH2PO4, 52.5 g K2HPO4, 2.5 g sodium citrate·2H2O. After autoclaving, this solution was diluted to 1× with sterile water and the following sterile solutions, per litre: 1 mL 1 M MgSO4·7H2O, 0.1 mL 0.5% thiamine plus the carbon source (10 mL of 40% succinate solution per litre).
4.3. Preparation of PANI and P3ABA Suspensions
PANI was finely ground using a mortar and pestle. This insoluble powder requires shaking at 200 rpm to stay in suspension. Reflecting the improved solubility of P3ABA, this polymer was suspended in broth by sonication (QSonica Q700 Sonicator, Newtown, CT, USA) at the following settings: amplitude 30, elapsed time 10 s, repeat 4×. Suspensions of PANI and P3ABA were prepared at 1% (w/v) for a final concentration of 0.5%.
4.4. Activity of PANI and P3ABA Suspensions against E. coli and S. aureus
Turbid overnight cultures of test bacteria were diluted to 106 CFU/mL in LB broth (E. coli 25922 lux and S. aureus 6538) or minimal A salts with 0.4% succinate (E. coli 25922 lux) [94]. The inocula were retrospectively enumerated on LB agar plates [64]. 500 μL of PANI suspension, P3ABA suspension, and growth media (untreated cells) were inoculated with 500 µL of diluted culture. At 0.5 h, 1 h, 2 h, and 4 h time points, each experimental sample was enumerated on LB agar plates. Following incubation, colonies were counted and CFU/mL was calculated. At least three biological replicates were obtained.
Linear regression analysis was used to compare the sensitivity of test strains to PANI or P3ABA suspensions. Specifically, the sensitivity of E. coli and S. aureus in LB broth to each suspension was compared and the sensitivity of E. coli in LB broth and in minimal media to each suspension was compared. Statistical analysis by linear regression was performed using GraphPad Prism software version 6 (GraphPad Software, Inc., La Jolla, CA, USA). Data was graphed in a scatter plot that was generated with viable cell counts post-treatment (CFU/mL) represented on the y-axis and time (h) represented on the x-axis. Linear regression was used to fit a straight line (regression line) through the data for the categorical factor (strain type or media type) generating the best-fit value of the slope and intercept. An analysis of covariance (ANCOVA) was used to compare the regression lines from the categorical factors to determine if there was a statistically significant difference in sensitivity.
4.5. Activity of Absorbent Surfaces Containing PANI and P3ABA against E. coli and S. aureus
Absorbent surfaces containing PANI or P3ABA can be modelled using agar, as drops of liquid containing bacteria will absorb into the agar surface [64]. Molten agar was mixed with varying amounts of PANI or P3ABA, which when left to set created absorbent surfaces containing the antimicrobial agents. PANI or P3ABA were established in agar at 1% and 2%; PANI was also established in agar at 8%. PANI and P3ABA containing absorbent surfaces were set up in triplicate in a 96 well plate by aliquoting 200 µL of each test agar and 200 µL of LB agar (for the untreated control) into individual wells. A turbid culture of test bacteria was diluted to 106 CFU/mL in broth and retrospectively enumerated. All of the test surfaces were inoculated with 10 µL diluted culture, resulting in 104 CFU in each well [55,56]. Agar samples for background readings received 10 µL LB broth.
At specified time points, bacterial cells were rescued in 200 µL fresh media in a 96 well plate [95]. Each type of absorbent surface was tested for the necessary time to achieve knockdown, therefore, highly active surfaces were tested only for the shorter treatment times. E. coli 25922 lux and S. aureus 6538 were challenged with PANI in agar for the following treatment times: 15 min, 30 min, 1 h, 2 h, 4 h, and 8 h. E. coli 25922 lux was exposed to P3ABA in agar for the following treatment times: 15 min, 30 min, 1 h, and 2 h. S. aureus 6538 was exposed to P3ABA in agar for the following treatment times: 15 min, 30 min, 1 h, 2 h, and 4 h. The viability of rescued E. coli 25922 lux was assessed after 16 h incubation by measuring bioluminescence using the VICTOR X Multilabel Plate Reader (Perkin Elmer, Foster City, CA, USA). The viability of rescued S. aureus 6538 was determined by measuring OD600 using the µQuant™ Microplate Spectrophotometer (BioTek Instruments, Winooski, VT, USA). Three biological replicates were obtained for each experiment.
The Friedman test was used to analyse the differences between untreated cells and those that were treated with PANI or P3ABA in agar. When a significant difference was identified (p value less than 0.05), specific groups were compared to each other using Dunn’s multiple comparison test. Dunn’s multiple comparison test was used to compare the treated and untreated cells at each time point, with a p value of less than 0.05 indicating a significant difference. Thus, comparisons were made between each treatment time for every concentration tested to identify a concentration-contact time combination that is associated with significant surface activity.
4.6. Activity of Non-Absorbent Surfaces Containing PANI and P3ABA against E. coli and S. aureus
Non-absorbent surface samples were prepared using SEBS films containing 5% PANI or 3% PANI or no additive (control film). The films were hole punched to generate ~5 mm diameter circles that fit into the wells of a 96 well plate. The film samples were disinfected by immersion in 70% ethanol for 10 min and dried in the Herasafe™ KS (NSF) Class II, Type A2 Biological Safety Cabinet (Thermo Scientific, Auckland, New Zealand) [25].
A turbid overnight culture of test bacteria was diluted to 106 CFU/mL in broth and enumerated. The activity of the PANI and P3ABA containing film samples was determined using the MSTA adapted from Japanese Industry Standard (JIS Z-2801) method [28,70]. A piece of film was placed in an empty well, inoculated with 10 µL of diluted culture, and a second piece of the same type of film was placed on top of the inoculum [28,70]. Film samples for background readings received 10 µL of LB broth. The film treatments were established in triplicate. At the specified time point(s) bacterial cells were rescued in 190 µL LB broth in a fresh 96 well plate [95]. The rescued cells were incubated at 37 °C in a sealed container with moist tissue for 16 h and the viability of rescued cells was determined [95]. Three biological replicates were obtained for each experiment.
For E. coli 25922 lux, cells were rescued after 2 h and 24 h treatments. The viability of cells post-treatment was assessed by using plate counts and measuring bioluminescence. To this end, a 100 µL aliquot of rescued cells was used to enumerate by drop counts and the remaining 100 µL of rescued cells was added to a dark OptiPlate-96 well microtitre plate containing 100 µL of LB broth for the measurement of bioluminescence using the VICTOR X Multilabel Plate Reader. For S. aureus 6538, cells were exposed to film treatments for only 24 h and the viability of cells post-treatment was assessed by incubating 200 µL of rescued cells in a 96 well plate for 16 h and measuring OD600 using the µQuant™ Microplate Spectrophotometer.
The activity of PANI and P3ABA in films against E. coli 25922 lux was analysed using a two-way repeated measures analysis of variation (2-way RM ANOVA). For both the plate counts and the bioluminescence data, the 2-way RM ANOVA determined how E. coli 25922 lux cell number was affected by two factors, treatment time (2 h and 24 h) and film type (PANI in film, P3ABA in film, no additive). A p value of less than 0.05 indicates that the cell number was significantly affected by at least one of the factors. When a significant difference was identified, treated cells were compared to the untreated control for each time point using Dunnett’s multiple comparison test with a p value of less than 0.05, indicating a significant difference.
The Friedman test was used to analyse the differences between untreated S. aureus 6538 cells and those that were treated with PANI or P3ABA in film. The Friedman test is a nonparametric test that compares three or more matched groups—cells treated with 5% PANI in film, 3% P3ABA in film, and control film. A p value of less than 0.05 indicates that at least one of the groups differs from the rest. When a significant difference was identified, specific groups were compared to each other using Dunn’s multiple comparison test. Dunn’s multiple comparison test was used to compare the treated and untreated cells at each time point, with a p value of less than 0.05 indicating a significant difference.
4.7. Characterisation of the Action of PANI and P3ABA Films against E. coli and S. aureus
4.7.1. Challenge of PANI and P3ABA Films with a Range of CFU Doses of E. coli 25922 lux and S. aureus 6538 in Saline
Film punches were prepared and decontaminated, as described above. The MSTA was performed with bacterial challenges (104 CFU, 103 CFU, 102 CFU, and 10 CFU) prepared in 10 µL saline. The 106 CFU/mL culture was enumerated. Following a 2 h treatment, cells were rescued in 190 µL LB broth and incubated in a fresh 96 well plate for 16 h. Viability of bacteria was assessed by measuring bioluminescence for E. coli 25922 lux and by measuring OD600 for S. aureus 6538. The activity of PANI and P3ABA in films against a range of CFU doses of E. coli 25922 lux and S. aureus 6538 was analysed using a 2-way RM ANOVA.
4.7.2. Assay to Evaluate the Influence of the Presence of Organic Matter on the Activity of PANI and P3ABA Films against E. coli 25922 lux
Film punches were prepared and decontaminated, as described above. MSTA was performed with bacterial challenges (104 CFU) in 10 µL saline or 10 µL LB broth. The inocula were enumerated. Following a 2 h treatment, cells were rescued in 190 µL LB broth and incubated in a fresh 96 well plate for 16 h. The viability of rescued cells was determined by measuring the bioluminescence.
4.8. Appendix A Methods
4.8.1. Validation of Utilisation of E. coli 25922 lux
To examine if E. coli 25922 lux can be used for the testing of surfaces containing PANI and P3ABA, in place of the non-tagged version, the MIC and MBC of both strains were determined [55]. A range of concentrations of PANI and P3ABA in suspension were tested (0.03125–4%). The suspensions were established at 2× the final desired concentration in 500 µL. The insolubility of PANI required each suspension to be set up separately by weighing the powder into 5 mL tubes and adding 500 µL LB broth. P3ABA suspensions were established from a stock solution using a doubling dilution series. 500 µL LB broth was aliquoted to set up an untreated control.
The PANI and P3ABA suspensions were inoculated with 500 µL of 106 CFU/mL of E. coli 25922 and E. coli 25922 lux. The MIC was defined as the lowest concentration of PANI or P3ABA that was able to inhibit the visible growth of test bacteria following a 24 h treatment [55,56]. Tubes that were observed by eye to have no visible growth were selected for MBC testing. For this, 20 µL of the experimental sample was spread onto six LB agar plates [55,56]. The spread plates were incubated at 37 °C for 16 h and the growth on these plates was determined. When countable colonies were present, the CFU/mL of the sample was calculated. The MBC was defined as the lowest concentration of PANI or P3ABA that either totally prevents growth or results in a ≥99.9% decrease in the initial inoculum following subculture on LB agar plates [55,96]. At least three biological replicates were obtained.
4.8.2. Determination of the Limit of Detection for E. coli 25922 lux and S. aureus 6538 Growing in a 96 Well Plate
The limit of detection of E. coli 25922 lux and S. aureus 6538 growing in a 96 well plate was examined by determining the lowest number of cells added to LB broth in a 96 well plate that can grow to detectable levels [95]. This was achieved by serially diluting an overnight culture in triplicate in a 96 well plate by transferring 20 µL of culture into wells containing 180 µL of LB broth. A range of inocula were established from ~109 CFU/mL to ~1 CFU/mL. The overnight culture was enumerated to confirm the cell numbers that were tested. The 96 well plate was incubated at 37 °C for 16 h in a sealed container with a moist tissue [95]. Growth of bacteria was assessed by measuring bioluminescence using the VICTOR X Multilabel Plate Reader for E. coli 25922 lux and by measuring OD600 using the µQuant™ Microplate Spectrophotometer for S. aureus 6538.
5. Conclusions
PANI and P3ABA both demonstrated bactericidal activity against E. coli and S. aureus in suspension and as part of an absorbent surface, with greater activity being observed with P3ABA. PANI in films was not active against E. coli or S. aureus, while P3ABA in films reduced the viability of E. coli after a 24 h treatment. The results that are presented in this work support the use of P3ABA to create contamination resistant surfaces.
Acknowledgments
The authors are grateful for research funding from both the New Zealand Ministry of Business, Innovation and Employment (MBIE) for research programmes UOAX0812 and UOAX1410, and the University of Auckland’s Vice Chancellors Strategic Development Fund, grant number 23563. The authors are grateful for funding (University of Auckland, SMS Publication Bursary) covering the costs to publish in open access. The authors thank Sudip Ray, Adeline Le Cocq, Chris Wilcox and Walt Wheelwright for purified PANI and P3ABA.
Author Contributions
Julia Robertson and Simon Swift conceived and designed the experiments; Julia Robertson performed the experiments and analysed the data; Marija Gizdavic-Nikolaidis contributed materials and advised on the chemistry aspects; Julia Robertson wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Appendix A
Appendix A.1. Validation of Utilisation of E. coli 25922 lux as a Proxy of E. coli 25922 for Investigation of PANI and P3ABA as Surface Antimicrobial Agents
Appendix A.1.1. PANI Has Similar Activity against E. coli 25922 and E. coli 25922 lux While P3ABA Is Less Active against the Latter
For E. coli 25922, a lux-tagged version was used as the released bioluminescence can be detected and serves as a marker of cell viability [28,92,93]. Utilisation of a bioluminescently-tagged strain is a practical alternative to enumeration by plate counts for future testing of these potential surface additives against slow growing bacteria, such as Mycobacterium tuberculosis [63]. E. coli 25922 lux has a chromosomal insertion of the bacterial luciferase (lux) operon (luxCDABE) into the 16S locus [93]. A cell expressing the lux operon (luxCDABE) will be in an altered state compared to the non-tagged version as cellular energy is diverted in order to generate the luminescence [28]. The bioluminescence reaction consumes reduced flavin mononucleotide (FMNH2) and a long chain fatty aldehyde, and tetradecanoic acid is diverted from the fatty acid biosynthesis pathway to regenerate the aldehyde substrate [97]. Therefore, it is possible that the lux-tagged version of E. coli 25922 may have different sensitivities to PANI and P3ABA compared to the non-bioluminescent version.
Following from this, the activity of PANI and P3ABA in LB broth was determined against E. coli 25922 and lux-tagged E. coli 25922. The measure of activity used was the standard minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) [55]. For PANI in suspension, E. coli 25922 and the lux-tagged version had similar sensitivities (Figure A1). P3ABA had a similar MIC against E. coli 25922 and E. coli 25922 lux (Figure A2); however, the MBC of P3ABA against E. coli 25922 (0.125%) was lower than that for E. coli 25922 lux (1–4%), which was statistically significant (Mann-Whitney test, p value: less than 0.05). The difference in activity of P3ABA observed against E. coli 25922 and E. coli 25922 lux may be reflective of the metabolic burden of light production. The lux-tagged E. coli 25922 may be less susceptible to the bactericidal action of P3ABA over a 24 h treatment time. Overall, these results support the use of lux-tagged E. coli for testing of PANI and P3ABA.
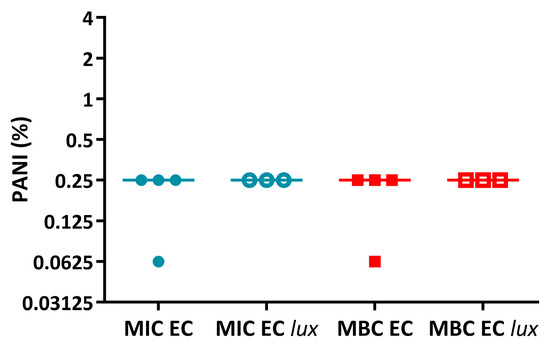
Figure A1.
Activity of PANI against E. coli 25922 and E. coli 25922 lux. The MIC (circles) and MBC (squares) of PANI against E. coli 25922 (EC) and E. coli 25922 lux (EC lux) in LB broth. Data obtained from E. coli 25922 is represented by filled data points while data obtained from E. coli 25922 lux is represented by unfilled data points.
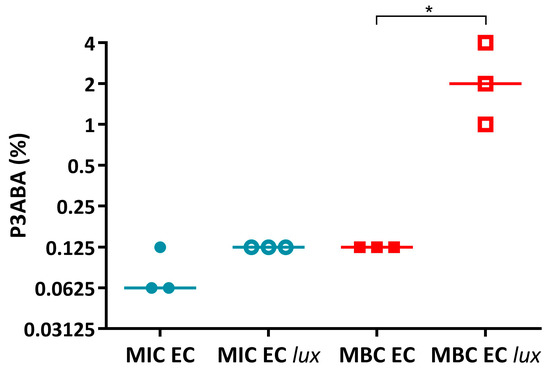
Figure A2.
Activity of P3ABA against E. coli 25922 and E. coli 25922 lux. The MIC (circles) and MBC (squares) of PANI against E. coli 25922 (EC) and E. coli 25922 lux (EC lux) in LB broth. Data obtained from E. coli 25922 is represented by filled data points while data obtained from E. coli 25922 lux is represented by unfilled data points. Statistical significance is represented by * (Mann-Whitney test, p value: less than 0.05).
Appendix A.1.2. The Limit of Detection for E. coli 25922 lux and S. aureus 6538 Recovered in a 96 Well Plate Is 100 CFU/mL
Examination of the activity of PANI and P3ABA in surfaces involved recovery of challenged cells in a 96 well plate, which facilitated high-throughput testing of many concentrations and treatment times against one inoculum [95]. Therefore, it was necessary to determine the limit of detection of E. coli 25922 lux and S. aureus 6538 growing in this manner to enable interpretation of surface testing results. The limit of detection was examined by determining the lowest number of cells added to 180 µL of LB broth in a 96 well plate that can grow to detectable levels [95]. The limit of detection is presented as CFU/mL to relate back to the initial inoculum concentration. The absolute number of cells present in the 180 µL volume in the wells would be roughly 5-fold lower than the CFU/mL value.
After a 16 h incubation of E. coli 25922 lux, bioluminescence 2 log or more above background levels was detected in the wells inoculated with ~109 to ~103 CFU/mL indicating bacterial growth (Figure A3). For the well inoculated with ~102 CFU/mL, the median bioluminescence level was 2 log above background levels; although, two points were only slightly above background levels (Figure A3). Therefore, the limit of detection of E. coli 25922 lux in a 96 well plate was 100 CFU/mL, which would correspond to ~20 CFU in the well.
After a 16 h incubation of S. aureus 6538, growth was detected in wells inoculated with ~109 to ~102 CFU/mL (with growth defined as OD600 above 0.05) (Figure A4). The median OD600 value from starting inoculum of ~10 CFU/mL was above the threshold for growth; however, two points were below background levels (Figure A4). Therefore, the limit of detection of S. aureus 6538 in 96 well plate was 100 CFU/mL, which would correspond to ~20 CFU in the well.
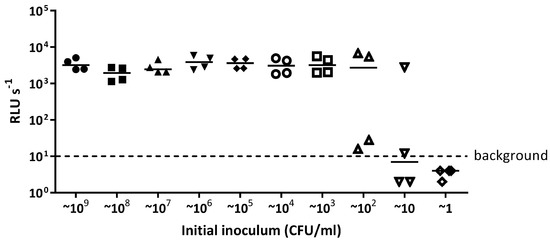
Figure A3.
The limit of detection of E. coli 25922 lux when grown in a 96 well plate. E. coli 25922 lux was serially diluted in LB broth from ~109 CFU/mL to ~1 CFU/mL in a 96 well plate and incubated at 37 °C for 16 h. Light release (RLU s−1) was measured from each dilution and detectable light levels above 10 RLU s−1 indicates growth. The data presented are from four independent experiments.
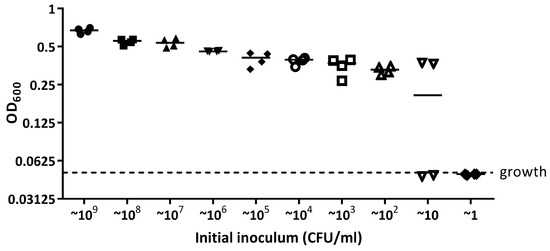
Figure A4.
The limit of detection of S. aureus 6538 when grown in a 96 well plate. S. aureus 6538 was serially diluted in LB broth from ~109 CFU/mL to ~1 CFU/mL in a 96 well plate and incubated at 37 °C for 16 h. OD600 was measured from each dilution and OD600 readings above 0.05 are considered as growth. The data presented are from four independent experiments.
References
- WHO. Antimicrobial Resistance: Global Report on Surveillance; WHO Press: Geneva, Switzerland, 2014. [Google Scholar]
- Dancer, S.J. Controlling hospital-acquired infection: Focus on the role of the environment and new technologies for decontamination. Clin. Microbiol. Rev. 2014, 27, 665–690. [Google Scholar] [CrossRef] [PubMed]
- Weber, D.J.; Anderson, D.; Rutala, W.A. The role of the surface environment in healthcare-associated infections. Curr. Opin. Infect. Dis. 2013, 26, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Schwebke, I.; Kampf, G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 2006, 6, 130. [Google Scholar] [CrossRef] [PubMed]
- Stiefel, U.; Cadnum, J.L.; Eckstein, B.C.; Guerrero, D.M.; Tima, M.A.; Donskey, C.J. Contamination of hands with methicillin-resistant Staphylococcus aureus after contact with environmental surfaces and after contact with the skin of colonized patients. Infect. Control Hosp. Epidemiol. 2011, 32, 185–187. [Google Scholar] [CrossRef] [PubMed]
- Page, K.; Wilson, M.; Parkin, I. Antimicrobial surfaces and their potential in reducing the role of the inanimate environment in the incidence of hospital-acquired infections. J. Mater. Chem. 2009, 19, 3819–3831. [Google Scholar] [CrossRef]
- Scott, E.; Bloomfield, S.F. The survival and transfer of microbial contamination via cloths, hands and utensils. J. Appl. Bacteriol. 1990, 68, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Rusin, P.; Maxwell, S.; Gerba, C. Comparative surface-to-hand and fingertip-to-mouth transfer efficiency of gram-positive bacteria, gram-negative bacteria, and phage. J. Appl. Microbiol. 2002, 93, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Gerhardts, A.; Hammer, T.R.; Balluff, C.; Mucha, H.; Hoefer, D. A model of the transmission of micro-organisms in a public setting and its correlation to pathogen infection risks. J. Appl. Microbiol. 2012, 112, 614–621. [Google Scholar] [CrossRef] [PubMed]
- Otter, J.A.; Yezli, S.; French, G.L. The role played by contaminated surfaces in the transmission of nosocomial pathogens. Infect. Control Hosp. Epidemiol. 2011, 32, 687–699. [Google Scholar] [CrossRef] [PubMed]
- French, G.L.; Otter, J.A.; Shannon, K.P.; Adams, N.M.T.; Watling, D.; Parks, M.J. Tackling contamination of the hospital environment by methicillin-resistant Staphylococcus aureus (MRSA): A comparison between conventional terminal cleaning and hydrogen peroxide vapour decontamination. J. Hosp. Infect. 2004, 57, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, A.; Pultz, N.J.; Gries, D.M.; Ray, A.J.; Eckstein, E.C.; Aron, D.C.; Donskey, C.J. Acquisition of nosocomial pathogens on hands after contact with environmental surfaces near hospitalized patients. Infect. Control Hosp. Epidemiol. 2004, 25, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.P.; Avery, L.M.; Killham, K.; Jones, D.L. Persistence of Escherichia coli O157 on farm surfaces under different environmental conditions. J. Appl. Microbiol. 2005, 98, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, B.; Cerf, O. Review—Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011, 145, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Otter, J.A.; Vickery, K.; Walker, J.T.; de Lancey Pulcini, E.; Stoodley, P.; Goldenberg, S.D.; Salkeld, J.A.G.; Chewins, J.; Yezli, S.; Edgeworth, J.D. Surface-attached cells, biofilms and biocide susceptibility: Implications for hospital cleaning and disinfection. J. Hosp. Infect. 2015, 89, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, F.H. Processes controlling the transmission of bacterial pathogens in the environment. Res. Microbiol. 2007, 158, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Van Houdt, R.; Michiels, C.W. Biofilm formation and the food industry, a focus on the bacterial outer surface. J. Appl. Microbiol. 2010, 109, 1117–1131. [Google Scholar] [CrossRef] [PubMed]
- Hota, B. Contamination, disinfection, and cross-colonization: Are hospital surfaces reservoirs for nosocomial infection? Clin. Infect. Dis. 2004, 39, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, D.M.; Nerandzic, M.M.; Jury, L.A.; Jinno, S.; Chang, S.; Donskey, C.J. Acquisition of spores on gloved hands after contact with the skin of patients with Clostridium difficile infection and with environmental surfaces in their rooms. Am. J. Infect. Control 2012, 40, 556–558. [Google Scholar] [CrossRef] [PubMed]
- Todd, E.C.D.; Greig, J.D.; Bartleson, C.A.; Michaels, B.S. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 5. Sources of contamination and pathogen excretion from infected persons. J. Food Prot. 2008, 71, 2582–2595. [Google Scholar] [CrossRef] [PubMed]
- Malik, R.E.; Cooper, R.A.; Griffith, C.J. Use of audit tools to evaluate the efficacy of cleaning systems in hospitals. Am. J. Infect. Control 2003, 31, 181–187. [Google Scholar] [CrossRef] [PubMed]
- French, G.L. The continuing crisis in antibiotic resistance. Int. J. Antimicrob. Agents 2010, 36, S3–S7. [Google Scholar] [CrossRef]
- Robine, E.; Boulangé-Petermann, L.; Derangère, D. Assessing bactericidal properties of materials: The case of metallic surfaces in contact with air. J. Microbiol. Methods 2002, 49, 225–234. [Google Scholar] [CrossRef]
- Escalada, M.G.; Russell, A.D.; Maillard, J.-Y.; Ochs, D. Triclosan-bacteria interactions: Single or multiple target sites? Lett. Appl. Microbiol. 2005, 41, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Ojeil, M.; Jermann, C.; Holah, J.; Denyer, S.P.; Maillard, J.-Y. Evaluation of new in vitro efficacy test for antimicrobial surface activity reflecting UK hospital conditions. J. Hosp. Infect. 2013, 85, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Suppakul, P.; Miltz, J.; Sonneveld, K.; Bigger, S.W. Active packaging technologies with an emphasis on antimicrobial packaging and its applications. J. Food Sci. 2003, 68, 408–420. [Google Scholar] [CrossRef]
- Bastarrachea, L.J.; Denis-Rohr, A.; Goddard, J.M. Antimicrobial food equipment coatings: Applications and challenges. Annu. Rev. Food Sci. Technol. 2015, 6, 97–118. [Google Scholar] [CrossRef] [PubMed]
- Green, J.-B.D.; Fulghum, T.; Nordhaus, M. Review of immobilized antimicrobial agents and methods for testing. Biointerphases 2011, 6, CL2–CL43. [Google Scholar] [CrossRef] [PubMed]
- Siedenbiedel, F.; Tiller, J.C. Antimicrobial polymers in solution and on surfaces: Overview and functional principles. Polymers 2012, 4, 46–71. [Google Scholar] [CrossRef]
- Cooksey, K. Effectiveness of antimicrobial food packaging materials. Food Addit. Contam. 2005, 22, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Hasan, J.; Crawford, R.J.; Ivanova, E.P. Antibacterial surfaces: The quest for a new generation of biomaterials. Trends Biotechnol. 2013, 31, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Bastarrachea, L.J.; Dhawan, S.; Sablani, S.S. Engineering Properties of Polymeric-Based Antimicrobial Films for Food Packaging: A Review. Food Eng. Rev. 2011, 3, 79–93. [Google Scholar] [CrossRef]
- Vasilev, K.; Cook, J.; Griesser, H.J. Antibacterial surfaces for biomedical devices. Expert Rev. Med. Devices 2009, 6, 553–567. [Google Scholar] [CrossRef] [PubMed]
- Fernández, A.; Soriano, E.; López-Carballo, G.; Picouet, P.; Lloret, E.; Gavara, R.; Hernández-Muñoz, P. Preservation of aseptic conditions in absorbent pads by using silver nanotechnology. Food Res. Int. 2009, 42, 1105–1112. [Google Scholar] [CrossRef]
- Humphreys, H. Self-disinfecting and microbiocide-impregnated surfaces and fabrics: What potential in interrupting the spread of healthcare-associated infection? Clin. Infect. Dis. 2014, 58, 848–853. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.; Spencer, M.; Edmiston, C. Role of healthcare apparel and other healthcare textiles in the transmission of pathogens: A review of the literature. J. Hosp. Infect. 2015, 90, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Weis, T.L.; Schurr, M.J.; Faith, N.G.; Czuprynski, C.J.; McAnulty, J.F.; Murphy, C.J.; Abbott, N.L. Surfaces modified with nanometer-thick silver-impregnated polymeric films that kill bacteria but support growth of mammalian cells. Biomaterials 2010, 31, 680–690. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Hu, Y.; Li, J.; Li, B. Chitosan/phosvitin antibacterial films fabricated via layer-by-layer deposition. Int. J. Biol. Macromol. 2014, 64, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Cen, L.; Neoh, K.G.; Kang, E.T. Antibacterial activity of cloth functionalized with N-alkylated poly(4-vinylpyridine). J. Biomed. Mater. Res. 2004, 71, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Ohl, M.; Schweizer, M.; Graham, M.; Heilmann, K.; Boyken, L.; Diekema, D. Hospital privacy curtains are frequently and rapidly contaminated with potentially pathogenic bacteria. Am. J. Infect. Control 2012, 40, 904–906. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, M.; Graham, M.; Ohl, M.; Heilmann, K.; Boyken, L.; Diekema, D. Novel Hospital Curtains with Antimicrobial Properties: A Randomized, Controlled Trial. Infect. Control Hosp. Epidemiol. 2012, 33, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Lemmen, S.; Scheithauer, S.; Haefner, H.; Yezli, S.; Mohr, M.; Otter, J.A. Evaluation of hydrogen peroxide vapor for the inactivation of nosocomial pathogens on porous and nonporous surfaces. Am. J. Infect. Control 2015, 43, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Gizdavic-Nikolaidis, M.R.; Ray, S.; Bennett, J.R.; Swift, S.; Bowmaker, G.A.; Easteal, A.J. Electrospun poly(aniline-co-ethyl 3-aminobenzoate)/poly(lactic acid) nanofibers and their potential in biomedical applications. J. Polym. Sci. Part A Polym. Chem. 2011, 49, 4902–4910. [Google Scholar] [CrossRef]
- Pandey, S.; Annapoorni, S.; Malhotra, B.D. Synthesis and Characterization of Poly(aniline-co-o-anisidine): A Processable Conducting Copolymer. Macromolecules 1993, 26, 3190–3193. [Google Scholar] [CrossRef]
- Gizdavic-Nikolaidis, M.R.; Bennett, J.R.; Swift, S.; Easteal, A.J.; Ambrose, M. Broad spectrum antimicrobial activity of functionalized polyanilines. Acta Biomater. 2011, 7, 4204–4209. [Google Scholar] [CrossRef] [PubMed]
- Gizdavic-Nikolaidis, M.R.; Bennett, J.R.; Zujovic, Z.; Swift, S.; Bowmaker, G.A. Characterization and antimicrobial efficacy of acetone extracted aniline oligomers. Synth. Met. 2012, 162, 1114–1119. [Google Scholar] [CrossRef]
- Dhand, C.; Das, M.; Datta, M.; Malhotra, B.D. Recent advances in polyaniline based biosensors. Biosens. Bioelectron. 2011, 26, 2811–2821. [Google Scholar] [CrossRef] [PubMed]
- Grennan, K.; Killard, A.J.; Hanson, C.J.; Cafolla, A.A.; Smyth, M.R. Optimisation and characterisation of biosensors based on polyaniline. Talanta 2006, 68, 1591–1600. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Jiang, T.; Hu, P.; Han, Z.; Lu, X.; Ye, P. Self-decontaminating properties of fluorinated copolymers integrated with ciprofloxacin for synergistically inhibiting the growth of Escherichia coli. J. Biomater. Sci. Polym. Ed. 2014, 25, 1920–1945. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.L.; Fan, W.J.; Shan, S.Y.; Hu, T.W.; Wang, Y.M.; Jia, Q.M. The Effects of Natural Dopant Acids on Morphologies and Antibacterial Activity of Polyaniline. Adv. Mater. Res. 2013, 650, 249–252. [Google Scholar] [CrossRef]
- Humpolicek, P.; Kasparkova, V.; Saha, P.; Stejskal, J. Biocompatibility of polyaniline. Synth. Met. 2012, 162, 722–727. [Google Scholar] [CrossRef]
- Qi, H.; Liu, M.; Xu, L.; Feng, L.; Tao, L.; Ji, Y.; Zhang, X.; Wei, Y. Biocompatibility evaluation of aniline oligomers with different end-functional groups. Toxicol. Res. 2013, 2, 427–433. [Google Scholar] [CrossRef]
- Seshadri, D.T.; Bhat, N.V. Use of polyaniline as an antimicrobial agent in textiles. Indian J. Fibre Text. Res. 2005, 30, 204–206. [Google Scholar]
- Gizdavic-Nikolaidis, M.R.; Pagnon, J.C.; Ali, N.; Sum, R.; Davies, N.; Roddam, L.F.; Ambrose, M. Functionalized polyanilines disrupt Pseudomonas aeruginosa and Staphylococcus aureus biofilms. Colloids Surf. B. Biointerfaces 2015, 136, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48 (Suppl. 1), 5–16. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Siani, H.; Maillard, J.-Y. Best practice in healthcare environment decontamination. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kram, K.E.; Finkel, S.E. Rich Medium Composition Affects Escherichia coli Survival, Glycation, and Mutation Frequency during Long-Term Batch Culture. Appl. Environ. Microbiol. 2015, 81, 4442–4450. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Bausch, C.; Richmond, C.; Blattner, F.R.; Conway, T. Functional genomics: Expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 1999, 181, 6425–6440. [Google Scholar] [PubMed]
- Zhang, J.; Greasham, R. Chemically defined media for commercial fermentations. Appl. Microbiol. Biotechnol. 1999, 51, 407–421. [Google Scholar] [CrossRef]
- Sezonov, G.; Joseleau-Petit, D.; D’Ari, R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Schellhorn, H.E. Control of RpoS in global gene expression of Escherichia coli in minimal media. Mol. Genet. Genom. 2009, 281, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Dalton, J.; Wiles, S.; Gizdavic-Nikolaidis, M.; Swift, S. The tuberculocidal activity of polyaniline and functionalised polyanilines. PeerJ 2016, 4, e2795. [Google Scholar] [CrossRef] [PubMed]
- Herigstad, B.; Hamilton, M.; Heersink, J. How to optimize the drop plate method for enumerating bacteria. J. Microbiol. Methods 2001, 44, 121–129. [Google Scholar] [CrossRef]
- Liu, J.; Liu, C.; Liu, Y.; Chen, M.; Hu, Y.; Yang, Z. Study on the grafting of chitosan-gelatin microcapsules onto cotton fabrics and its antibacterial effect. Colloids Surf. B Biointerfaces 2013, 109, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Andreu, N.; Zelmer, A.; Fletcher, T.; Elkington, P.T.; Ward, T.H.; Ripoll, J.; Parish, T.; Bancroft, G.J.; Schaible, U.; Robertson, B.D.; et al. Optimisation of bioluminescent reporters for use with mycobacteria. PLoS ONE 2010, 5, e10777. [Google Scholar] [CrossRef] [PubMed]
- Andreu, N.; Zelmer, A.; Sampson, S.L.; Ikeh, M.; Bancroft, G.J.; Schaible, U.E.; Wiles, S.; Robertson, B.D. Rapid in vivo assessment of drug efficacy against Mycobacterium tuberculosis using an improved firefly luciferase. J. Antimicrob. Chemother. 2013, 68, 2118–2127. [Google Scholar] [CrossRef] [PubMed]
- Andreu, N.; Zelmer, A.; Wiles, S. Noninvasive biophotonic imaging for studies of infectious disease. FEMS Microbiol. Rev. 2011, 35, 360–394. [Google Scholar] [CrossRef] [PubMed]
- Appendini, P.; Hotchkiss, J.H. Review of antimicrobial food packaging. Innov. Food Sci. Emerg. Technol. 2002, 3, 113–126. [Google Scholar] [CrossRef]
- Japanese Standards Association. JIS Z 2801:2000 Antimicrobial Products—Test for Antimicrobial Activity and Efficacy; Japanese Standards Association: Tokyo, Japan, 2000. [Google Scholar]
- Humphreys, P.N. Testing standards for sporicides. J. Hosp. Infect. 2011, 77, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Warnes, S.L.; Keevil, C.W. Lack of Involvement of Fenton Chemistry in Death of Methicillin-Resistant and Methicillin-Sensitive Strains of Staphylococcus aureus and Destruction of Their Genomes on Wet or Dry Copper Alloy Surfaces. Appl. Environ. Microbiol. 2016, 82, 2132–2136. [Google Scholar] [CrossRef] [PubMed]
- Lobritz, M.A.; Belenky, P.; Porter, C.B.M.; Gutierrez, A.; Yang, J.H.; Schwarz, E.G.; Dwyer, D.J.; Khalil, A.S.; Collins, J.J. Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. USA 2015, 112, 8173–8180. [Google Scholar] [CrossRef] [PubMed]
- Gizdavic-Nikolaidis, M.R.; Easteal, A.J.; Stepanovic, S. Bioactive Aniline Copolymers. International Patent Application No. PCT/NZ2008/000254, 26 September 2008. [Google Scholar]
- Gizdavic-Nikolaidis, M.R.; Bowmaker, G.A.; Zujovic, Z. The Synthesis, Physical Properties, Bioactivity and Potential Applications of Polyanilines; Cambridge Scholars Publishing: Newcastle, UK, 2018; in press. [Google Scholar]
- Fröls, S.; Dyall-Smith, M.; Pfeifer, F. Biofilm formation by haloarchaea. Environ. Microbiol. 2012, 14, 3159–3174. [Google Scholar] [CrossRef] [PubMed]
- Shi, N.; Guo, X.; Jing, H.; Gong, J.; Sun, C.; Yang, K. Antibacterial effect of the conducting polyaniline. J. Mater. Sci. Technol. 2006, 22, 289–290. [Google Scholar]
- Tan, C.; Smith, R.P.; Srimani, J.K.; Riccione, K.A.; Prasada, S.; Kuehn, M.; You, L. The inoculum effect and band-pass bacterial response to periodic antibiotic treatment. Mol. Syst. Biol. 2012, 8, 617. [Google Scholar] [CrossRef] [PubMed]
- Warnes, S.L.; Green, S.M.; Michels, H.T.; Keevil, C.W. Biocidal efficacy of copper alloys against pathogenic enterococci involves degradation of genomic and plasmid DNAs. Appl. Environ. Microbiol. 2010, 76, 5390–5401. [Google Scholar] [CrossRef] [PubMed]
- Oie, S.; Hosokawa, I.; Kamiya, A. Contamination of room door handles by methicillin-sensitive/methicillin-resistant Staphylococcus aureus. J. Hosp. Infect. 2002, 51, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Oie, S.; Suenaga, S.; Sawa, A.; Kamiya, A. Association between isolation sites of methicillin-resistant Staphylococcus aureus (MRSA) in patients with MRSA-positive body sites and MRSA contamination in their surrounding environmental surfaces. Jpn. J. Infect. Dis. 2007, 60, 367–369. [Google Scholar] [PubMed]
- Schmidt, M.G.; Attaway, H.H.; Sharpe, P.A.; John, J.F.; Sepkowitz, K.A.; Morgan, A.; Fairey, S.E.; Singh, S.; Steed, L.L.; Cantey, J.R.; et al. Sustained reduction of microbial burden on common hospital surfaces through introduction of copper. J. Clin. Microbiol. 2012, 50, 2217–2223. [Google Scholar] [CrossRef] [PubMed]
- O’Gorman, J.; Humphreys, H. Application of copper to prevent and control infection. Where are we now? J. Hosp. Infect. 2012, 81, 217–223. [Google Scholar] [CrossRef]
- Noyce, J.O.; Michels, H.T.; Keevil, C.W. Use of copper cast alloys to control Escherichia coli O157 cross-contamination during food processing. Appl. Environ. Microbiol. 2006, 72, 4239–4244. [Google Scholar] [CrossRef] [PubMed]
- Rabie, A.J.; McLaren, I.M.; Breslin, M.F.; Sayers, R.; Davies, R.H. Assessment of anti-Salmonella activity of boot dip samples. Avian Pathol. 2015, 44, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Fraise, A. Currently available sporicides for use in healthcare, and their limitations. J. Hosp. Infect. 2011, 77, 210–212. [Google Scholar] [CrossRef] [PubMed]
- Honney, C. Colonisation Resistant Surfaces. MSc Thesis, University of Auckland, Auckland, New Zealand, 2014. [Google Scholar]
- Møretrø, T.; Høiby-pettersen, G.S.; Habimana, O.; Heir, E.; Langsrud, S. Assessment of the antibacterial activity of a triclosan-containing cutting board. Int. J. Food Microbiol. 2011, 146, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Cutter, C.N. The effectiveness of triclosan-incorporated plastic against bacteria on beef surfaces. J. Food Prot. 1999, 62, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Kalyon, B.D.; Olgun, U. Antibacterial efficacy of triclosan-incorporated polymers. Am. J. Infect. Control 2001, 29, 124–125. [Google Scholar] [CrossRef] [PubMed]
- Connolly, P.; Bloomfield, S.F.; Denyer, S.P. The use of impedance for preservative efficacy testing of pharmaceuticals and cosmetic products. J. Appl. Bacteriol. 1994, 76, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.T. Development of a Series of Bioluminescent Sensor Strains for Rapid Screening of Antimicrobials. Ph.D. Thesis, University of Auckland, Auckland, New Zealand, 2013. [Google Scholar]
- Riedel, C.U.; Casey, P.G.; Mulcahy, H.; O’Gara, F.; Gahan, C.G.M.; Hill, C. Construction of p16Slux, a novel vector for improved bioluminescent labeling of gram-negative bacteria. Appl. Environ. Microbiol. 2007, 73, 7092–7095. [Google Scholar] [CrossRef] [PubMed]
- Seaver, L.C.; Imlay, J.A. Are respiratory enzymes the primary sources of intracellular hydrogen peroxide? J. Biol. Chem. 2004, 279, 48742–48750. [Google Scholar] [CrossRef] [PubMed]
- Brewster, J.D. A simple micro-growth assay for enumerating bacteria. J. Microbiol. Methods 2003, 53, 77–86. [Google Scholar] [CrossRef]
- Pankey, G.A.; Sabath, L.D. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of Gram-positive bacterial infections. Clin. Infect. Dis. 2004, 38, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Meighen, E. Genetics of bacterial bioluminescence. Annu. Rev. Genet. 1994, 28, 117–139. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).