Comparison of Nitrogen Depletion and Repletion on Lipid Production in Yeast and Fungal Species
Abstract
:1. Introduction
2. Results and Discussion
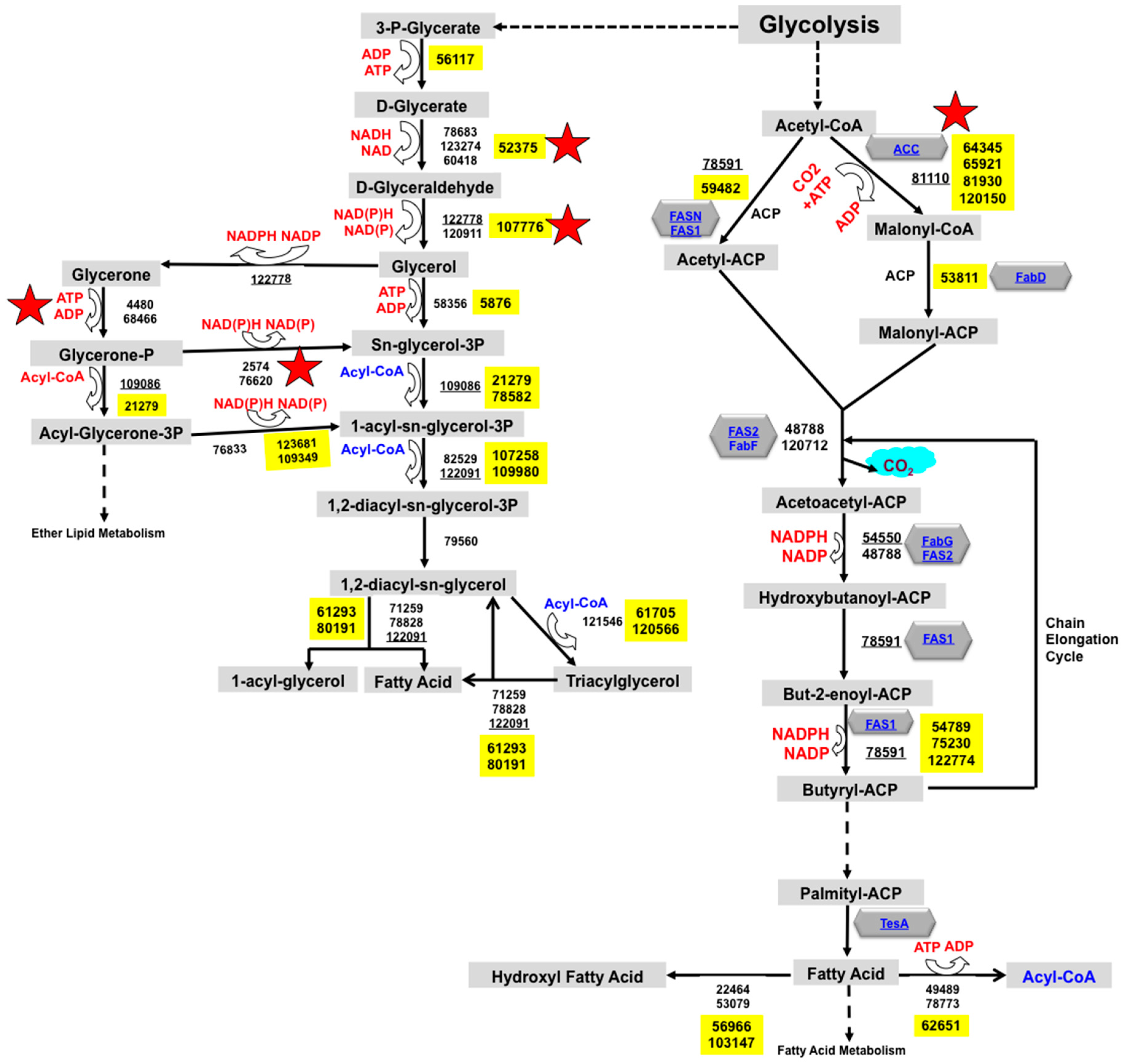
2.1. Reconstruction of TAG Biosynthesis Pathway in Trichoderma reesei by Genomic Metabolic Pathway Analysis
2.2. Lipid Production by Yarrowia lipolytica in Different Media
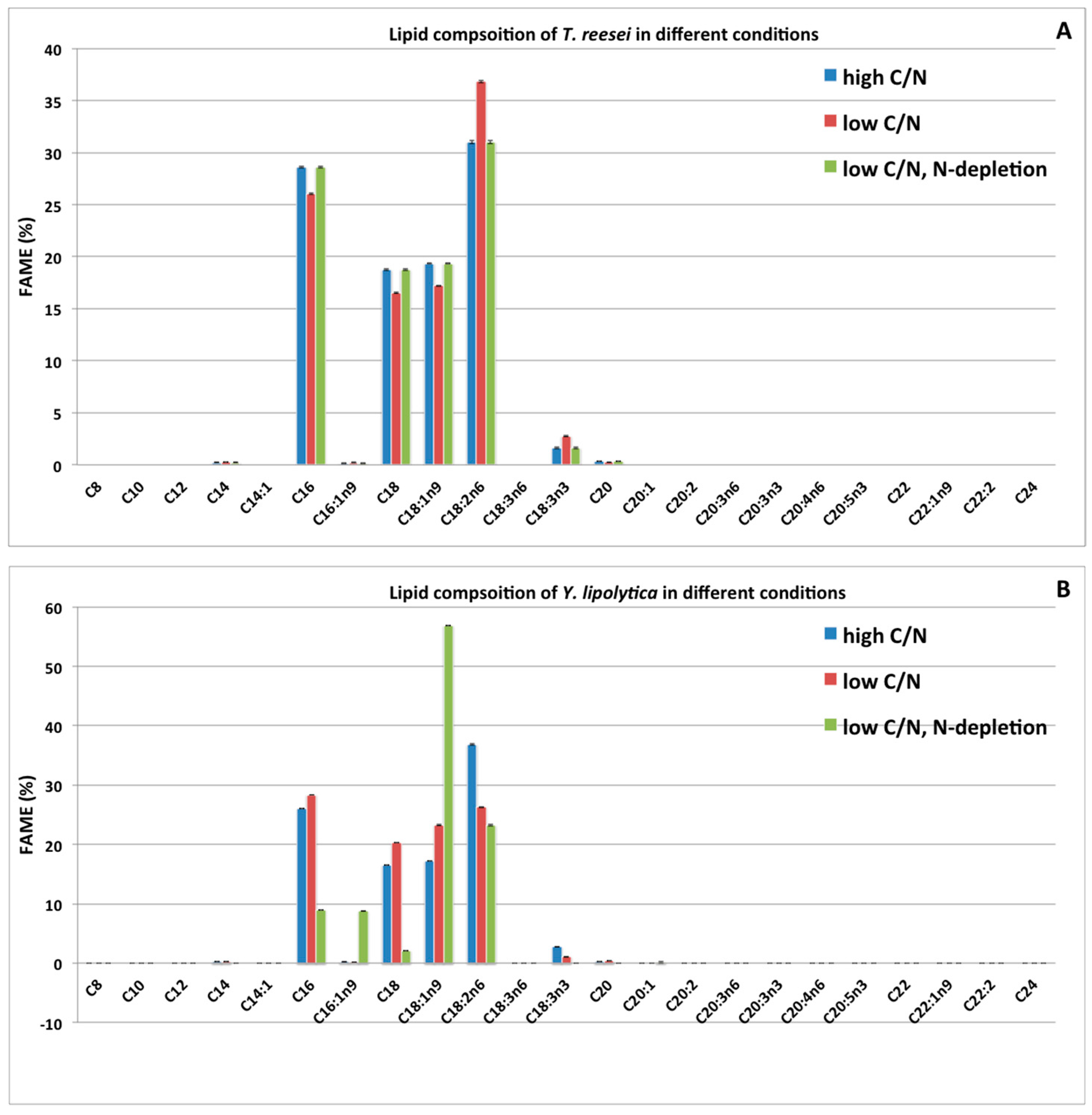
2.3. Effects of Two Nitrogen Deficiency Strategies on Lipid Production Yield
2.4. Impacts of Two Nitrogen Deficiency Strategies on the Lipid Composition Profile
3. Materials and Methods
3.1. Trichoderma reesei Fatty Acid Metabolism and TAG Biosynthesis Pathway Reconstruction
3.2. Strain and Growth Conditions
3.3. Lipid Determination and Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CBP | consolidated bioprocessing |
| TAG | triacylglycerol |
| C/N ratio | carbon/nitrogen ratio |
| FAME | fatty acid methyl ester |
References
- Peralta-Yahya, P.P.; Ouellet, M.; Chan, R.; Mukhopadhyay, A.; Keasling, J.D.; Lee, T.S. Identification and microbial production of a terpene-based advanced biofuel. Nat. Commun. 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Peralta-Yahya, P.P.; Keasling, J.D. Advanced biofuel production in microbes. Biotechnol. J. 2010, 5, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Connor, M.R.; Liao, J.C. Microbial production of advanced transportation fuels in non-natural hosts. Curr. Opin. Biotechnol. 2009, 20, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, S.; Hanai, T.; Liao, J.C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 2008, 451, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Hasunuma, T.; Okazaki, F.; Okai, N.; Hara, K.Y.; Ishii, J.; Kondo, A. A review of enzymes and microbes for lignocellulosic biorefinery and the possibility of their application to consolidated bioprocessing technology. Bioresour. Technol. 2013, 135, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Lynd, L.R.; Zyl, W.H.; McBride, J.E.; Laser, M. Consolidated bioprocessing of cellulosic biomass: An update. Curr. Opin. Biotechnol. 2005, 16, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Singh, A.; Himmel, M.E. Perspectives and new directions for the production of bioethanol using consolidated bioprocessing of lignocellulose. Curr. Opin. Biotechnol. 2009, 20, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Barth, G.; Gaillardin, C. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol. Rev. 1997, 19, 219–237. [Google Scholar] [CrossRef] [PubMed]
- Dujon, B.; Sherman, D.; Fischer, G.; Durrens, P.; Casaregola, S.; Lafontaine, I.; Montigny, J.; Marck, C.; Neuvéglise, C.; Talla, E. Genome evolution in yeasts. Nature 2004, 430, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Sherman, D.; Durrens, P.; Iragne, F.; Beyne, E.; Nikolski, M.; Souciet, J.L. Genolevures complete genomes provide data and tools for comparative genomics of hemiascomycetous yeasts. Nucleic Acids Res. 2006, 34, 432–435. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.C.; Yang, B.C.; Kuo, T.T. One-step transformation of yeast in stationary phase. Curr. Genet. 1992, 21, 83–84. [Google Scholar] [CrossRef]
- Wang, J.H.; Hung, W.P.; Tsai, S.H. High efficiency transformation by electroporation of Yarrowia lipolytica. J. Microbiol. 2011, 49, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Davidow, L.S.; Apostolakos, D.; O’Donnell, M.M.; Proctor, A.R.; Ogrydziak, D.M.; Wing, R.A.; Stasko, I.; DeZeeuw, J.R. Integrative transformation of the yeast Yarrowia lipolytica. Curr. Genet. 1985, 10, 39–48. [Google Scholar] [CrossRef]
- Juretzek, T.; Dall, M.T.; Mauersberger, S.; Gaillardin, C.; Barth, G.; Nicaud, J.M. Vectors for gene expression and amplification in the yeast Yarrowia lipolytica. Yeast 2000, 18, 97–113. [Google Scholar] [CrossRef]
- Dall, M.T.; Nicaud, J.M.; Gaillardin, C. Multiple-copy integration in the yeast Yarrowia lipolytica. Curr. Genet. 1994, 26, 38–44. [Google Scholar] [CrossRef]
- Ratledge, C.; Wynn, J.P. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv. Appl. Microbiol. 2002, 51, 1–51. [Google Scholar] [PubMed]
- Beopoulos, A.; Cescut, J.; Haddouche, R.; Uribelarrea, J.L.; Molina-Jouve, C.; Nicaud, J.M. Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 2009, 48, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Tai, M.; Stephanopoulos, G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 2013, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Hill, A.; Liu, L.; Knight, R.; Miller, J.; Pan, A.; Otoupal, P.; Alper, H.S. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 2014, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Madzak, C. Yarrowia lipolytica: Recent achievements in heterologous protein expression and pathway engineering. Appl. Microbiol. Biotechnol. 2015, 99, 4559–4577. [Google Scholar] [CrossRef] [PubMed]
- Boonvitthya, N.; Bozonnet, S.; Burapatana, V.; O’Donohue, M.J.; Chulalaksananukul, W. Comparison of the heterologous expression of Trichoderma reesei endoglucanase II and cellobiohydrolase II in the yeasts Pichia pastoris and Yarrowia lipolytica. Mol. Biotechnol. 2013, 54, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wang, W.; Alahuhta, M.; Wall, T.V.; Baker, J.O.; Taylor, L.E.; Decker, S.R.; Himmel, M.E.; Zhang, M. Engineering towards a complete heterologous cellulase secretome in Yarrowia lipolytica reveals its potential for consolidated bioprocessing. Biotechnol. Biofuels 2014, 7, 148. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wei, H.; Alahuhta, M.; Chen, X.; Hyman, D.; Johnson, D.K.; Zhang, M.; Himmel, M.E. Heterologous expression of xylanase enzymes in lipogenic yeast Yarrowia lipolytica. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.P.; Duquesne, S.; Bozonnet, S.; Cioci, G.; Nicaud, J.M.; Marty, A.; O’Donohue, M.J. Development of cellobiose-degrading ability in Yarrowia lipolytica strain by overexpression of endogenous genes. Biotechnol. Biofuels 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Schuster, A.; Schmoll, M. Biology and biotechnology of Trichoderma. Appl. Microbiol. Biotechnol. 2010, 87, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Hakkinen, M.; Arvas, M.; Oja, M.; Aro, N.; Penttila, M.; Saloheimo, M.; Pakula, T.M. Re-annotation of the CAZy genes of Trichoderma reesei and transcription in the presence of lignocellulosic substrates. Microb. Cell Fact 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, J.; Coutinho, P.; Mathis, H.; Quenot, A.; Record, E.; Asther, M.; Heiss-Blanquet, S. Transcriptional profiling of cellulase and expansin-related genes in a hypercellulolytic Trichoderma reesei. Biotechnol. Lett. 2009, 31, 1399–1405. [Google Scholar] [CrossRef] [PubMed]
- Martinez, D.; Berka, R.M.; Henrissat, B.; Saloheimo, M.; Arvas, M.; Baker, S.E.; Chapman, J.; Chertkov, O.; Coutinho, P.M.; Cullen, D. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 2008, 26, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Dowzer, C.E.; Kelly, J.M. Cloning of the creA gene from Aspergillus nidulans: A gene involved in carbon catabolite repression. Curr. Genet. 1989, 15, 457–459. [Google Scholar] [CrossRef]
- Ilmen, M.; Thrane, C.; Penttilä, M. The glucose repressor genecre1 of Trichoderma: Isolation and expression of a full-length and a truncated mutant form. Mol. Gen. Genet. MGG 1996, 251, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Portnoy, T.; Margeot, A.; Linke, R.; Atanasova, L.; Fekete, E.; Sandor, E.; Hartl, L.; Karaffa, L.; Druzhinina, I.S.; Seiboth, B. The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei: A master regulator of carbon assimilation. BMC Genom. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Hynes, M. Studies on the role of the are a gene in the regulation of nitrogen catabolism in Aspergillus nidulans. Aust. J. Biol. Sci. 1975, 28, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Stricker, A.R.; Grosstessner-Hain, K.; Würleitner, E.; Mach, R.L. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and D-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 2006, 5, 2128–2137. [Google Scholar] [CrossRef] [PubMed]
- Aro, N.; Ilmén, M.; Saloheimo, A.; Penttilä, M. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl. Environ. Microbiol. 2003, 69, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Aro, N.; Saloheimo, A.; Ilmén, M.; Penttilä, M. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 2001, 276, 24309–24314. [Google Scholar] [CrossRef] [PubMed]
- Nitta, M.; Furukawa, T.; Shida, Y.; Mori, K.; Kuhara, S.; Morikawa, Y.; Ogasawara, W. A new Zn(II)(2)Cys(6)-type transcription factor BglR regulates beta-glucosidase expression in Trichoderma reesei. Fungal. Genet. Biol. 2012, 49, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Schmoll, M.; Kubicek, C.P. Regulation of Trichoderma cellulase formation: Lessons in molecular biology from an industrial fungus: A review. Acta Microbiol. Immunol. Hung. 2003, 50, 125–145. [Google Scholar] [CrossRef] [PubMed]
- Nogawa, M.; Goto, M.; Okada, H.; Morikawa, Y. L-Sorbose induces cellulase gene transcription in the cellulolytic fungus Trichoderma reesei. Curr. Genet. 2001, 38, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Bhanja, A.; Minde, G.; Magdum, S.; Kalyanraman, V. Comparative studies of oleaginous fungal strains (Mucor circinelloides and Trichoderma reesei) for effective wastewater treatment and Bio-Oil production. Biotechnol. Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.E.; Hasan, M.; Lepe-Casillas, M.; Thornton, A.J. Effect of temperature and pH on lipid accumulation by Trichoderma reesei. Appl. Microbiol. Biotechnol. 1990, 34, 335–339. [Google Scholar] [CrossRef]
- Brown, D.E.; Hasan, M.; Thornton, A.J. Fat production by Trichoderma reesei. Biotechnol. Lett. 1988, 10, 249–254. [Google Scholar] [CrossRef]
- Serrano-Carreon, L.; Hathout, Y.; Bensoussan, M.; Belin, J.M. Lipid accumulation in Trichoderma species. FEMS Microbiol. Lett. 1992, 93, 181–187. [Google Scholar] [CrossRef]
- Dey, P.; Mall, N.; Chattopadhyay, A.; Chakraborty, M.; Maiti, M.K. Enhancement of lipid productivity in oleaginous Colletotrichum fungus through genetic transformation using the yeast CtDGAT2b gene under model-optimized growth condition. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.H.; Jiang, J.G. Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology. Prog. Lipid Res. 2013, 52, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Courchesne, N.M.D.; Parisien, A.; Wang, B.; Lan, C.Q. Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J. Biotechnol. 2009, 141, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Castanha, R.F.; Mariano, A.P.; Morais, L.A.S.; Scramini, S.; Monteiro, R.T.R. Optimization of lipids production by Cryptococcus laurentii 11 using cheese whey with molasses. Braz. J. Microbiol. 2014, 45, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Papanikolaou, S.; Aggelis, G. Lipid production by Yarrowia lipolytica growing on industrial glycerol in a single-stage continuous culture. Bioresour. Technol. 2002, 82, 43–49. [Google Scholar] [CrossRef]
- Huang, C.; Zong, M.H.; Wu, H.; Liu, Q.P. Microbial oil production from rice straw hydrolysate by Trichosporon fermentans. Bioresour. Technol. 2009, 100, 4535–4538. [Google Scholar] [CrossRef] [PubMed]
- Kavscek, M.; Bhutada, G.; Madl, T.; Natter, K. Optimization of lipid production with a genome-scale model of Yarrowia lipolytica. BMC Syst. Biol. 2015, 9. [Google Scholar] [CrossRef] [PubMed]
- Braunwald, T.; Schwemmlein, L.; Graeff-Honninger, S.; French, W.T.; Hernandez, R.; Holmes, W.E.; Claupein, W. Effect of different C/N ratios on carotenoid and lipid production by Rhodotorula glutinis. Appl. Microbiol. Biotechnol. 2013, 97, 6581–6588. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wu, H.; Li, R.F.; Zong, M.H. Improving lipid production from bagasse hydrolysate with Trichosporon fermentans by response surface methodology. New Biotechnol. 2012, 29, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Chaisawang, M.; Verduyn, C.; Chauvatcharin, S.; Suphantharika, M. Metabolic networks and bioenergetics of Aurantiochytrium sp. B-072 during storage lipid formation. Braz. J. Microbiol. 2012, 43, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Hirooka, S.; Higuchi, S.; Uzuka, A.; Nozaki, H.; Miyagishima, S.Y. Acidophilic green alga Pseudochlorella sp. YKT1 accumulates high amount of lipid droplets under a nitrogen-depleted condition at a low-pH. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Horsman, M.; Wang, B.; Wu, N.; Lan, C.Q. Effects of nitrogen sources on cell growth and lipid accumulation of green alga Neochloris oleoabundans. Appl. Microbiol. Biotechnol. 2008, 81, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Guarnieri, M.T.; Nag, A.; Yang, S.; Pienkos, P.T. Proteomic analysis of Chlorella vulgaris: potential targets for enhanced lipid accumulation. J. Proteom. 2013, 93, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Guarnieri, M.T.; Nag, A.; Smolinski, S.L.; Darzins, A.; Seibert, M.; Pienkos, P.T. Examination of triacylglycerol biosynthetic pathways via de novo transcriptomic and proteomic analyses in an unsequenced microalga. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Seip, J.; Jackson, R.; He, H.; Zhu, Q.; Hong, S.P. Snf1 is a regulator of lipid accumulation in Yarrowia lipolytica. Appl. Environ. Microbiol. 2013, 79, 7360–7370. [Google Scholar] [CrossRef] [PubMed]
- Sitepu, I.R.; Sestric, R.; Ignatia, L.; Levin, D.; German, J.B.; Gillies, L.A.; Almada, L.A.; Boundy-Mills, K.L. Manipulation of culture conditions alters lipid content and fatty acid profiles of a wide variety of known and new oleaginous yeast species. Bioresour. Technol. 2013, 144, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Tsigie, Y.A.; Wang, C.Y.; Truong, C.T.; Ju, Y.H. Lipid production from Yarrowia lipolytica Po1g grown in sugarcane bagasse hydrolysate. Bioresour. Technol. 2011, 102, 9216–9222. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Jackson, E.N. Metabolic engineering of Yarrowia lipolytica for industrial applications. Curr. Opin. Biotechnol. 2015, 36, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Morin, N.; Cescut, J.; Beopoulos, A.; Lelandais, G.; Le, V.; Uribelarrea, J.L.; Molina-Jouve, C.; Nicaud, J.M. Transcriptomic analyses during the transition from biomass production to lipid accumulation in the oleaginous yeast Yarrowia lipolytica. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, W.; Wei, H.; Himmel, M.E.; Zhang, M. Identification of genetic targets to improve lignocellulosic hydrocarbon production in Trichoderma reesei using public genomic and transcriptomic datasets. In Direct Microbial Conversion of Biomass to Advanced Biofuels; Himmel, M.E., Waltham, M.A., Eds.; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Cakmak, T.; Angun, P.; Ozkan, A.D.; Cakmak, Z.; Olmez, T.T.; Tekinay, T. Nitrogen and sulfur deprivation differentiate lipid accumulation targets of Chlamydomonas reinhardtii. Bioengineered 2012, 3, 343–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madzak, C.; Treton, B.; Blanchin-Roland, S. Strong hybrid promoters and integrative expression/secretion vectors for quasi-constitutive expression of heterologous proteins in the yeast Yarrowia lipolytica. J. Mol. Microbiol. Biotechnol. 2000, 2, 207–216. [Google Scholar] [PubMed]
- Laurens, L.M.; Quinn, M.; Wychen, S.; Templeton, D.W.; Wolfrum, E.J. Accurate and reliable quantification of total microalgal fuel potential as fatty acid methyl esters by in situ transesterification. Anal. Bioanal. Chem. 2012, 403, 167–178. [Google Scholar] [CrossRef] [PubMed]


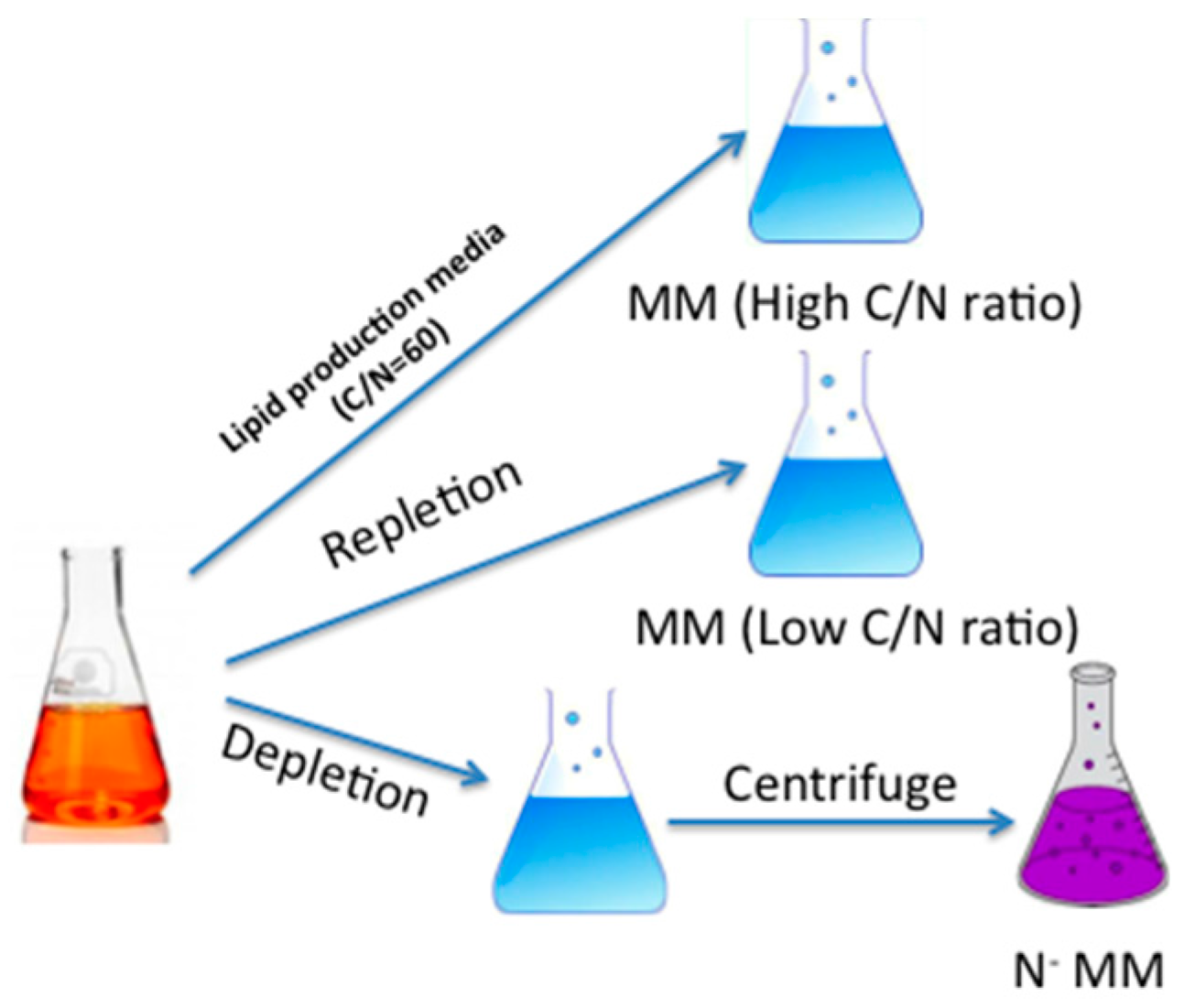
| Chemical | MM | High C/N Ratio MM (g/L) | Low C/N Ratio MM (g/L) | No Nitrogen MM (N-MM, g/L) |
|---|---|---|---|---|
| Glucose | 30 | 30 | 30 | 30 |
| (NH4)2SO4 | 0.5 | 0.5 | 5 | 0 |
| Yeast extract | 0.5 | 1.5 | 1.5 | 0 |
| KH2PO4 | 7 | 7 | 7 | 7 |
| Na2HPO4 | 2 | 2 | 2 | 2 |
| MgSO4·7H2O | 1.5 | 1.5 | 1.5 | 1.5 |
| CaCl2·6H2O | 0.1 | 0.1 | 0.1 | 0.1 |
| FeCl3·6H2O | 0.008 | 0.008 | 0.008 | 0.008 |
| ZnSO4·7H2O | 0.0001 | 0.0001 | 0.0001 | 0.0001 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Wang, W.; Wei, H.; Van Wychen, S.; Pienkos, P.T.; Zhang, M.; Himmel, M.E. Comparison of Nitrogen Depletion and Repletion on Lipid Production in Yeast and Fungal Species. Energies 2016, 9, 685. https://doi.org/10.3390/en9090685
Yang S, Wang W, Wei H, Van Wychen S, Pienkos PT, Zhang M, Himmel ME. Comparison of Nitrogen Depletion and Repletion on Lipid Production in Yeast and Fungal Species. Energies. 2016; 9(9):685. https://doi.org/10.3390/en9090685
Chicago/Turabian StyleYang, Shihui, Wei Wang, Hui Wei, Stefanie Van Wychen, Philip T. Pienkos, Min Zhang, and Michael E. Himmel. 2016. "Comparison of Nitrogen Depletion and Repletion on Lipid Production in Yeast and Fungal Species" Energies 9, no. 9: 685. https://doi.org/10.3390/en9090685
APA StyleYang, S., Wang, W., Wei, H., Van Wychen, S., Pienkos, P. T., Zhang, M., & Himmel, M. E. (2016). Comparison of Nitrogen Depletion and Repletion on Lipid Production in Yeast and Fungal Species. Energies, 9(9), 685. https://doi.org/10.3390/en9090685








