Biogas Production and Microbial Communities of Mesophilic and Thermophilic Anaerobic Co-Digestion of Animal Manures and Food Wastes in Costa Rica
Abstract
:1. Introduction
2. Materials and Methods
2.1. Feedstocks
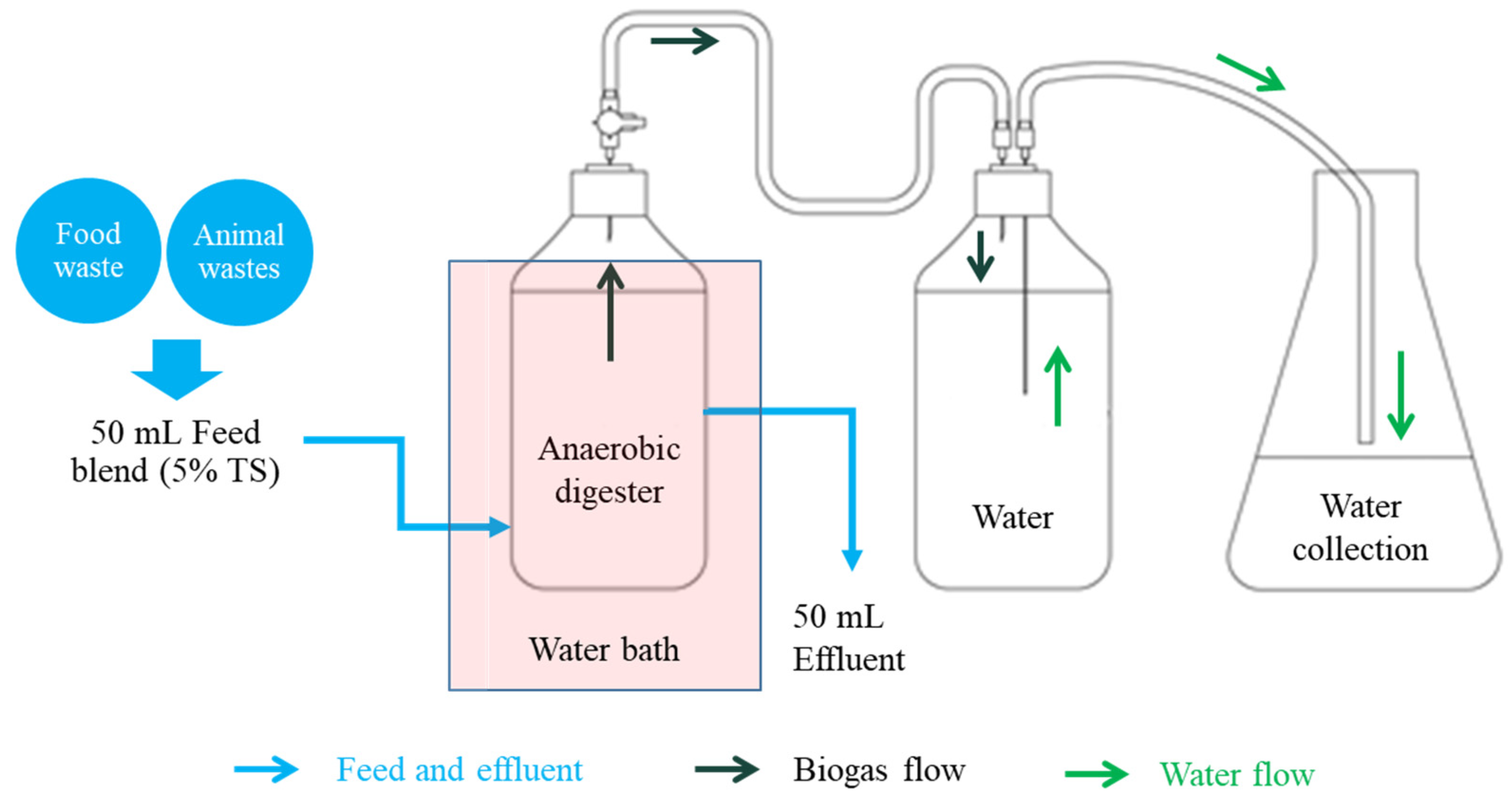
2.2. Design and Operation of Bioreactors
2.3. Analytical Methods
2.4. DNA Extraction
2.5. Microbial Community Analysis
2.6. Statistical Analysis
3. Results
3.1. Feedstock Composition
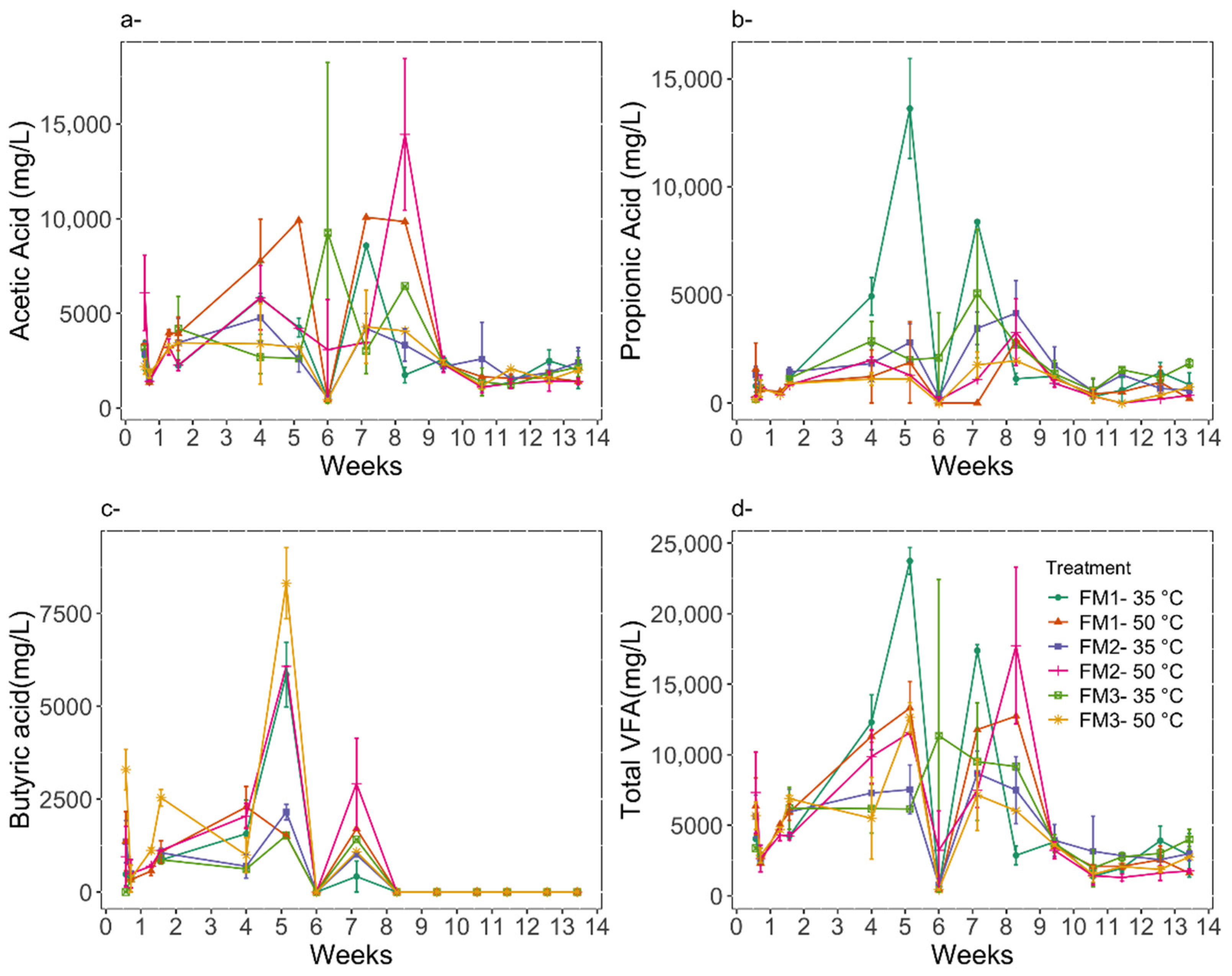
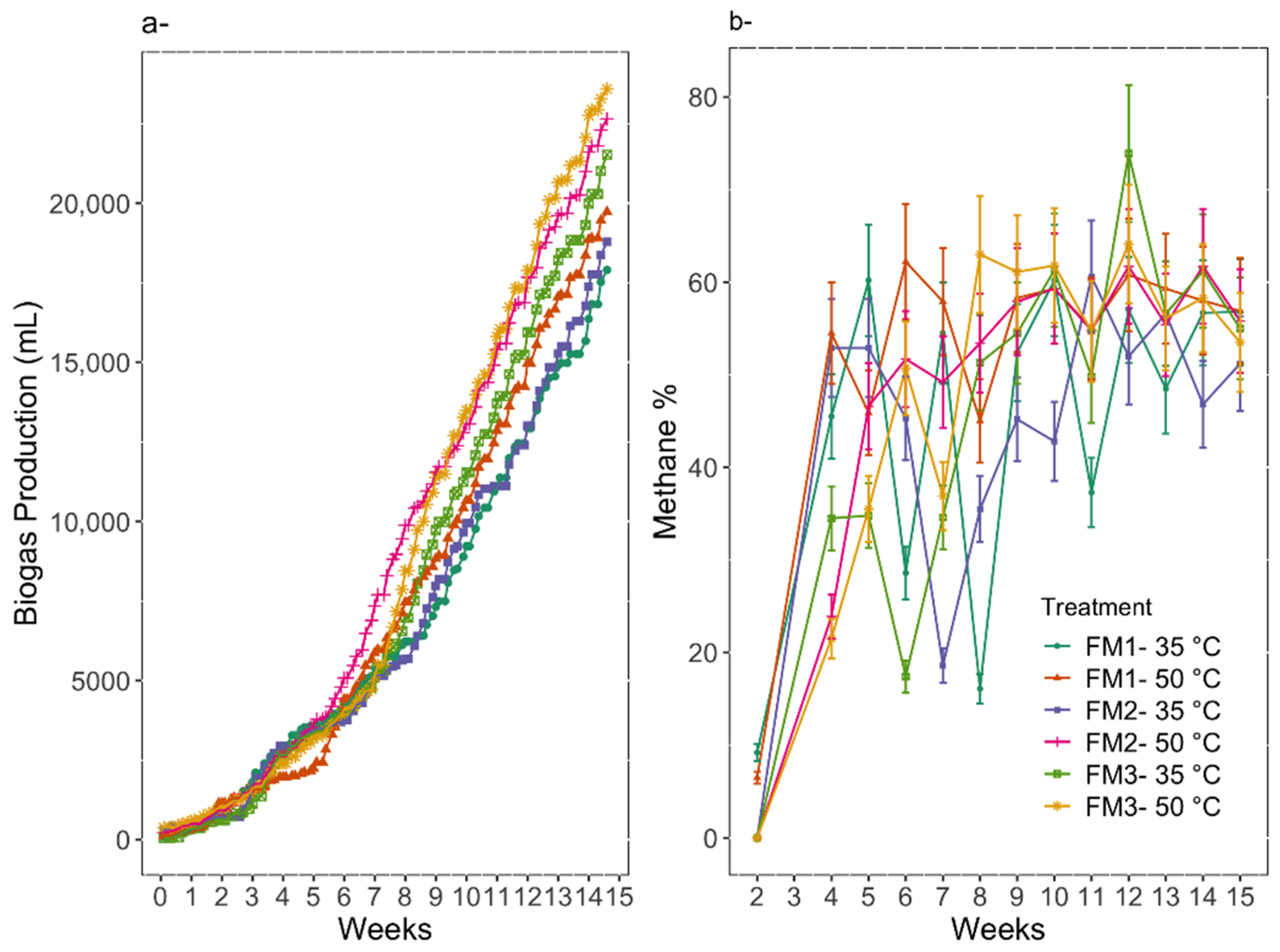
3.2. Digestion Performance
| Treatment | Acetic Acid (mg/L) | Propanoic Acid (mg/L) | Butyric Acid (mg/L) | Total VFA (mg/L) |
|---|---|---|---|---|
| FM1 50 °C | 3069 ± 919 | 1019 ± 284 | 0 | 4089 ± 1186 |
| FM2 50 °C | 3666 ± 1544 | 840 ± 391 | 0 | 4506 ± 1924 |
| FM3 50 °C | 2205 ± 282 | 768 ± 200 | 0 | 2974 ± 465 |
| FM1 35 °C | 1879 ± 227 | 917 ± 170 | 0 | 2796 ± 373 |
| FM2 35 °C | 2326 ± 331 | 1500 ± 445 | 0 | 3826 ± 680 |
| FM3 35 °C | 2568 ± 548 | 1532 ± 226 | 0 | 4099 ± 740 |
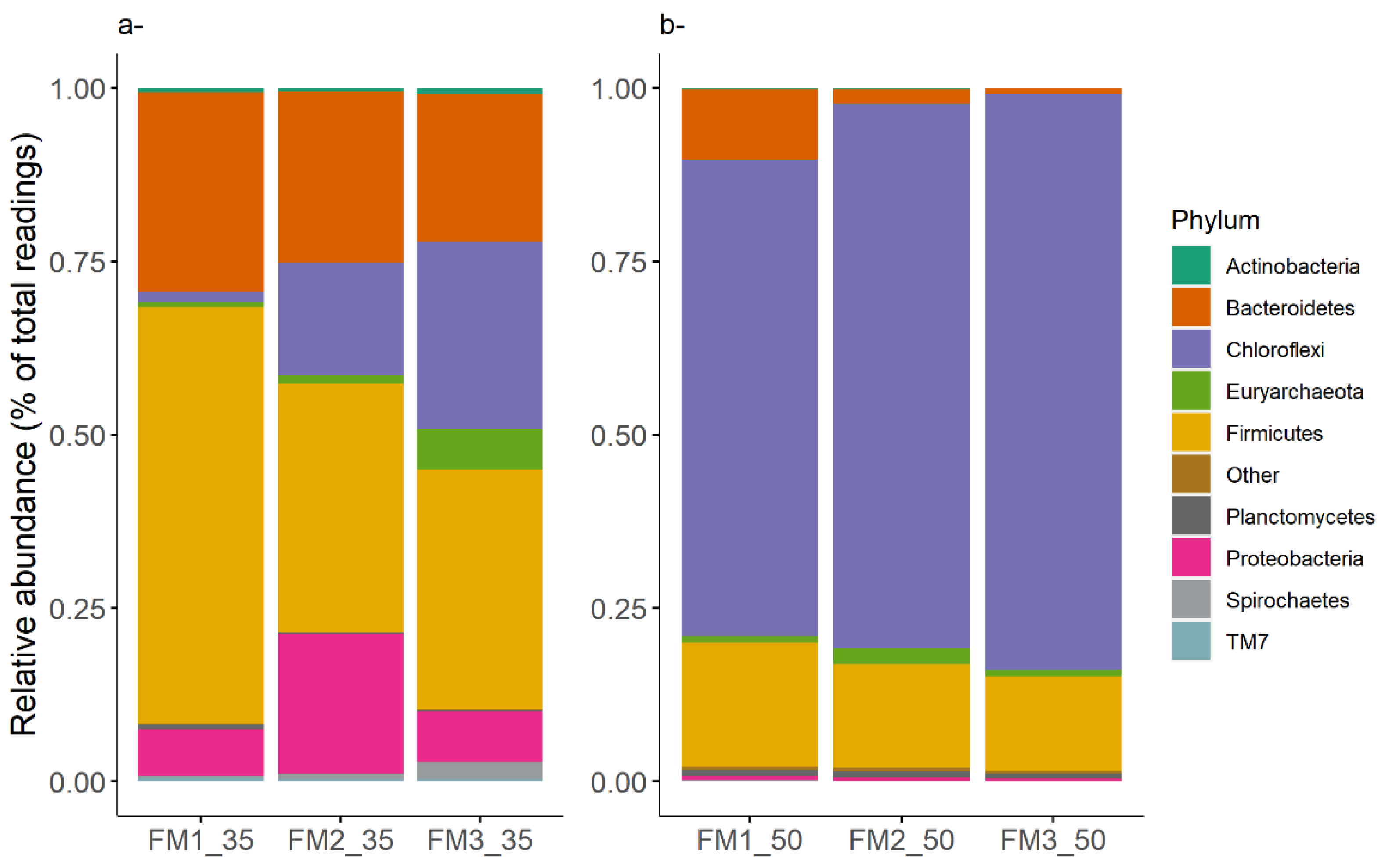
3.3. Prokaryotic Community
3.3.1. Core Communities
3.3.2. Effects of Culture Conditions on Microbial Communities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fulbright, N.R. Renewable Energy in Latin America, LLP. Available online: http://large.stanford.edu/courses/2017/ph240/pinilla2/docs/nrf-feb17.pdf (accessed on 15 January 2022).
- Aguilar, R.E.; Bustamante, M.; Kirk, D.; Miranda, J.A.; Baudrit, D.; Aguilar, J.F.; Rodriguez, W.; Reinhold, D.; Liao, W. Technical and economic feasibility of a solar-bio-powered waste utilization and treatment system in Central America. J. Environ. Manag. 2016, 184, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Coto, C.O. Uso de los Residuos Agrícolas Orgánicos como Fuente de Energía: Evaluación de la Generación de Residuos Agrícolas Orgánicos (RAO) en Costa Rica e Identificación de Sector Prioritario, FITTACORI, San José, Costa Rica. November 2012. Available online: https://www.mag.go.cr/proyectos/proy-residuos-agricolas-org/productos/Informe%20RAO%20CR%20Producto%201%20Final.pdf (accessed on 15 January 2022).
- Khan, I.U.; Othman, M.H.D.; Hashim, H.; Matsuura, T.; Ismail, A.F.; Rezaei-DashtArzhandi, M.; Azelee, I.W. Biogas as a renewable energy fuel–A review of biogas upgrading, utilisation and storage. Energy Convers. Manag. 2017, 150, 277–294. [Google Scholar] [CrossRef]
- Piechota, G. Removal of siloxanes from biogas upgraded to biomethane by Cryogenic Temperature Condensation System. J. Clean. Prod. 2021, 308, 127404. [Google Scholar] [CrossRef]
- Tang, Y.Q.; Ji, P.; Hayashi, J.; Koike, Y.; Wu, X.L.; Kida, K. Characteristic microbial community of a dry thermophilic methanogenic digester: Its long-term stability and change with feeding. Appl. Microbiol. Biotechnol. 2011, 91, 1447–1461. [Google Scholar] [CrossRef] [PubMed]
- Yue, Z.; Chen, R.; Yang, F.; MacLellan, J.; Marsh, T.; Liu, Y.; Liao, W. Effects of dairy manure and corn stover co-digestion on anaerobic microbes and corresponding digestion performance. Bioresour. Technol. 2013, 128, 65–71. [Google Scholar] [CrossRef]
- Wang, S.; Hou, X.; Su, H. Exploration of the relationship between biogas production and microbial community under high salinity conditions. Sci. Rep. 2017, 7, 1149. [Google Scholar] [CrossRef] [Green Version]
- Mei, R.; Nobu, M.K.; Narihiro, T.; Kuroda, K.; Munoz, S.J.; Wu, Z.; Ye, L.; Lee, P.K.H.; Lee, P.H.; Van Lier, J.B.; et al. Operation-driven heterogeneity and overlooked feed-associated populations in global anaerobic digester microbiome. Water Res. 2017, 124, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Rivière, D.; Desvignes, V.; Pelletier, E.; Chaussonnerie, S.; Guermazi, S.; Weissenbach, J.; Li, T.; Camacho, P.; Sghir, A. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 2009, 3, 700–714. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Kang, H.-J.; Lee, Y.H.; Lee, T.J.; Han, K.; Choi, Y.; Park, H.-D. Monitoring bacterial community structure and variability in time scale in full-scale anaerobic digesters. J. Environ. Monit. 2012, 14, 1893–1905. [Google Scholar] [CrossRef]
- St-Pierre, B.; Wright, A.D. Comparative metagenomic analysis of bacterial populations in three full-scale mesophilic anaerobic manure digesters. Appl. Microbiol. Biotechnol. 2014, 98, 2709–2717. [Google Scholar] [CrossRef]
- Freilich, S.; Kreimer, A.; Meilijson, I.; Gophna, U.; Sharan, R.; Ruppin, E. The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res. 2010, 38, 3857–3868. [Google Scholar] [CrossRef] [PubMed]
- Rui, J.; Li, J.; Zhang, S.; Yan, X.; Wang, Y.; Li., X. The core populations and co-occurrence patterns of prokaryotic communities in household biogas digesters. Biotechnol. Biofuels 2015, 8, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rezadehbashi, M.; Baldwin, S.A. Core sulphate-reducing microorganisms in metal-removing semi-passive biochemical reactors and the co-occurrence of methanogens. Microorganisms 2018, 6, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carballa, M.; Regueiro, L.; Lema, J.M. Microbial management of anaerobic digestion: Exploiting the microbiome-functionality nexus. Curr. Opin. Biotechnol. 2015, 33, 103–111. [Google Scholar] [CrossRef]
- Regueiro, L.; Lema, J.M.; Carballa, M. Key microbial communities steering the functioning of anaerobic digesters during hydraulic and organic overloading shocks. Bioresour. Technol. 2015, 197, 208–216. [Google Scholar] [CrossRef]
- Ortseifen, V.; Stolze, Y.; Maus, I.; Sczyrba, A.; Bremges, A.; Albaum, S.P.; Jaenicke, S.; Fracowiak, J.; Pühler, A.; Schlüter, A. An integrated metagenome and -proteome analysis of the microbial community residing in a biogas production plant. J. Biotechnol. 2016, 231, 268–279. [Google Scholar] [CrossRef] [Green Version]
- Ju, F.; Lau, F.; Zhang, T. Linking microbial community, environmental variables, and methanogenesis in anaerobic biogas digesters of chemically enhanced primary treatment sludge. Environ. Sci. Technol. 2017, 51, 3982–3992. [Google Scholar] [CrossRef]
- El-Mashad, H.M.; van Loon, W.K.P.; Zeeman, G. A model of solar energy utilisation in the anaerobic digestion of cattle manure. Biosyst. Eng. 2003, 84, 231–238. [Google Scholar] [CrossRef]
- Ward, A.J.; Hobbs, P.J.; Holliman, P.J.; Jones, D.L. Optimisation of the anaerobic digestion of agricultural resources. Bioresour. Technol. 2008, 99, 7928–7940. [Google Scholar] [CrossRef]
- Gebreeyessus, G.D.; Jenicek, P. Thermophilic versus mesophilic anaerobic digestion of sewage sludge: A comparative review. Bioengineering 2016, 3, 15. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Rodríguez, J.; Pérez, M.; Romero, L.I. Comparison of mesophilic and thermophilic dry anaerobic digestion of OFMSW: Kinetic analysis. Chem. Eng. J. 2013, 232, 59–64. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater, 23rd ed.; American Public Health Association (APHA): Washington, DC, USA, 2017. [Google Scholar]
- Stewart, B.A.; Porter, L.K.; Beard, W.E. Determination of Total Nitrogen and Carbon in Soils by a Commercial Dumas Apparatus. Soil Sci. Soc. Am. J. 1964, 28, 366–368. [Google Scholar] [CrossRef]
- Bertsch, F.; Henríquez, C.; Salas, R. Fertilidad de Suelos: Manual de Laboratorio, 1st ed.; Asociación Costarricense de la Ciencia del Suelo: San José, Costa Rica, 1995; Volume 39, p. 45. [Google Scholar]
- Sluiter, A.; Hames, B.; Ruiz, R.; Scarlata, C.; Sluiter, J.; Templeton, D.; Crocker, D. Determination of structural carbohydrates and lignin in biomass. Lab. Anal. Proced. 2008, 1617, 1–16. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Knight, R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [Green Version]
- Clarke, K.R.; Gorley, R.N. PRIMER v7: User Manual/Tutorial. PRIMER-EPlymouth; Plymouth Marine Laboratory: Plymouth, UK, 2015. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6. Mol. Biol. Evol. 2013, 12, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Verma, S. Anaerobic Digestion of Biodegradable Organics in Municipal Solid Wastes. Master’s Thesis, Department of Earth & Environmental Engineering, Columbia University, New York, NY, USA, 4 May 2002. [Google Scholar]
- Chen, Y.; Cheng, J.J.; Creamer, K.S. Inhibition of anaerobic digestion process: A review. Bioresour. Technol. 2008, 99, 4044–4064. [Google Scholar] [CrossRef]
- Sievers, D.M.; Brune, D.E. Carbon/nitrogen ratio and anaerobic digestion of swine waste. Trans. ASAE 1978, 21, 537–0541. [Google Scholar] [CrossRef]
- Maclellan, J.; Chen, R.; Kraemer, R.; Zhong, Y.; Liu, Y.; Liao, W. Anaerobic treatment of lignocellulosic material to co-produce methane and digested fiber for ethanol biorefining. Bioresour. Technol. 2013, 130, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.G.; Yoo, S.; Hwang, K.; Song, M.; Kim, W.; Han, G.; Hwang, S. Dynamics of transitional acidogenic community along with methanogenic population during anaerobic digestion of swine wastewater. Process. Biochem. 2011, 46, 1607–1613. [Google Scholar] [CrossRef]
- Bouallagui, H.; Touhami, Y.; Cheikh, R.B.; Hamdi, M. Bioreactor performance in anaerobic digestion of fruit and vegetable wastes. Process. Biochem. 2005, 40, 989–995. [Google Scholar] [CrossRef]
- Page, L.H.; Ni, J.Q.; Heber, A.J.; Mosier, N.S.; Liu, X.; Joo, H.S.; Ndegwa, P.M.; Harrison, J.H. Characteristics of volatile fatty acids in stored dairy manure before and after anaerobic digestion. Biosyst. Eng. 2014, 118, 16–28. [Google Scholar] [CrossRef]
- Callaghan, F.J.; Wase, D.A.J.; Thayanithy, K.; Forster, C.F. Continuous co-digestion of cattle slurry with fruit and vegetable wastes and chicken manure. Biomass Bioenergy 2002, 22, 71–77. [Google Scholar] [CrossRef]
- Wang, X.; Yang, G.; Feng, Y.; Ren, G.; Han, X. Optimizing feeding composition and carbon–nitrogen ratios for improved methane yield during anaerobic co-digestion of dairy, chicken manure and wheat straw. Bioresour. Technol. 2012, 120, 78–83. [Google Scholar] [CrossRef]
- Mata-Alvarez, J.; Mace, S.; Llabres, P. Anaerobic digestion of organic solid wastes. An overview of research achievements and perspectives. Bioresour. Technol. 2000, 74, 3–16. [Google Scholar] [CrossRef]
- Neves, L.; Oliveira, R.; Alves, M.M. Co-digestion of cow manure, food waste and intermittent input of fat. Bioresour. Technol. 2009, 100, 1957–1962. [Google Scholar] [CrossRef] [Green Version]
- El-Mashad, H.M.; Zhang, R. Biogas production from co-digestion of dairy manure and food waste. Bioresour. Technol. 2010, 101, 4021–4028. [Google Scholar] [CrossRef]
- Molinuevo-Salces, B.; González-Fernández, C.; Gómez, X.; García-González, M.C.; Morán, A. Vegetable processing wastes addition to improve swine manure anaerobic digestion: Evaluation in terms of methane yield and SEM characterization. Appl. Energy 2012, 91, 36–42. [Google Scholar] [CrossRef]
- Zhang, C.; Xiao, G.; Peng, L.; Su, H.; Tan, T. The anaerobic co-digestion of food waste and cattle manure. Bioresour. Technol. 2013, 129, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, X.; Li, F.; Yang, G. Effects of temperature and carbon-nitrogen (C/N) ratio on the performance of anaerobic co-digestion of dairy manure, chicken manure and rice straw: Focusing on ammonia inhibition. PLoS ONE 2014, 9, e97265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Zuo, J.; Gan, L.; Li, P.; Liu, F.; Wang, K.; Chen, L.; Gan, H. Effects of mixture ratio on anaerobic co-digestion with fruit and vegetable waste and food waste of China. Res. J. Environ. Sci. 2011, 23, 1403–1408. [Google Scholar] [CrossRef]
- Shi, J.; Wang, Z.; Stiverson, J.A.; Yu, Z.; Li, Y. Reactor performance and microbial community dynamics during solid-state anaerobic digestion of corn stover at mesophilic and thermophilic conditions. Bioresour. Technol. 2013, 136, 574–581. [Google Scholar] [CrossRef]
- Venkiteshwaran, K.; Bocher, B.; Maki, J.; Zitomer, D. Relating anaerobic digestion microbial community and process function: Supplementary issue: Water microbiology. Microbiol. Insights 2015, 8, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Joyce, A.; Ijaz, U.Z.; Nzeteu, C.; Vaughan, A.; Shirran, S.L.; Botting, C.H.; Quince, C.; O’Flaherty, V.; Abram, F. Linking microbial community structure and function during the acidified anaerobic digestion of grass. Front. Microbiol. 2018, 9, 540. [Google Scholar] [CrossRef]
- Lim, J.W.; Park, T.; Tong, Y.W.; Yu, Z. The microbiome driving anaerobic digestion and microbial analysis. Adv. Bioenergy 2020, 5, 1–61. [Google Scholar]
- Shaw, G.T.W.; Liu, A.C.; Weng, C.Y.; Chou, C.-Y.; Wang, D. Inferring microbial interactions in thermophilic and mesophilic anaerobic digestion of hog waste. PLoS ONE 2017, 12, e0181395. [Google Scholar] [CrossRef]
- Wan, S.; Sun, L.; Douieb, Y.; Sun, J.; Luo, W. Anaerobic digestion of municipal solid waste composed of food waste, wastepaper, and plastic in a single-stage system: Performance and microbial community structure characterization. Bioresour. Technol. 2013, 146, 619–627. [Google Scholar] [CrossRef]
- Ward, L.M.; Hemp, J.; Shih, P.M.; McGlynn, S.E.; Fischer, W.W. Evolution of phototrophy in the Chloroflexi phylum driven by horizontal gene transfer. Front. Microbiol. 2018, 9, 260. [Google Scholar] [CrossRef] [PubMed]
- Overmann, J. Green nonsulfur bacteria. eLS 2008. [Google Scholar] [CrossRef]
- Sun, L.; Toyonaga, M.; Ohashi, A.; Tourlousse, D.M.; Matsuura, N.; Meng, X.-Y.; Tamaki, H.; Hanada, S.; Cruz, R.; Yamaguchi, T.; et al. Lentimicrobium saccharophilum gen. nov., sp. nov., a strictly anaerobic bacterium representing a new family in the phylum Bacteroidetes, and proposal of Lentimicrobiaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 2635–2642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabari, L.; Gannoun, H.; Khelifi, E.; Cayol, J.-L.; Godon, J.-J.; Hamdi, M.; Fardeau, M.-L. Bacterial ecology of abattoir wastewater treated by an anaerobic digestor. Braz. J. Microbiol. 2016, 47, 73–84. [Google Scholar] [CrossRef]
- Chen, R.; Murillo, M.; Zhong, Y.; Marsh, T.; Roman, M.B.; Hernández, W.; Uribe, L.; Uribe Lorio, L.; Kirk, D.; Reinhold, D.M.; et al. Responses of anaerobic microorganisms to different culture conditions and corresponding effects on biogas production and solid digestate quality. Biomass Bioenergy 2016, 85, 84–93. [Google Scholar] [CrossRef]
- Zhong, Y.; Chen, R.; Rojas-Sossa, J.-P.; Isaguirre, C.; Mashburn, A.; Marsh, T.; Liu, Y.; Liao, W. Anaerobic co-digestion of energy crop and agricultural wastes to prepare uniform-format cellulosic feedstock for biorefining. Renew. Energy 2020, 147, 1358–1370. [Google Scholar] [CrossRef]
- Pagaling, E.; Grant, W.D.; Cowan, D.A.; Jones, B.E.; Ma, Y.; Ventosa, A.; Heaphy, S. Bacterial and archaeal diversity in two hot spring microbial mats from the geothermal region of Tengchong, China. Extremophiles 2012, 16, 607–618. [Google Scholar] [CrossRef]
- Tank, M.; Thiel, V.; Ward, D.M.; Bryant, D.A. A panoply of phototrophs: An overview of the thermophilic Chlorophototrophs of the microbial mats of alkaline siliceous hot springs in Yellowstone National Park, WY, USA. In Modern Topics in the Phototrophic Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2017; pp. 87–137. [Google Scholar]
- Lin, Q.; He, G.; Rui, J.; Fang, X.; Tao, Y.; Li, J.; Li, X. Microorganism-regulated mechanisms of temperature effects on the performance of anaerobic digestion. Microb. Cell Fact. 2016, 15, 96. [Google Scholar] [CrossRef] [Green Version]
- Westerholm, M.; Isaksson, S.; Karlsson Lindsjö, O.; Schnürer, A. Microbial community adaptability to altered temperature conditions determines the potential for process optimisation in biogas production. Appl. Energy 2018, 226, 838–848. [Google Scholar] [CrossRef]
- Hessler, T.; Harrison, S.T.; Huddy, R.J. Stratification of microbial communities throughout a biological sulphate reducing up-flow anaerobic packed bed reactor, revealed through 16S metagenomics. Res. Microbiol. 2018, 169, 543–551. [Google Scholar] [CrossRef]
- Zhang, J.; Wei, Y.; Xiao, W.; Zhou, Z.; Yan, X. Performance and spatial community succession of an anaerobic baffled reactor treating-acetone-butanol-ethanol fermentation wastewater. Bioresour. Technol. 2011, 102, 7407–7414. [Google Scholar] [CrossRef] [PubMed]
- Allison, M.J.; Mayberry, W.R.; McSweeney, C.R.; Stahl, D.A. Synergistes jonesii, gen. nov., sp. nov: A rumen bacterium that degrades toxic pyridinediols. Syst. Appl. Microbiol. 1992, 15, 522–529. [Google Scholar] [CrossRef]
- McSweeney, C.S.; Padmanabha, J.; Halliday, M.J.; Hubbard, B.; Dierens, L.; Denman, S.E.; Shelton, H. Detection of Synergistes jonesii and genetic variants in ruminants from different geographical locations. Trop. Grassl.—Forrajes Trop. 2019, 7, 154–163. [Google Scholar] [CrossRef] [Green Version]
- Tajima, K.; Nonaka, I.; Higuchi, K.; Takusari, N.; Kurihara, M.; Takenaka, A.; Mitsumori, M.; Kajikawa, H.; Aminov, R.I. Influence of high temperature and humidity on rumen bacterial diversity in Holstein heifers. Anaerobe 2007, 13, 57–64. [Google Scholar] [CrossRef]
- Ozbayram, E.G.; Ince, O.; Ince, B.; Harms, H.; Kleinsteuber, S. Comparison of rumen and manure microbiomes and implications for the inoculation of anaerobic digesters. Microorganisms 2018, 6, 15. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.H.; Wang, S.B.; Zhang, J.S.; Zhang, J.; Yan, X.; Zhou, H.K.; Zhao, G.P.; Zhou, Z.H. The structure of the bacterial and archaeal community in a biogas digester as revealed by denaturing gradient gel electrophoresis and 16S rDNA sequencing analysis. J. Appl. Microbiol. 2009, 106, 952–966. [Google Scholar] [CrossRef]
- Li, L.; He, Q.; Ma, Y.; Wang, X.; Peng, X. Dynamics of microbial community in a mesophilic anaerobic digester treating food waste: Relationship between community structure and process stability. Bioresour. Technol. 2015, 189, 113–120. [Google Scholar] [CrossRef]
- Li, Y.F.; Nelson, M.C.; Chen, P.H.; Graf, J.; Li, Y.; Yu, Z. Comparison of the microbial communities in solid-state anaerobic digestion (SS-AD) reactors operated at mesophilic and thermophilic temperatures. Appl. Microbiol. Biotechnol. 2015, 99, 969–980. [Google Scholar] [CrossRef]
- Jia, Y.; Wilkins, D.; Lu, H.; Cai, M.; Lee, P.K. Long-term enrichment on cellulose or xylan causes functional and taxonomic convergence of microbial communities from anaerobic digesters. Appl. Environ. Microbiol. 2016, 82, 1519–1529. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Banaszak, J.E.; Parameswaran, P.; Alder, J.; Krajmalnik-Brown, R.; Rittmann, B.E. Focused-pulsed sludge pre-treatment increases the bacterial diversity and relative abundance of acetoclastic methanogens in a full-scale anaerobic digester. Water Res. 2009, 43, 4517–4526. [Google Scholar] [CrossRef]
- Hook, S.E.; Steele, M.A.; Northwood, K.S.; Dijkstra, J.; France, J.; Wright, A.D.G.; McBride, B.W. Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol. Ecol. 2011, 78, 275–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, B.; Wang, L.Y.; Mbadinga, S.M.; Liu, J.F.; Yang, S.Z.; Gu, J.D.; Mu, B.Z. Anaerolineaceae and Methanosaeta turned to be the dominant microorganisms in alkanes-dependent methanogenic culture after long-term of incubation. AMB Express 2015, 5, 37. [Google Scholar] [CrossRef] [Green Version]
- Breznak, J.A. The Genus Sporomusa. Prokaryotes 2006, 4, 991–1001. [Google Scholar]
- Liu, Y.; Qiao, J.T.; Yuan, X.Z.; Guo, R.B.; Qiu, Y.L. Hydrogenispora ethanolica gen. nov., sp. nov., an anaerobic carbohydrate-fermenting bacterium from anaerobic sludge. Int. J. Syst. Evol. Microbiol. 2014, 64, 1756–1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueki, A.; Ohtaki, Y.; Kaku, N.; Ueki, K. Descriptions of Anaerotaenia torta gen. nov., sp. nov. and Anaerocolumna cellulosilytica gen. nov., sp. nov. isolated from a methanogenic reactor of cattle waste and reclassification of Clostridium aminovalericum, Clostridium jejuense and Clostridium xylanovorans as Anaerocolumna species. Int. J. Syst. Evol. Microbiol. 2016, 66, 2936–2943. [Google Scholar]
- Wang, L.; Hatem, A.; Catalyurek, U.V.; Morrison, M.; Yu, Z. Metagenomic insights into the carbohydrate-active enzymes carried by the microorganisms adhering to solid digesta in the rumen of cows. PLoS ONE 2013, 8, e78507. [Google Scholar] [CrossRef] [Green Version]
- Koeck, D.E.; Ludwig, W.; Wanner, G.; Zverlov, V.V.; Liebl, W.; Schwarz, W.H. Herbinix hemicellulosilytica gen. nov., sp. nov., a thermophilic cellulose-degrading bacterium isolated from a thermophilic biogas reactor. Int. J. Syst. Evol. Microbiol. 2015, 65, 2365–2371. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Wang, C.; Sun, F.; Zhu, W.; Wu, W. A comparison of microbial characteristics between the thermophilic and mesophilic anaerobic digesters exposed to elevated food waste loadings. Bioresour. Technol. 2014, 152, 420–428. [Google Scholar] [CrossRef]
- De Vrieze, J.; Hennebel, T.; Boon, N.; Verstraete, W. Methanosarcina: The rediscovered methanogen for heavy duty biomethanation. Bioresour. Technol. 2012, 112, 1–9. [Google Scholar] [CrossRef]
- Doloman, A.; Soboh, Y.; Walters, A.J.; Sims, R.C.; Miller, C.D. Qualitative analysis of microbial dynamics during anaerobic digestion of microalgal biomass in a UASB reactor. Int. J. Microbiol. 2017, 2017, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Gagliano, M.; Braguglia, C.M.; Gianico, A.; Mininni, G.; Nakamura, K.; Rossetti, S. Thermophilic anaerobic digestion of thermal pretreated sludge: Role of microbial community structure and correlation with process performances. Water Res. 2015, 68, 498–509. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.; Vítězová, M.; Vítěz, T.; Bartoš, M. Production of biogas: Relationship between methanogenic and sulfate-reducing microorganisms. Open Life Sci. 2017, 12, 82–91. [Google Scholar] [CrossRef]
- Tian, J.; Wang, Y.; Dong, X. Methanoculleus hydrogenitrophicus sp. nov., a methanogenic archaeon isolated from wetland soil. Int. J. Syst. Evol. Microbiol. 2010, 60, 2165–2169. [Google Scholar] [CrossRef] [PubMed]


| Parameter * | Dairy Manure | Chicken Litter | Fruits and Vegetables | Post-Consumer Food Wastes |
|---|---|---|---|---|
| Total solids (%) | 13.2 | 85 | 8.3 | 33.6 |
| C (%TS) | 36.9 | 38.9 | 42.0 | 45.7 |
| N (%TS) | 2.3 | 3.9 | 2.6 | 3.3 |
| C/N ratio | 16.0 | 9.9 | 16.2 | 13.8 |
| Glucan (%TS) | 14.7 | 25.3 | 18.6 | 55.9 |
| Xylan (%TS) | 12.6 | 9.4 | 7.5 | 2.9 |
| Lignin (%TS) | 27.3 | 6.79 | 21.8 | 11.4 |
| Treatment | Total Solids (%) | Volatile Solids (%) | Glucan (%TS) | Xylan (%TS) | Lignin (%TS) |
|---|---|---|---|---|---|
| FM1 50 °C | 2.9 ± 0.04 | 2.1 ± 0.04 | 10.7 ± 0.5 | 9.2 ± 0.2 | 28.5 ± 0.7 |
| FM2 50 °C | 2.6 ± 0.04 | 1.9 ± 0.04 | 10.9 ± 0.2 | 8.7 ± 0.04 | 27.8 ± 0.5 |
| FM3 50 °C | 2.5 ± 0.1 | 1.8 ± 0.1 | 14.0 ± 0.5 | 9.9 ± 0.1 | 27.1 ± 0.7 |
| FM1 35 °C | 2.9 ± 0.04 | 2.2 ± 0.04 | 10.8 ± 0.7 | 9.0 ± 0.5 | 28.8 ± 0.1 |
| FM2 35 °C | 2.5 ± 0.2 | 1.8 ± 0.2 | 14.0 ± 0.1 | 10.3 ± 0.04 | 26.4 ± 0.1 |
| FM3 35 °C | 2.4 ± 0.1 | 1.7 ± 0.1 | 14.4 ± 0.6 | 10.2 ± 0.3 | 28.0 ± 1.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murillo-Roos, M.; Uribe-Lorío, L.; Fuentes-Schweizer, P.; Vidaurre-Barahona, D.; Brenes-Guillén, L.; Jiménez, I.; Arguedas, T.; Liao, W.; Uribe, L. Biogas Production and Microbial Communities of Mesophilic and Thermophilic Anaerobic Co-Digestion of Animal Manures and Food Wastes in Costa Rica. Energies 2022, 15, 3252. https://doi.org/10.3390/en15093252
Murillo-Roos M, Uribe-Lorío L, Fuentes-Schweizer P, Vidaurre-Barahona D, Brenes-Guillén L, Jiménez I, Arguedas T, Liao W, Uribe L. Biogas Production and Microbial Communities of Mesophilic and Thermophilic Anaerobic Co-Digestion of Animal Manures and Food Wastes in Costa Rica. Energies. 2022; 15(9):3252. https://doi.org/10.3390/en15093252
Chicago/Turabian StyleMurillo-Roos, Mariana, Lorena Uribe-Lorío, Paola Fuentes-Schweizer, Daniela Vidaurre-Barahona, Laura Brenes-Guillén, Ivannia Jiménez, Tatiana Arguedas, Wei Liao, and Lidieth Uribe. 2022. "Biogas Production and Microbial Communities of Mesophilic and Thermophilic Anaerobic Co-Digestion of Animal Manures and Food Wastes in Costa Rica" Energies 15, no. 9: 3252. https://doi.org/10.3390/en15093252
APA StyleMurillo-Roos, M., Uribe-Lorío, L., Fuentes-Schweizer, P., Vidaurre-Barahona, D., Brenes-Guillén, L., Jiménez, I., Arguedas, T., Liao, W., & Uribe, L. (2022). Biogas Production and Microbial Communities of Mesophilic and Thermophilic Anaerobic Co-Digestion of Animal Manures and Food Wastes in Costa Rica. Energies, 15(9), 3252. https://doi.org/10.3390/en15093252







