Long-Term, Simultaneous Impact of Antimicrobials on the Efficiency of Anaerobic Digestion of Sewage Sludge and Changes in the Microbial Community
Abstract
1. Introduction
2. Materials and Methods
2.1. Substrate and Inoculum
2.2. Methane Fermen Tation
2.3. Sampling
2.4. Analytical Procedures
2.5. Isolation of Genomic DNAs
2.6. Library Preparation and Sequencing
2.7. Bioinformatics Analysis
2.8. Statistical Analysis
3. Results and Discussion
3.1. The Impact of Increasing Antibiotic Concentrations on the Efficiency of Anaerobic Digestion
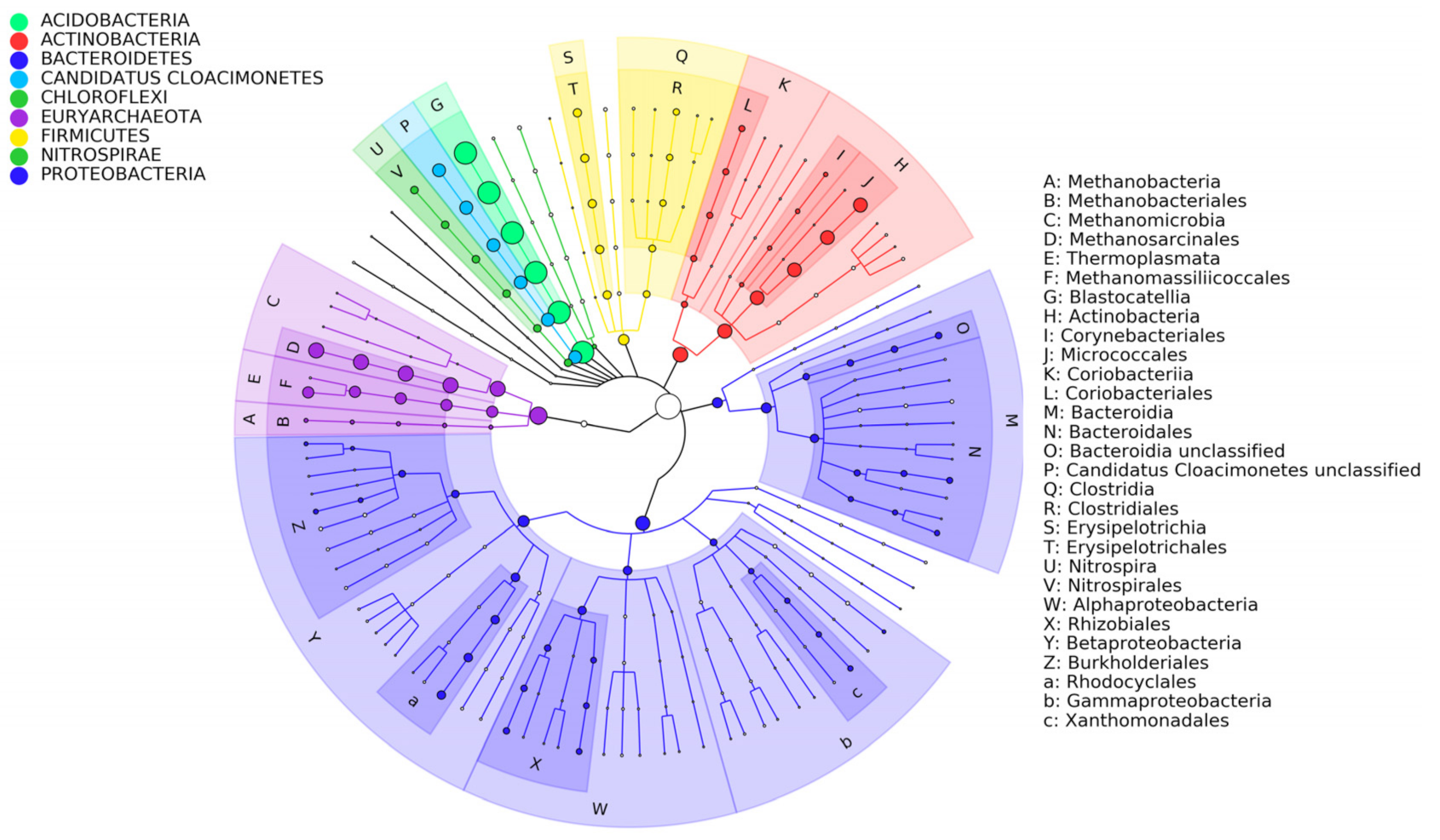
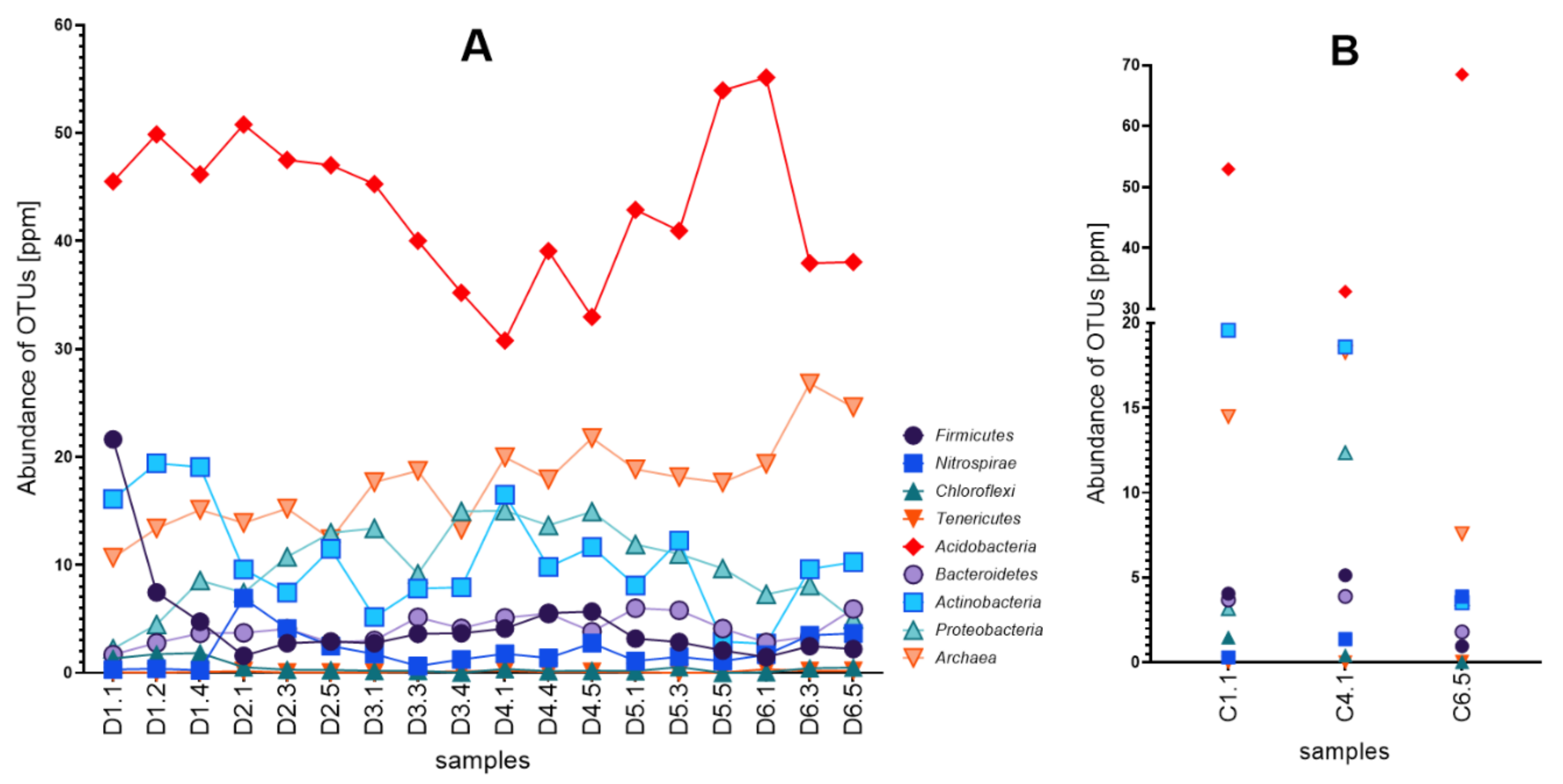
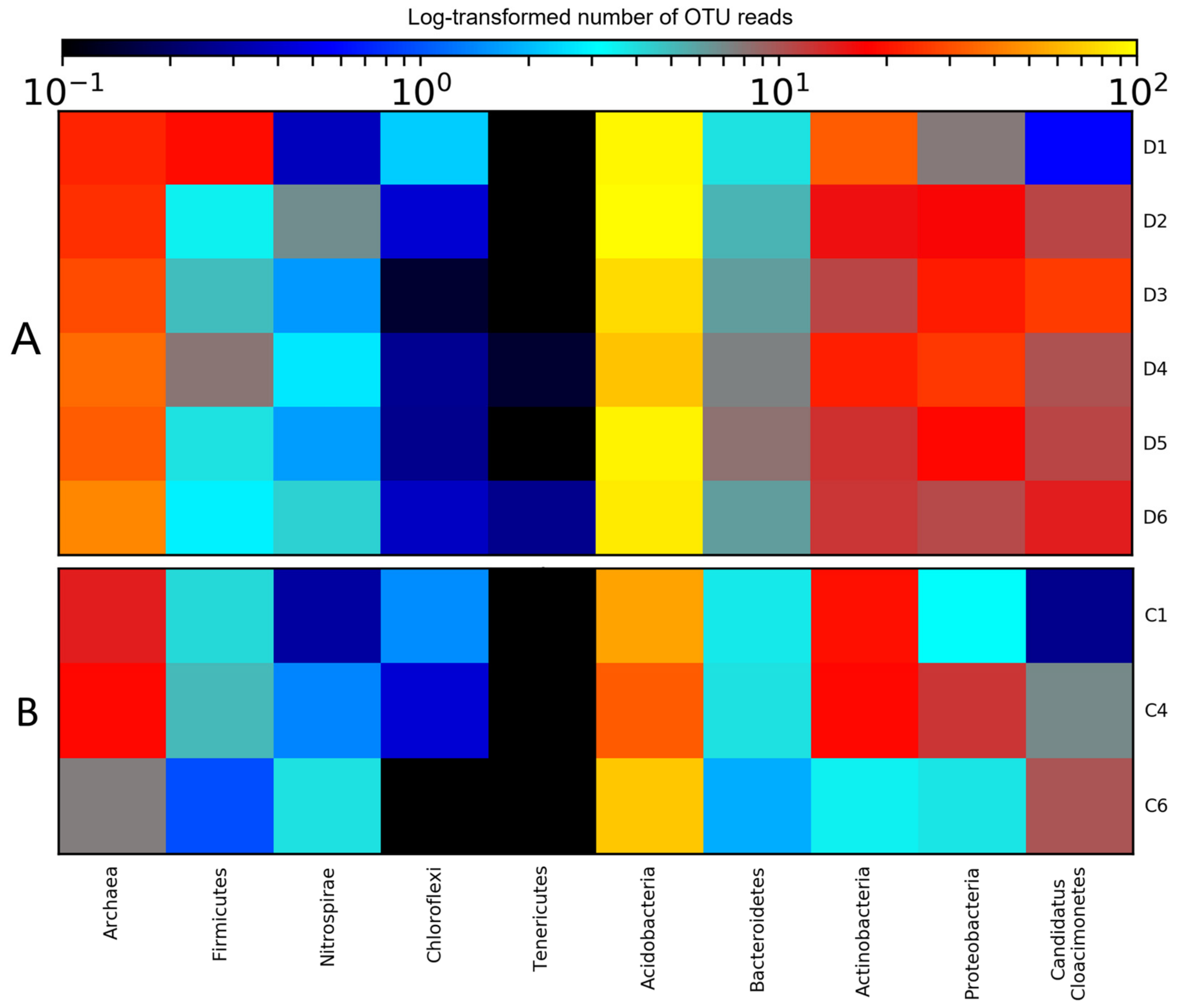
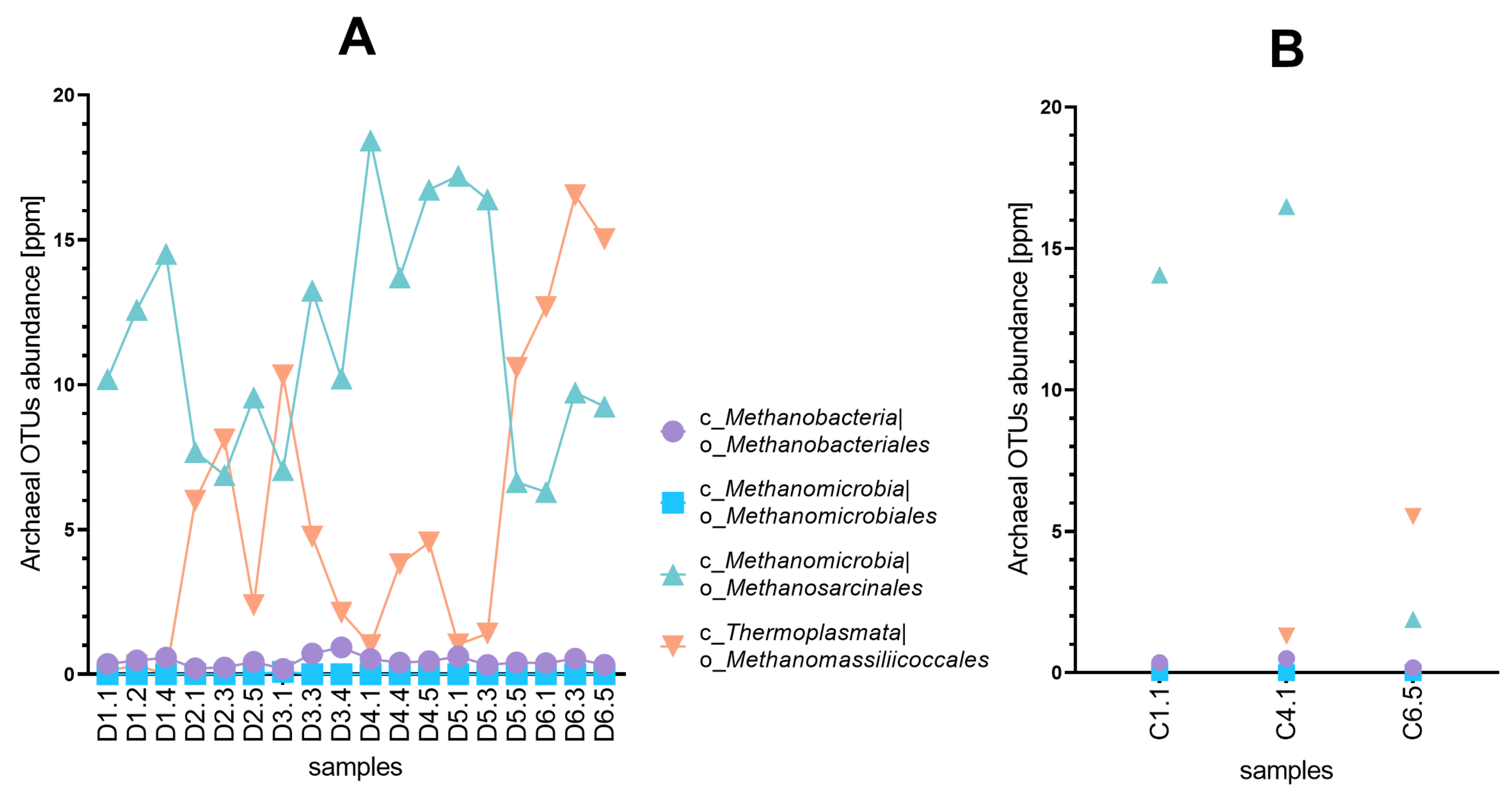
3.2. Diversity of Microbial Consortia
3.3. The Prevalence of Antibiotic Resistance Genes in Anaerobic Digesters
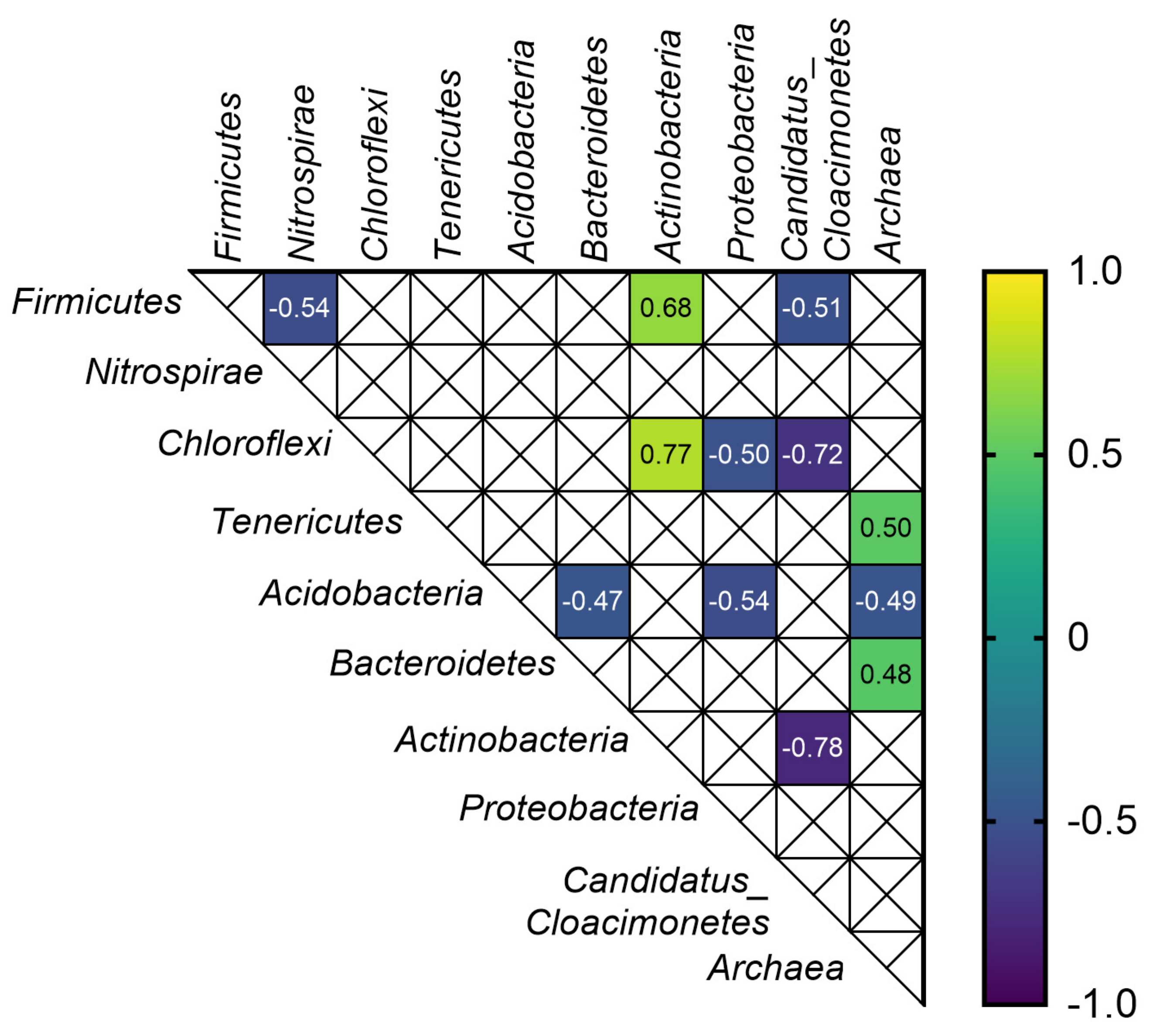
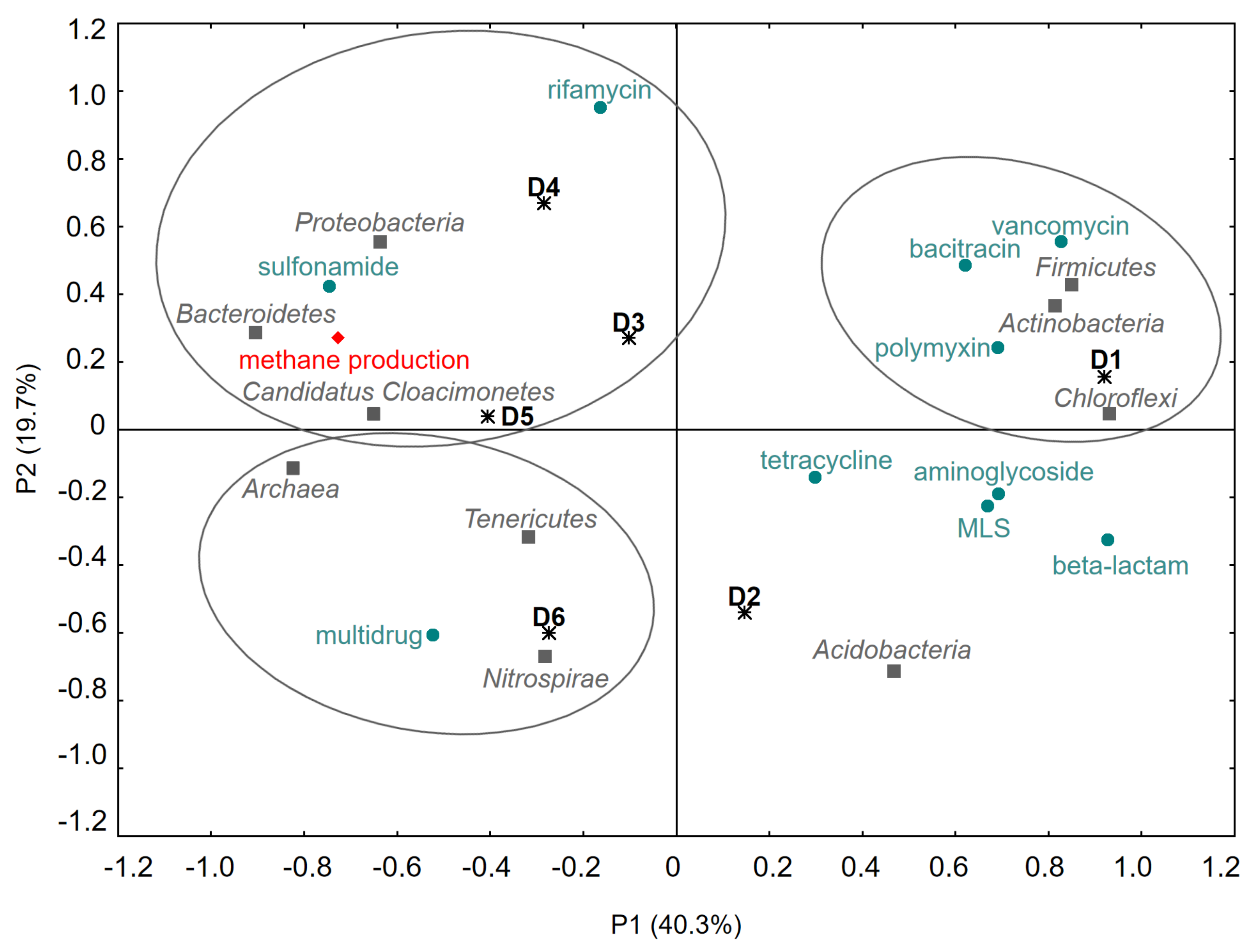
3.4. Correlations between Anaerobic Digestion Parameters, Microbial Biodiversity and Selected ARGs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carvalho, I.T.; Santos, L. Antibiotics in the aquatic environments: A review of the European scenario. Environ. Int. 2016, 94, 736–757. [Google Scholar] [CrossRef] [PubMed]
- Al-Riyami, I.M.; Ahmed, M.; Al-Busaidi, A.; Choudri, B.S. Antibiotics in wastewaters: A review with focus on Oman. Appl. Water Sci. 2018, 8, 199. [Google Scholar] [CrossRef]
- Bound, J.P.; Voulvoulis, N. Household Disposal of Pharmaceuticals as a Pathway for Aquatic Contamination in the United Kingdom. Environ. Health Perspect. 2005, 113, 1705–1711. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.M.; Ullman, J.L.; Teel, A.; Watts, R.J.; Frear, C. The effects of the antibiotics ampicillin, florfenicol, sulfamethazine, and tylosin on biogas production and their degradation efficiency during anaerobic digestion. Bioresour. Technol. 2013, 149, 244–252. [Google Scholar] [CrossRef]
- Czatzkowska, M.; Harnisz, M.; Korzeniewska, E.; Rusanowska, P.; Bajkacz, S.; Felis, E.; Jastrzębski, J.P.; Paukszto, Ł.; Koniuszewska, I. The impact of antimicrobials on the efficiency of methane fermentation of sewage sludge, changes in microbial biodiversity and the spread of antibiotic resistance. J. Hazard. Mater. 2021, 416, 125773. [Google Scholar] [CrossRef]
- Grobelak, A.; Grosser, A.; Kacprzak, M.; Kamizela, T. Sewage sludge processing and management in small and medium-sized municipal wastewater treatment plant-new technical solution. J. Environ. Manag. 2019, 234, 90–96. [Google Scholar] [CrossRef]
- Ju, F.; Li, B.; Ma, L.; Wang, Y.; Huang, D.; Zhang, T. Antibiotic resistance genes and human bacterial pathogens: Co-occurrence, removal, and enrichment in municipal sewage sludge digesters. Water Res. 2016, 91, 1–10. [Google Scholar] [CrossRef]
- Shin, J.; Rhee, C.; Shin, J.; Jang, H.M.; Shin, S.G.; Kim, Y.M. Determining the composition of bacterial community and relative abundance of specific antibiotics resistance genes via thermophilic anaerobic digestion of sewage sludge. Bioresour. Technol. 2020, 311, 123510. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, Q.; Meng, F.; Ruan, L.; Sun, T.; Liu, X.; Liu, W.; Zhu, Y.; Li, W.; Meng, F. Deciphering the role of calcium peroxide on the fate of antibiotic resistance genes and mobile genetic elements during bioelectrochemically-assisted anaerobic composting of excess dewatered sludge. Chem. Eng. J. 2020, 397, 125355. [Google Scholar] [CrossRef]
- Xiao, Y.; Raheem, A.; Ding, L.; Chen, W.-H.; Chen, X.; Wang, F.; Lin, S.-L. Pretreatment, modification and applications of sewage sludge-derived biochar for resource recovery—A review. Chemosphere 2021, 287, 131969. [Google Scholar] [CrossRef]
- Chen, Q.; An, X.; Li, H.; Su, J.; Ma, Y.; Zhu, Y.-G. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 2016, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.; Ying, G.-G.; Singer, A.; Zhu, Y.-G. Review of antibiotic resistance in China and its environment. Environ. Int. 2018, 110, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Alonso, L.L.; Demetrio, P.M.; Capparelli, A.L.; Marino, D.J. Behavior of ionophore antibiotics in aquatic environments in Argentina: The distribution on different scales in water courses and the role of wetlands in depuration. Environ. Int. 2019, 133, 105144. [Google Scholar] [CrossRef]
- Świątczak, P.; Cydzik-Kwiatkowska, A.; Rusanowska, P. Microbiota of anaerobic digesters in a full-scale wastewater treatment plant. Arch. Environ. Prot. 2017, 43, 53–60. [Google Scholar] [CrossRef]
- Czatzkowska, M.; Harnisz, M.; Korzeniewska, E.; Koniuszewska, I. Inhibitors of the methane fermentation process with particular emphasis on the microbiological aspect: A review. Energy Sci. Eng. 2020, 8, 1880–1897. [Google Scholar] [CrossRef]
- Chen, Y.; Cheng, J.J.; Creamer, K.S. Inhibition of anaerobic digestion process: A review. Bioresour. Technol. 2008, 99, 4044–4064. [Google Scholar] [CrossRef] [PubMed]
- Scarlat, N.; Dallemand, J.-F.; Fahl, F. Biogas: Developments and perspectives in Europe. Renew. Energy 2018, 129, 457–472. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef]
- Schmidt, V.M.; Pinchbeck, G.; McIntyre, K.M.; Nuttall, T.; McEwan, N.; Dawson, S.; Williams, N.J. Routine antibiotic therapy in dogs increases the detection of antimicrobial-resistant faecal Escherichia coli. J. Antimicrob. Chemother. 2018, 73, 3305–3316. [Google Scholar] [CrossRef]
- Rusanowska, P.; Zieliński, M.; Dębowski, M.; Harnisz, M.; Korzeniewska, E.; Amenda, E. Inhibition of Methane Fermentation by Antibiotics Introduced to Municipal Anaerobic Sludge. Proceedings 2018, 2, 1274. [Google Scholar] [CrossRef]
- Amani, T.; Nosrati, M.; Sreekrishnan, T.R. Anaerobic digestion from the viewpoint of microbiological, chemical, and operational aspects—A review. Environ. Rev. 2010, 18, 255–278. [Google Scholar] [CrossRef]
- Alvarado, A.; Montañez-Hernández, L.E.; Palacio-Molina, S.L.; Oropeza-Navarro, R.; Luévanos-Escareño, M.P.; Balagurusamy, N. Microbial trophic interactions and mcrA gene expression in monitoring of anaerobic digesters. Front. Microbiol. 2014, 5, 597. [Google Scholar] [CrossRef] [PubMed]
- Becerra-Castro, C.; Lopes, A.R.; Vaz-Moreira, I.; Silva, E.F.; Manaia, C.M.; Nunes, O.C. Wastewater reuse in irrigation: A microbiological perspective on implications in soil fertility and human and environmental health. Environ. Int. 2015, 75, 117–135. [Google Scholar] [CrossRef]
- Xu, Y.-B.; Hou, M.-Y.; Li, Y.-F.; Huang, L.; Ruan, J.-J.; Zheng, L.; Qiao, Q.-X.; Du, Q.-P. Distribution of tetracycline resistance genes and AmpC β-lactamase genes in representative non-urban sewage plants and correlations with treatment processes and heavy metals. Chemosphere 2017, 170, 274–281. [Google Scholar] [CrossRef]
- Barancheshme, F.; Munir, M. Development of Antibiotic Resistance in Wastewater Treatment Plants. In Antimicrobial Resistance—A Global Threat; IntechOpen: London, UK, 2019; pp. 75–92. [Google Scholar] [CrossRef]
- Sun, C.; Li, W.; Chen, Z.; Qin, W.; Wen, X. Responses of antibiotics, antibiotic resistance genes, and mobile genetic elements in sewage sludge to thermal hydrolysis pre-treatment and various anaerobic digestion conditions. Environ. Int. 2019, 133, 105156. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Zhang, Y.; Yang, M. Chronic impacts of oxytetracycline on mesophilic anaerobic digestion of excess sludge: Inhibition of hydrolytic acidification and enrichment of antibiotic resistome. Environ. Pollut. 2018, 238, 1017–1026. [Google Scholar] [CrossRef]
- Zhang, X.; Gu, J.; Wang, X.; Zhang, K.; Yin, Y.; Zhang, R.; Zhang, S. Effects of tylosin, ciprofloxacin, and sulfadimidine on mcrA gene abundance and the methanogen community during anaerobic digestion of cattle manure. Chemosphere 2019, 221, 81–88. [Google Scholar] [CrossRef]
- Wang, M.; Li, R.; Zhao, Q. Distribution and removal of antibiotic resistance genes during anaerobic sludge digestion with alkaline, thermal hydrolysis and ultrasonic pretreatments. Front. Environ. Sci. Eng. 2019, 13, 43. [Google Scholar] [CrossRef]
- Redhead, S.; Nieuwland, J.; Esteves, S.; Lee, D.-H.; Kim, D.-W.; Mathias, J.; Cha, C.-J.; Toleman, M.; Dinsdale, R.; Guwy, A.; et al. Fate of antibiotic resistant E. coli and antibiotic resistance genes during full scale conventional and advanced anaerobic digestion of sewage sludge. PLoS ONE 2020, 15, e0237283. [Google Scholar] [CrossRef]
- Koniuszewska, I.; Czatzkowska, M.; Harnisz, M.; Korzeniewska, E. The Impact of Antimicrobial Substances on the Methanogenic Community during Methane Fermentation of Sewage Sludge and Cattle Slurry. Appl. Sci. 2021, 11, 369. [Google Scholar] [CrossRef]
- Hernández Ceruelos, A.; Romero-Quezada, L.C.; Ruvalcaba Ledezma, J.C.; López Contreras, L. Therapeutic uses of metronidazole and its side effects: An update. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 397–401. [Google Scholar] [PubMed]
- Davis, J.L.; Papich, M.G. Chapter 65—Antimicrobial Therapy. In Equine Infectious Diseases, 2nd ed.; Sellon, D.C., Long, M.T., Eds.; Saunders WB: St. Louis, MO, USA, 2014; pp. 571–584.e5. [Google Scholar] [CrossRef]
- Kong, K.-F.; Schneper, L.; Mathee, K. Beta-lactam antibiotics: From antibiosis to resistance and bacteriology. APMIS 2010, 118, 1–36. [Google Scholar] [CrossRef] [PubMed]
- Pandey, N.; Cascella, M. Beta Lactam Antibiotics. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Thai, T.; Salisbury, B.H.; Zito, P.M. Ciprofloxacin. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- D’addazio, L.B.; Moraes, S.R. Microrganismos isolados de infecção do trato urinário da comunidade. Rev. Saúde Pública 2015, 6, 11–13. [Google Scholar] [CrossRef]
- Reis, A.C.C.; Santos, S.R.D.S.; de Souza, S.C.; Saldanha, M.G.; Pitanga, T.N.; Oliveira, R.R. Ciprofloxacin resistance pattern among bacteria isolated from patients with community-acquired urinary tract infection. Rev. Inst. Med. Trop. São Paulo 2016, 58, 53. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mustapha, N.A.; Sakai, K.; Shirai, Y.; Maeda, T. Impact of different antibiotics on methane production using waste-activated sludge: Mechanisms and microbial community dynamics. Appl. Microbiol. Biotechnol. 2016, 100, 9355–9364. [Google Scholar] [CrossRef]
- Giebułtowicz, J.; Nałęcz-Jawecki, G.; Harnisz, M.; Kucharski, D.; Korzeniewska, E.; Płaza, G. Environmental Risk and Risk of Resistance Selection Due to Antimicrobials’ Occurrence in Two Polish Wastewater Treatment Plants and Receiving Surface Water. Molecules 2020, 25, 1470. [Google Scholar] [CrossRef]
- Kisielewska, M.; Debowski, M.; Zieliński, M. Improvement of biohydrogen production using a reduced pressure fermentation. Bioprocess Biosyst. Eng. 2015, 38, 1925–1933. [Google Scholar] [CrossRef]
- APHA. Standard Methods for Water and Wastewater Examination, 19th ed.; American Public Health Association: Washington, DC, USA, 1995. [Google Scholar]
- Kneaddata v. 0.7.6 Software. Available online: https://github.com/biobakery/kneaddata (accessed on 12 November 2021).
- Franzosa, E.A.; McIver, L.J.; Rahnavard, G.; Thompson, L.R.; Schirmer, M.; Weingart, G.; Lipson, K.S.; Knight, R.; Caporaso, J.G.; Segata, N.; et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 2018, 15, 962–968. [Google Scholar] [CrossRef]
- Segata, N.; Waldron, L.; Ballarini, A.; Narasimhan, V.; Jousson, O.; Huttenhower, C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 2012, 9, 811–814. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed]
- Asnicar, F.; Weingart, G.; Tickle, T.; Huttenhower, C.; Segata, N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 2015, 3, e1029. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Jiang, X.-T.; Chai, B.; Li, L.; Yang, Y.; Cole, J.R.; Tiedje, J.M.; Zhang, T. ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics 2018, 34, 2263–2270. [Google Scholar] [CrossRef]
- Hclust2 Python Script. Available online: https://github.com/SegataLab/hclust2 (accessed on 15 November 2021).
- The Custom R Function. Available online: https://raw.githubusercontent.com/BioinfoCoreFacility/metaGen/main/R/cor.OTUvsGENES.R (accessed on 15 November 2021).
- Álvarez, J.; Otero, L.; Lema, J.; Omil, F. The effect and fate of antibiotics during the anaerobic digestion of pig manure. Bioresour. Technol. 2010, 101, 8581–8586. [Google Scholar] [CrossRef] [PubMed]
- Lagator, M.; Uecker, H.; Neve, P. Adaptation at different points along antibiotic concentration gradients. Biol. Lett. 2021, 17, 20200913. [Google Scholar] [CrossRef]
- Xiong, L.; Yang, Y.; Ye, Y.-N.; Teng, J.L.L.; Chan, E.; Watt, R.M.; Guo, F.-B.; Lau, S.K.P.; Woo, P.C.Y. Laribacter hongkongensisanaerobic adaptation mediated by arginine metabolism is controlled by the cooperation of FNR and ArgR. Environ. Microbiol. 2017, 19, 1266–1280. [Google Scholar] [CrossRef]
- Lallai, A.; Mura, G.; Onnis, N. The effects of certain antibiotics on biogas production in the anaerobic digestion of pig waste slurry. Bioresour. Technol. 2002, 82, 205–208. [Google Scholar] [CrossRef]
- Spielmeyer, A.; Breier, B.; Groißmeier, K.; Hamscher, G. Elimination patterns of worldwide used sulfonamides and tetracyclines during anaerobic fermentation. Bioresour. Technol. 2015, 193, 307–314. [Google Scholar] [CrossRef]
- Harb, M.; Wei, C.-H.; Wang, N.; Amy, G.; Hong, P.-Y. Organic micropollutants in aerobic and anaerobic membrane bioreactors: Changes in microbial communities and gene expression. Bioresour. Technol. 2016, 218, 882–891. [Google Scholar] [CrossRef]
- Lu, X.; Zhen, G.; Liu, Y.; Hojo, T.; Estrada, A.L.; Li, Y.-Y. Long-term effect of the antibiotic cefalexin on methane production during waste activated sludge anaerobic digestion. Bioresour. Technol. 2014, 169, 644–651. [Google Scholar] [CrossRef]
- Shah, F.A.; Mahmood, Q.; Maroof Shah, M.; Pervez, A.; Ahmad Asad, S. Retracted: Microbial Ecology of Anaerobic Digesters: The Key Players of Anaerobiosis. Sci. World J. 2017, 2017, 3852369. [Google Scholar] [CrossRef]
- De Chaves, M.G.; Silva, G.G.Z.; Rossetto, R.; Edwards, R.A.; Tsai, S.M.; Navarrete, A.A. Acidobacteria Subgroups and Their Metabolic Potential for Carbon Degradation in Sugarcane Soil Amended with Vinasse and Nitrogen Fertilizers. Front. Microbiol. 2019, 10, 1680. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, J.M.; Singleton, C.; Clegg, L.-A.; Petriglieri, F.; Nielsen, P.H. High Diversity and Functional Potential of Undescribed “Acidobacteriota” in Danish Wastewater Treatment Plants. Front. Microbiol. 2021, 12, 643950. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, C.; Al-Soud, W.A.; Larsson, M.; Alm, E.; Yekta, S.S.; Svensson, B.H.; Sørensen, S.J.; Karlsson, A. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol. Ecol. 2013, 85, 612–626. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, K.; Xia, Y.; Lau, F.T.K.; Tang, D.T.W.; Fung, W.C.; Fang, H.H.P.; Zhang, T. Metagenomic analysis of sludge from full-scale anaerobic digesters operated in municipal wastewater treatment plants. Appl. Microbiol. Biotechnol. 2014, 98, 5709–5718. [Google Scholar] [CrossRef]
- Walter, A.; Probst, M.; Franke-Whittle, I.H.; Ebner, C.; Podmirseg, S.M.; Etemadi-Shalamzari, M.; Hupfauf, S.; Insam, H. Microbiota in anaerobic digestion of sewage sludge with and without co-substrates. Water Environ. J. 2018, 33, 214–222. [Google Scholar] [CrossRef]
- Talavera-Caro, A.G.; Lira, I.O.H.-D.; Cruz, E.R.; Sánchez-Muñoz, M.A.; Balagurusamy, N. The Realm of Microorganisms in Biogas Production: Microbial Diversity, Functional Role, Community Interactions, and Monitoring the Status of Biogas Plant. In Biogas Production; Balagurusamy, N., Chandel, A.K., Eds.; Springer: Cham, Switzerland, 2020. [Google Scholar] [CrossRef]
- Lim, S.D.; Mayer, J.A.; Yim, W.C.; Cushman, J.C. Plant tissue succulence engineering improves water-use efficiency, water-deficit stress attenuation and salinity tolerance in Arabidopsis. Plant J. 2020, 103, 1049–1072. [Google Scholar] [CrossRef]
- Müller, B.; Sun, L.; Westerholm, M.; Schnürer, A. Bacterial community composition and fhs profiles of low- and high-ammonia biogas digesters reveal novel syntrophic acetate-oxidising bacteria. Biotechnol. Biofuels 2016, 9, 48. [Google Scholar] [CrossRef]
- Klang, J.; Szewzyk, U.; Bock, D.; Theuerl, S. Nexus between the microbial diversity level and the stress tolerance within the biogas process. Anaerobe 2019, 56, 8–16. [Google Scholar] [CrossRef]
- Braz, G.H.; Fernandez-Gonzalez, N.; Lema, J.M.; Carballa, M. Organic overloading affects the microbial interactions during anaerobic digestion in sewage sludge reactors. Chemosphere 2019, 222, 323–332. [Google Scholar] [CrossRef]
- Poirier, S.; Déjean, S.; Midoux, C.; Cao, K.-A.L.; Chapleur, O. Integrating independent microbial studies to build predictive models of anaerobic digestion inhibition by ammonia and phenol. Bioresour. Technol. 2020, 316, 123952. [Google Scholar] [CrossRef]
- Singh, A.; Müller, B.; Schnürer, A. Profiling temporal dynamics of acetogenic communities in anaerobic digesters using next-generation sequencing and T-RFLP. Sci. Rep. 2021, 11, 13298. [Google Scholar] [CrossRef]
- Veloo, A.; Baas, W.; Haan, F.; Coco, J.; Rossen, J. Prevalence of antimicrobial resistance genes in Bacteroides spp. and Prevotella spp. Dutch clinical isolates. Clin. Microbiol. Infect. 2019, 25, 1156.e9–1156.e13. [Google Scholar] [CrossRef] [PubMed]
- Duranti, S.; Mangifesta, M.; Lugli, G.A.; Turroni, F.; Anzalone, R.; Milani, C.; Mancabelli, L.; Ossiprandi, M.C.; Ventura, M. Bifidobacterium vansinderenii sp. nov., isolated from faeces of emperor tamarin (Saguinus imperator). Int. J. Syst. Evol. Microbiol. 2017, 67, 3987–3995. [Google Scholar] [CrossRef]
- Rozman, V.; Lorbeg, P.M.; Accetto, T.; Matijašić, B.B. Characterization of antimicrobial resistance in lactobacilli and bifidobacteria used as probiotics or starter cultures based on integration of phenotypic and in silico data. Int. J. Food Microbiol. 2019, 314, 108388. [Google Scholar] [CrossRef] [PubMed]
- Burkovski, A. Cell Envelope of Corynebacteria: Structure and Influence on Pathogenicity. ISRN Microbiol. 2013, 2013, 935736. [Google Scholar] [CrossRef]
- Bernard, K.A.; Pacheco, A.L.; Loomer, C.; Burdz, T.; Wiebe, D.; Huynh, C.; Kaplen, B.; Olson, A.B.; Cnockaert, M.; Eguchi, H.; et al. Corynebacterium lowii sp. nov. and Corynebacterium oculi sp. nov., derived from human clinical disease and an emended description of Corynebacterium mastitidis. Int. J. Syst. Evol. Microbiol. 2016, 66, 2803–2812. [Google Scholar] [CrossRef]
- Liu, Y.; Whitman, W.B. Metabolic, Phylogenetic, and Ecological Diversity of the Methanogenic Archaea. Ann. N. Y. Acad. Sci. 2008, 1125, 171–189. [Google Scholar] [CrossRef] [PubMed]
- Dridi, B.; Fardeau, M.-L.; Ollivier, B.; Raoult, D.; Drancourt, M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2012, 62, 1902–1907. [Google Scholar] [CrossRef] [PubMed]
- Lackner, N.; Hintersonnleitner, A.; Wagner, A.O.; Illmer, P. Hydrogenotrophic Methanogenesis and Autotrophic Growth of Methanosarcina thermophila. Archaea 2018, 2018, 4712608. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Peng, Y.; Ni, B.-J.; Han, X.; Fan, L.; Yuan, Z. Dissecting microbial community structure and methane-producing pathways of a full-scale anaerobic reactor digesting activated sludge from wastewater treatment by metagenomic sequencing. Microb. Cell Factories 2015, 14, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, H.; Zhang, Y.; Si, D.; Chen, Q. Evolution of microbial community along with increasing solid concentration during high-solids anaerobic digestion of sewage sludge. Bioresour. Technol. 2016, 216, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Enzmann, F.; Mayer, F.; Rother, M.; Holtmann, D. Methanogens: Biochemical background and biotechnological applications. AMB Express 2018, 8, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Miura, Y.; Hiraiwa, M.N.; Ito, T.; Itonaga, T.; Watanabe, Y.; Okabe, S. Bacterial community structures in MBRs treating municipal wastewater: Relationship between community stability and reactor performance. Water Res. 2007, 41, 627–637. [Google Scholar] [CrossRef]
- Song, M.; Shin, S.G.; Hwang, S. Methanogenic population dynamics assessed by real-time quantitative PCR in sludge granule in upflow anaerobic sludge blanket treating swine wastewater. Bioresour. Technol. 2010, 101, S23–S28. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-J.; Pan, J.; Liu, Y.; Duan, C.-H.; Li, M. Genomic and transcriptomic insights into methanogenesis potential of novel methanogens from mangrove sediments. Microbiome 2020, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Yang, Z.-H.; Zheng, Y.; Wang, Q.-P.; Bai, Y.; Liu, J.-B.; Zhang, Y.-R.; Xiong, W.-P.; Lu, Y.; Fan, C.-Z. Metagenomic analysis reveals the effects of long-term antibiotic pressure on sludge anaerobic digestion and antimicrobial resistance risk. Bioresour. Technol. 2019, 282, 179–188. [Google Scholar] [CrossRef]
- Ni, B.-J.; Zeng, S.; Wei, W.; Dai, X.; Sun, J. Impact of roxithromycin on waste activated sludge anaerobic digestion: Methane production, carbon transformation and antibiotic resistance genes. Sci. Total Environ. 2019, 703, 134899. [Google Scholar] [CrossRef]
- Zou, Y.; Qian, X.; Liu, X.; Song, Y.; Song, C.; Wu, S.; An, Y.; Yuan, R.; Wang, Y.; Xie, Y. The effect of antibiotic resistance onHelicobacter pylorieradication efficacy: A systematic review and meta-analysis. Helicobacter 2020, 25, e12714. [Google Scholar] [CrossRef]
- Shi, Y.; Zhang, H.; Tian, Z.; Yang, M.; Zhang, Y. Characteristics of ARG-carrying plasmidome in the cultivable microbial community from wastewater treatment system under high oxytetracycline concentration. Appl. Microbiol. Biotechnol. 2018, 102, 1847–1858. [Google Scholar] [CrossRef]
- Lamm, A.; Gozlan, I.; Rotstein, A.; Avisar, D. Detection of amoxicillin-diketopiperazine-2′, 5′ in wastewater samples. J. Environ. Sci. Health Part A 2009, 44, 1512–1517. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Larsson, D.J. Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environ. Int. 2016, 86, 140–149. [Google Scholar] [CrossRef] [PubMed]



| Antibiotic Concentration (µg/mL) | |||
|---|---|---|---|
| Dose ID | AMO | CIP | MET |
| D1 | 1 | 0.25 | 0.25 |
| D2 | 2 | 0.5 | 0.5 |
| D3 | 4 | 1 | 1 |
| D4 | 8 | 4 | 4 |
| D5 | 16 | 8 | 8 |
| D6 | 36 | 16 | 16 |
| Methane and VFA Analyses | Sequencing | ||||
|---|---|---|---|---|---|
| Experimental series 1 | Sample | Process Bioreactor | Control Bioreactor | Process Bioreactor | Control Bioreactor |
| Sample ID | |||||
| 1 | D1.1 | C1.1 | D1.1 | C1.1 | |
| 2 | D1.2 | C1.2 | D1.2 | na | |
| 3 | D1.3 | C1.3 | na | na | |
| 4 | D1.4 | C1.4 | D1.4 | na | |
| Experimental series 2 | Sample | Process bioreactor | Control bioreactor | Process bioreactor | Control bioreactor |
| Sample ID | |||||
| 1 | D2.1 | C2.1 | D2.1 | na | |
| 2 | D2.2 | C2.2 | na | na | |
| 3 | D2.3 | C2.3 | D2.3 | na | |
| 4 | D2.4 | C2.4 | na | na | |
| 5 | D2.5 | C2.5 | D2.5 | na | |
| Experimental series 3 | Sample | Process bioreactor | Control bioreactor | Process bioreactor | Control bioreactor |
| Sample ID | |||||
| 1 | D3.1 | C3.1 | D3.1 | na | |
| 2 | D3.2 | C3.2 | na | na | |
| 3 | D3.3 | C3.3 | D3.3 | na | |
| 4 | D3.4 | C3.4 | D3.4 | na | |
| Experimental series 4 | Sample | Process bioreactor | Control bioreactor | Process bioreactor | Control bioreactor |
| Sample ID | |||||
| 1 | D4.1 | C4.1 | D4.1 | C4.1 | |
| 2 | D4.2 | C4.2 | na | na | |
| 3 | D4.3 | C4.3 | na | na | |
| 4 | D4.4 | C4.4 | D4.4 | na | |
| 5 | D4.5 | C4.5 | D4.5 | na | |
| Experimental series 5 | Sample | Process bioreactor | Control bioreactor | Process bioreactor | Control bioreactor |
| Sample ID | |||||
| 1 | D5.1 | C5.1 | D5.1 | na | |
| 2 | D5.2 | C5.2 | na | na | |
| 3 | D5.3 | C5.3 | D5.3 | na | |
| 4 | D5.4 | C5.4 | na | na | |
| 5 | D5.5 | C5.5 | D5.5 | na | |
| Experimental series 6 | Sample | Process bioreactor | Control bioreactor | Process bioreactor | Control bioreactor |
| Sample ID | |||||
| 1 | D6.1 | C6.1 | D6.1 | na | |
| 2 | D6.2 | C6.2 | na | na | |
| 3 | D6.3 | C6.3 | D6.3 | na | |
| 4 | D6.4 | C6.4 | na | na | |
| 5 | D6.5 | C6.5 | D6.5 | C6.5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czatzkowska, M.; Harnisz, M.; Korzeniewska, E.; Wolak, I.; Rusanowska, P.; Paukszto, Ł.; Jastrzębski, J.P.; Bajkacz, S. Long-Term, Simultaneous Impact of Antimicrobials on the Efficiency of Anaerobic Digestion of Sewage Sludge and Changes in the Microbial Community. Energies 2022, 15, 1826. https://doi.org/10.3390/en15051826
Czatzkowska M, Harnisz M, Korzeniewska E, Wolak I, Rusanowska P, Paukszto Ł, Jastrzębski JP, Bajkacz S. Long-Term, Simultaneous Impact of Antimicrobials on the Efficiency of Anaerobic Digestion of Sewage Sludge and Changes in the Microbial Community. Energies. 2022; 15(5):1826. https://doi.org/10.3390/en15051826
Chicago/Turabian StyleCzatzkowska, Małgorzata, Monika Harnisz, Ewa Korzeniewska, Izabela Wolak, Paulina Rusanowska, Łukasz Paukszto, Jan P. Jastrzębski, and Sylwia Bajkacz. 2022. "Long-Term, Simultaneous Impact of Antimicrobials on the Efficiency of Anaerobic Digestion of Sewage Sludge and Changes in the Microbial Community" Energies 15, no. 5: 1826. https://doi.org/10.3390/en15051826
APA StyleCzatzkowska, M., Harnisz, M., Korzeniewska, E., Wolak, I., Rusanowska, P., Paukszto, Ł., Jastrzębski, J. P., & Bajkacz, S. (2022). Long-Term, Simultaneous Impact of Antimicrobials on the Efficiency of Anaerobic Digestion of Sewage Sludge and Changes in the Microbial Community. Energies, 15(5), 1826. https://doi.org/10.3390/en15051826








