Abstract
The microbial enhanced oil recovery (MEOR) method is an eco-friendly and economical alternative technology. The technology involves a variety of uncertainties, and its success depends on controlling microbial growth and metabolism. Though a few numerical studies have been carried out to reduce the uncertainties, no attempt has been made to consider temperature, pressure, and salinity in an integrated manner. In this study, a new modeling method incorporating these environmental impacts was proposed, and MEOR analysis was performed. As a result, accurate modeling was possible to prevent overestimating the performance of MEOR. In addition, oil recovery was maximized through sensitivity analysis and optimization based on an integrative model. Finally, applying MEOR to an actual reservoir model showed a 7% increase in oil recovery compared to waterflooding. This result proved the practical applicability of the method.
1. Introduction
Among various methods used to increase the productivity of the reservoir, chemical flooding, the gas injection method, and thermal methods are the most widely used processes for enhanced oil recovery (EOR) [1,2,3,4,5,6,7]. In addition to these methods, there is an alternative technology called microbial enhanced oil recovery (MEOR). The MEOR method is a technology that uses microbial metabolism and products to improve oil production [8,9]. This technology is eco-friendly in that the materials used are biodegradable, and it is economical in that the cost to produce them is low. The MEOR process changes the petrophysical and/or petrochemical properties of the reservoir system depending on the microorganisms and the microbial materials [10]. Among the microorganisms used in MEOR, Leuconostoc mesenteroiedes is a microbial species that produces a biopolymer called dextran. It is used to block the high permeability zone, which decreases productivity due to early water breakthrough after waterflooding. Dextran improves productivity by the bypass effect for injected water by selective plugging in the reservoir.
Since the 1950s, MEOR has been applied in many countries in various ways, among which selective plugging has also been used as a major mechanism to increase oil productivity. The North Burbank Unit in Oklahoma [11] and North Blowhorn Creek Oil Unit in Alabama [12], USA, showed increased oil production through selective plugging. In Canada, plugging by Leuconostoc was applied to the MEOR [13], and a similar method was attempted for the fractured reservoir [14]. Dutch researchers also showed the result of increasing oil productivity and improving water-oil ratio through the selective plugging effect by Betacoccus dextranicus [15]. In addition, MEORs using selective plugging have been applied in various fields in China, UK, Saudi Arabia, etc. [16]. However, for the result of applying MEOR, one out of ten was evaluated as ineffective [17] because proper application failed due to uncertainty in the method.
Various environmental factors affect the growth of microorganisms [10,18,19,20]. Temperature has the greatest influence on the growth of microbes [21,22,23,24,25,26,27,28,29], and there are a range of possible growth temperatures and an optimum growth temperature depending on the microbial species. The effect of temperature on the rate of microbial growth is similar to the activity of an enzyme. The growth rate increases as the temperature increases to a certain point where protein denaturation occurs. The growth rate begins to decrease from the moment the temperature exceeds that temperature [30]. Pressure affects microbial survival [31,32,33,34], and higher pressure conditions adversely affect microbial survival. The destruction of microbial cells depends on the level of pressure and the time of exposure to the pressure [35]. Salinity is also known to affect microbial growth [36,37,38,39,40,41,42,43]. Salt concentration has a great influence on enzymatic catalysis [44]. In general, the salinity range of the reservoir is known to be 100 mg/L to 300 g/L or more [45]. According to Maudgalya et al. [46], successful cases of MEOR have been reported in a reservoir where the temperature is below 200 °F and the salinity is above 1000 ppm.
A few numerical studies on selective plugging by bacteria have been conducted. Early models described microbial growth based on the Monod equation, and oil recovery was calculated using a simple fluid flow model [47,48,49]. Delshad et al. [50] depicted MEOR using UTCHEM, and Stewart and Kim [51] simulated the selective plugging effect using a biofilm evolution-removal model. Vilcáez et al. [52] and Surasani et al. [53] studied Leconostoc mesenteroides growth and plugging phenomena using a model called CrunchFlow. Recently, selective plugging simulation and oil productivity analysis have been performed using CMG STARS [54,55]. These previous studies have not been able to simultaneously reflect the effects of temperature, pressure, and salinity on the growth of microorganisms, and have not performed optimization considering these environmental factors.
Most of the earliest studies were limited to describing microbial metabolism in simple transport models. They were not suitable for analyzing oil productivity for complex reservoir conditions in filed scale. Only a few recent studies have attempted to analyze MEOR effects in the reservoir scale. However, studies that have evaluated and optimized considering the subsurface environment are not available yet. A typical reservoir environment is high temperature, high pressure, and high salinity, which are fatal to the growth of microorganisms. If these factors are not considered, it is impossible to accurately evaluate the MEOR performance. Therefore, this study attempted to perform an MEOR simulation considering three environmental variables at the same time.
The first objective of this study was to develop a microbial growth model that comprehensively considers the effects of temperature, pressure, and salinity through the Arrhenius equation. Experimental results were used to verify the proposed model, and comparisons with previously developed models were made. Oil recovery was compared between the models with and without reflecting the environmental impact to accurately predict the MEOR result. Optimization was performed to maximize oil recovery. This process employed a response surface methodology, and the injection scenarios were set as design parameters. In addition, the sensitivity of each parameter to oil recovery was analyzed. Finally, the MEOR results for the actual heterogeneous reservoir were shown to represent the applicability of the technology.
2. Methods
2.1. Fluid Flow in Permeable Media
In this study, the multiphase and multicomponent flow in permeable media is calculated through the conservation equations as follows [56]:
Here, subscript is the component, subscript is the phase, subscript is the stationary phase, is the number of phases, is the number of components, is the porosity, is the saturation, is the mass fraction, is the superficial velocity, is the dispersion tensor, and is the kinetic reaction rate. The first term on the left side is the accumulation, the second term is the flux, and the right side is the source term.
Under spatially discretized conditions, CMG STARS used in this study calculates the conservation equation of flowing component as follows:
Here, subscripts , , , are water phase, oil phase, gas phase, and layer k, respectively. The , , are mole fractions of component i in water phase, oil phase, and gas phase, respectively. is the number of neighboring regions or grid block faces, is the transmissibility, is the potential, is the component dispersibility, and is the phase rate in layer k.
2.2. Stoichiometric Equations
The model bacteria used in this study were Leuconostoc mesenterioides. They generate a polysaccharidic biopolymer known as dextran under sucrose-rich conditions. Their metabolic mechanisms are well known, and modelling in this study was performed based on experimental results [57,58]. Reactions that happen during microbial metabolism were divided into four steps: (1) microbial growth, (2) sucrose hydrolysis, (3) dextran production, and (4) microbial decay.
The stoichiometric equation for microbial growth is described as follows [52]:
Here, is sucrose, is the model bacteria, is fructose, is lactate, is acetate, is mannitol, and is ethanol.
The sucrose is hydrolyzed into glucose and fructose by microbial enzymatic reactions or is converted into dextran by the linking of glucosidic chains. The hydrolysis reaction is as follows:
and the dextran formation is as follows [59]:
The molecular weight of dextran measured in this study was 10,053 lb/mole, and based on this, was assumed to be 6.2.
To account for the microbial decay reaction, we followed Bültemeier et al. [60], who assumed that the dead microbes did not participate in the chemical reactions but were changed to water as follows:
2.3. Microbial Reaction Rate
In this study, the rate of microbial reactions was described using the Arrhenius equation. It is mainly used in chemical reactions instead of the Monod equation, which is widely used in environmental engineering. By adding a division factor to the general Arrhenius equation, the role of sucrose as a limiting factor is described as follows:
Here, is reaction rate, is the frequency factor, is the division factor, is the activation energy, is the gas constant, is temperature, is the number of components, is the concentration of component , is the sucrose concentration, and and are constants.
The microbial decay rate is expressed as a function related to the frequency factor and the microbial population as follows:
where is the reaction rate for bacterial decay.
2.4. Environmental Factor Effects on Reaction Rate
2.4.1. Temperature
The cardinal temperature model proposed by Rosso et al. [61] is expressed as follows:
Here, is the reaction rate as a function of temperature, is optimum reaction rate, is the optimum temperature for microbial growth, is the upper limit for growth temperature, and is the lower limit for growth temperature.
This temperature model is described by modifying the exponential term of the Arrhenius equation as follows [62]:
where is the constant of the j-th step, is the activation energy of the j-th step, and is the temperature of the j-th step. The final form of the equation reflecting the temperature effect is given as:
2.4.2. Pressure
Basak et al. [35] developed a decay model of L. mesenteroides as a function of pressure. The population of microbes followed first-order kinetics during the pressure-hold time as follows:
where is the initial number of microbes, is the number of surviving microbes, is the rate constant, and is the pressure-hold time. The time taken to reduce the number of microbes to one tenth is defined as a decimal reduction time (), and Equation (13) is as . The relationship between pressure and decimal time is as follows:
where is the decimal time of the j-th step, is the pressure of the j-th step, and is the negative reciprocal slope of vs. . Experiments showed that and have a linear relationship. Through this relationship, in Equation (13) can be estimated and microbial destruction due to pressure can be calculated.
In this study, the frequency factor in Equation (7) is determined as a constant by pressure as follows:
where is the reaction rate as affected by pressure, and is the frequency factor for each pressure step. is then used as a matching parameter for history match, and the value for each pressure step is derived.
2.4.3. Salinity
Leroi et al. [63] presented the reaction rate as a function of NaCl concentration based on the cardinal model as follows:
where is the reaction rate as a function of NaCl concentration, is NaCl concentration, is the optimum NaCl concentration for microbial growth, and is the upper limit of NaCl concentration for microbial growth. Since the optimal NaCl concentration of the model bacteria was close to zero [64,65,66], Equation (16) can be defined as follows:
In this study, the growth inhibition by NaCl is described as the backward reaction of Equation (2), and the rate is expressed as follows:
where is the backward reaction rate by NaCl, and is a constant.
2.5. Permeability Reduction Model
The generated dextran is insoluble, and it is adsorbed or trapped in the pores, which lowers the permeability of the reservoir. In this study, dextran was assumed to be a solid phase, and the change in pore volume caused by the produced dextran affected the change in permeability as follows:
where is the changed porosity, is the initial porosity, is the concentration of the solid phase in pore space, is the density of the solid phase, is the changed permeability, is the initial permeability, and is the multiplier factor. represents the volume of the solid phase in the pore space. When the volume occupied by the solid phase in the pores increases, the porosity decreases, which leads to a decrease in permeability.
2.6. History Matching Error
The history matching error means the relative difference between the measured data and simulation results for each objective function. The matching error can be estimated as follows [62]:
Here, is the history matching error, and are time and elapsed time, respectively, is the simulated result, is the measured result, and is the normalization scale. In this study, the normalization scale is the maximum of the following quantities:
where is the measured maximum change, and is the measurement error. The value of means that if the simulated result is between (measured value − ) and (measured value + ), the result of this matching is satisfactory. That is, is half the absolute error range.
2.7. Proxy Model for Sensitivity Analysis and Optimization
In this study, an optimization process was performed using the response surface methodology (RSM) to maximize the oil recovery. The RSM is a method to search the correlation between input parameter and response (objective function). The main idea is to use a proxy model to represent the original simulation result. Linear or quadratic form is mainly used to make a proxy model, and the latter is applied in this study. This proxy model is also used to analyze the sensitivity of each parameter to the objective function. A quadratic polynomial method was applied to build a proxy model. The quadratic model is expressed as follows [62,67]:
Here, is the objective function (response), expresses the linear effect of parameters, are the quadratic effects of the parameters, indicate the interaction effects of parameters, is the intercept, are the coefficients of linear terms, are the coefficients of quadratic terms, and are the coefficients of interaction terms.
The sensitivity indicates how much a parameter change affects the objective function change. The greater the value of coefficient (effect estimate), the greater the sensitivity of the parameter to the objective function. At this time, each parameter must have scale invariance. All parameters have an average value of zero and are set to vary from −1 to 1.
3. Results
3.1. Batch and Sandpack Model Simulation
The results of microbial growth, dextran formation, and the subsequent decrease in permeability were taken from previous studies [54,58]. Verification of the microbial growth and dextran production was performed through the batch model shown in Table A1. Verification of the permeability reduction was performed using the sandpack simulation shown in Table A2. The batch simulation was performed under the conditions of 1 atm and 30 °C for a total volume of 1 L. The initial sucrose concentration in the batch model was 15 g/L, and the growth was carried out for 24 h. In the sandpack simulation, sucrose injection and soaking were conducted 20 times for 44 h. The concentration of the injection was 15 g/L, and the flow rate was 0.018 bbl/day. The detailed process of the simulations was referenced from Jeong et al. [54].
The reaction rates of the four types of microbial metabolism are calculated using Equations (6) and (7), and the history matching for the experiment was performed with matching parameters , , and . The values of the variables minimizing the matching error are shown in Table 1. Figure 1a,b describes the microbial growth and dextran production, respectively. The pressure difference measured in the pressure measurements shown in Table A2 is shown in Figure 1c, and the simulation results are matched with the experimental data. The matching parameter in this simulation was in Equation (21), and it was calculated to be 244.8. The history match errors measured in the batch model and sandpack model are 1.6% and 4.6%, respectively.

Table 1.
Batch model for bacterial growth and dextran generation.
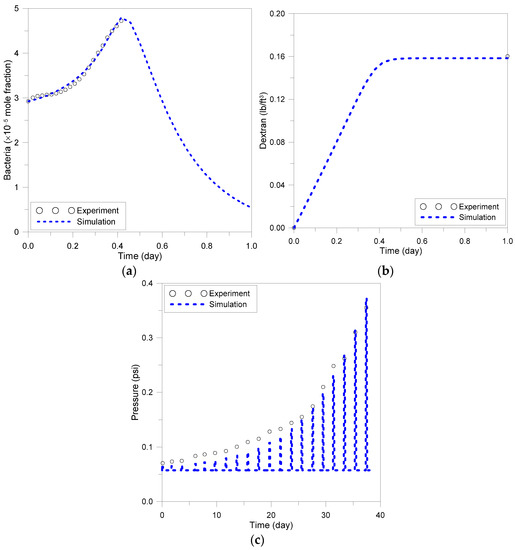
Figure 1.
Results of batch and sandpack model simulation: (a) bacterial growth, (b) dextran generation, (c) pressure increase due to decrease in permeability.
3.2. Verification of Models Reflecting Environmental Factors
The previous batch model was simulated under constant temperature, pressure, and salinity conditions. Comparisons with existing methods were conducted after applying the proposed environmental factor reflection model to this model. The reaction rate calculated by the proposed model was compared with the result calculated by the cardinal model, and the results are depicted in Figure 2a. Microbial growth temperature conditions for calculating Equation (9) were referenced in previous studies [55]. It was shown that in Equation (10) could accurately calculate the of the cardinal model. Figure 2b shows the result of calculating the microbial decay as a function of the pressure step using Equations (13) and (14). This graph shows the microbial decay rate according to the pressure used in this study, and the same result is calculated with Equation (15). Microbial growth behavior as a function of NaCl concentration is depicted in Figure 2c. As shown, the salinity effect on bacterial growth was described using Equation (19). Like the existing model, the proposed model was able to derive results that well reflected microbial growth and decay reactions according to temperature, pressure, and salinity.
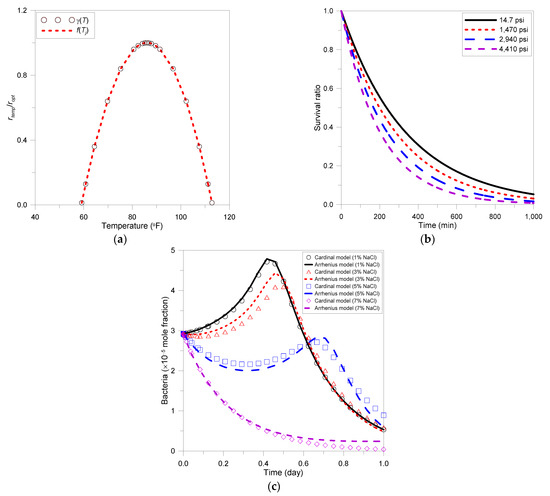
Figure 2.
Results of environmental factor reflection model: (a) temperature effect, (b) pressure effect, (c) salinity effect.
3.3. Optimization of MEOR
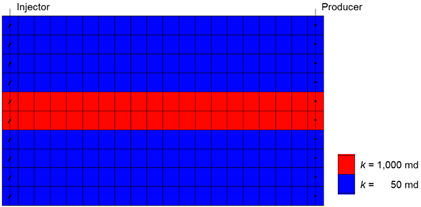
The conditions of the reservoir used for the MEOR process are indicated in Table A3. The reservoir had a high probability of a conformance problem because of the high transmittance channels. Production was assumed to continue for three years. After implementing waterflooding during the first year, MEOR treatment was performed for eight weeks, and then waterflooding was implemented again. The model that did not reflect the environmental impact was named the “ideal case”, and the model accounting for the environmental impact was named the “real case”. Before the optimization, the injection rate in both cases was 50 bbl/day, and the sucrose concentration was 0.005 (mole fraction). In the real case, the temperature of the injected water and the NaCl concentration were set to 75 °F and 1%, respectively. These no-optimization models were used as a base case for each case. As shown in Figure 3, the model considering environmental factors tended to decrease the productivity of oil by 21% compared to the ideal case. Nevertheless, it could be seen that the oil recovery of MEOR in the real case was 25% higher than that of waterflooding.
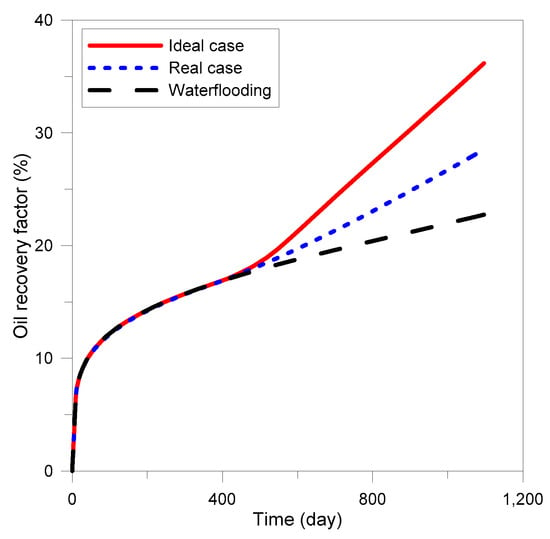
Figure 3.
Oil recovery prediction results for the ideal case and real case.
To maximize oil recovery, the ideal and real case injection scenarios were optimized using a proxy model created by the RSM. The proxy model set the oil recovery as the objective function and calculated design parameters that maximize the oil recovery. The design parameters for optimum injection included sucrose concentration, treatment period, injection rate, temperature, and salinity. The detailed ranges of all parameters and the optimum designs calculated by the proxy model are listed in Table 2. The optimal injection design differs in the two cases, especially in sucrose concentration and injection temperature. In addition, the optimum temperature for injection was measured somewhat lower than the optimum temperature for microbial growth, i.e., 86 °F [55]. The influence of each design parameter on oil recovery could be explained based on the sensitivity analysis.

Table 2.
Range of design parameters and optimization results.
Figure 4 shows a tornado plot resulting from the sensitivity analysis of the design parameters. The values in figures called effect estimates are the expected change of the oil recovery when the scaled parameters travel from −1 to 1. In other words, the larger these values are, the greater the influence on oil recovery change. The sensitivity analysis showed that there were differences in the priority of parameters affecting oil recovery in both cases. In both cases, the quadratic effect of the treatment period had the greatest effect on recovery. However, the quadratic effect of injection temperature had the second largest effect on recovery in the real case. The maximum oil production predicted through this optimization is shown in Figure 5. The maximum oil recovery for the ideal case was 48.3%, which was a 33% increase over the no-optimized model. The recovery of the optimized real case was 41.8%, which was a 47% increase over the no-optimized model. These results showed that it was important to consider environmental effects for the accurate prediction and optimization of productivity. The actual proxy models used for optimization are the same as Equations (A1) and (A2).
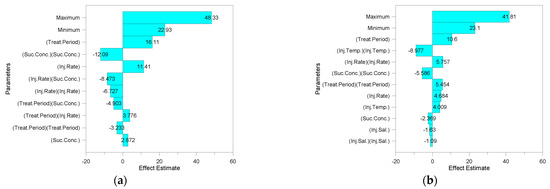
Figure 4.
Sensitivity analysis for microbial enhanced oil recovery (MEOR): (a) ideal case and (b) real case.
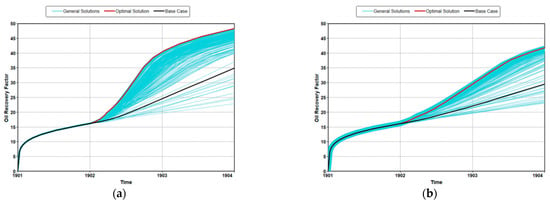
Figure 5.
Optimization results for oil recovery: (a) ideal case and (b) real case.
3.4. Field Application
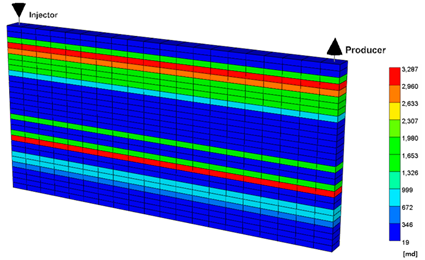
To evaluate the effectiveness of MEOR through the developed model, the process was applied to the highly-heterogeneous actual field model based on logging data from King Island [53]. The reservoir conditions are shown in Table 3. Since the reservoir depth was approximately 5000 ft, the reservoir pressure was assumed to be 2500 psi, which was calculated using a typical pressure gradient of 0.5 psi/ft. The reservoir pressure was also assumed to be 100 °F. Waterflooding and MEOR treatment were applied in the same way as the previous models, and the injection temperature and salinity were 80 °F and 1% NaCl, respectively. Figure 6 represents the influence of the MEOR process on the oil recovery and water-oil ratio (WOR). The recovery factor of MEOR was 36.4%, which was improved by 2.6% compared with waterflooding. WOR was also reduced by the MEOR, which was decreased by 11%. The distribution of dextran produced in the reservoir and the resulting permeability reduction in the highly permeable zone are shown in Figure 7. Most of the injected fluid flowed through the highly permeable layers, and it was found that most of the dextran was generated near the injection well. At this time, even if dextran was formed in the high permeability layers as shown in Figure 7a, the effect of reducing the permeability was not very large, as shown in Figure 7b.

Table 3.
Initial conditions of the King Island reservoir.
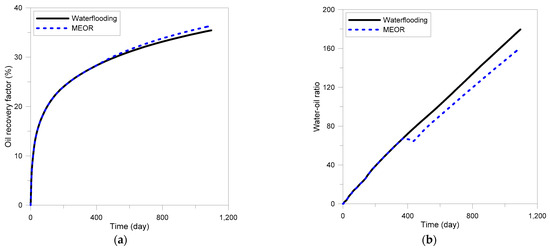
Figure 6.
Improvements of recovery efficiency by MEOR in field applications: (a) recovery factor, (b) water-oil ratio.
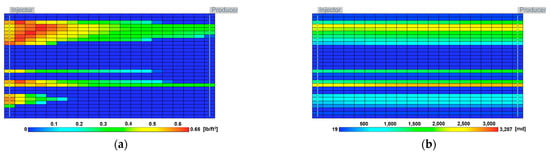
Figure 7.
Distributions of (a) dextran and (b) permeability of the field case at the end of the simulation.
Particularly in layered systems, the results of 2D and 3D models will not show a significant difference. However, some differences may occur due to the curvilinear flowline in the 3D model. To confirm this, the extension of the 2D model to the 3D heterogeneous model was shown in Figure 8a. Though the average permeability of each layer was the same as in the previous 2D model, the permeability distributions in the horizontal direction were geostatistically generated to have a Dykstra–Parsons coefficient of 0.4 (Figure 8b). This coefficient can be calculated as follows [68]:
where is the Dykstra–Parsons coefficient, is the mean permeability at 50% probability, and is the permeability at 84.1% probability. This coefficient is the most widely used to describe permeability variability in the petroleum industry. When it is zero, it means a homogeneous reservoir, and the closer to 1, the higher the heterogeneity.
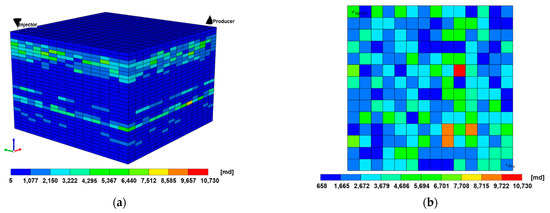
Figure 8.
(a) 3D reservoir model and (b) areal heterogeneity of the fourth layer.
The injection conditions were also the same as the previous model, except that the injection rate increased by 9.8 times to 490 bbl/day in consideration of the increase in the reservoir volume. The results are shown in Figure 9. The oil recovery increased 3.2% in MEOR compared to waterflooding (Figure 9a). This result explained that when the reservoir was heterogeneous even in the horizontal direction, the effect of MEOR also increased compared to waterflooding. In addition, it was confirmed that a lot of dextran was generated in the vicinity of the injector in high permeable zones (Figure 9b). There was a slight difference between 2D and 3D results. Since the overall trend of the results was the same, subsequent optimization analysis was conducted on a 2D layered model that was advantageous in terms of computation time.
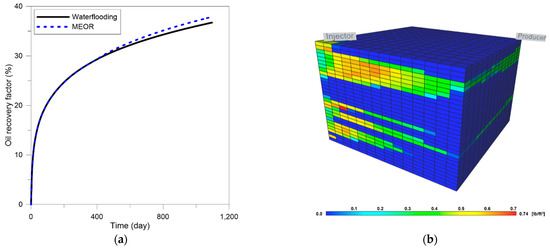
Figure 9.
Results for 3D model: (a) oil recovery factor, (b) dextran generation.
The optimization process was also applied to maximize oil recovery in this field. The range of design parameters for optimal injection and results are listed in Table 4 (the proxy model used is shown in Equation (A3)). Optimization increased the oil recovery to 37.8%, which was a 7% increase over waterflooding (Figure 10). Figure 11a shows the permeability reduction due to the use of MEOR. The permeability reduction in the high permeability zone was up to 18% in the no-optimization model and up to 41% in the optimized model. The oil saturation distribution of each layer is depicted in Figure 11b. Comparing the waterflooding and optimized models, oil saturation was reduced by up to 16% in some high permeability layers and up to 10% in some low permeability layers. Due to very large permeability contrast, the permeability of the high permeable zones was still larger than that of the low permeable zones even after plugging. This explains why the oil in the low permeable region was not swept sufficiently.

Table 4.
Range of design parameters and optimization results for actual field.
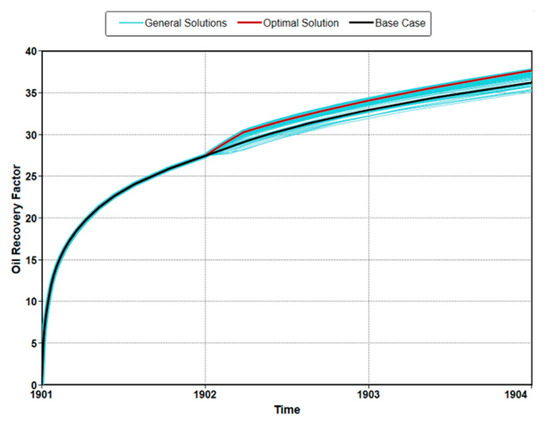
Figure 10.
Oil recovery improvement by optimization.
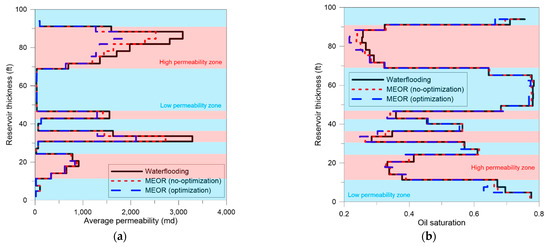
Figure 11.
Distributions of (a) permeability and (b) oil saturation at the end of simulations.
4. Conclusions
In this study, a new MEOR model incorporating environmental influences was developed to describe microbial metabolism in actual field conditions. An MEOR optimization method was discussed based on this model. The existing microbial growth models for temperature, pressure, and salinity were integrated using the Arrhenius method. To verify the proposed method, the batch model and sandpack model were employed, and the environmental impacts on microbial growth were successfully accounted for using the proposed method. The importance of these environmental models was that they could accurately analyze MEOR productivity. Based on our analysis of ideal and real cases for the hypothetical reservoir, the productivity of MEOR was overestimated when the environmental impact was not considered. Additionally, a sensitivity analysis showed that the temperature effect had a great influence on the oil recovery in a real case. In particular, it was found that the injection temperature should consider both the reservoir temperature and the optimum temperature for microbial growth. Optimization showed that the optimal design of nutrient injection could further increase oil recovery. The King Island model, which was an actual reservoir, also showed that it was possible to predict the improved oil recovery accurately using the MEOR method that reflected environmental factors. This study showed that the MEOR analysis could be performed more accurately through the proposed method, and optimization for improved recovery efficiency was also essential.
Author Contributions
Conceptualization, M.S.J. and K.S.L.; methodology, M.S.J. and K.S.L.; software, M.S.J.; validation, M.S.J. and K.S.L.; formal analysis, M.S.J.; investigation, M.S.J., Y.W.L. and H.S.L.; resources, M.S.J. and K.S.L.; data curation, M.S.J.; writing—original draft preparation, M.S.J.; writing—review and editing, M.S.J. and K.S.L.; visualization, M.S.J., Y.W.L. and H.S.L.; supervision, K.S.L.; project administration, K.S.L.; funding acquisition, K.S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Korea Institute of Energy Technology Evaluation and Planning (KETEP) grant funded by the Korea Government (MOTIE) (No. 20208550000040). This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2020R1F1A1070406).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Nomenclature
| A | constant | a0 | intercept |
| aij | coefficients of interaction terms | Aj | constant of the j-th step |
| aj | coefficients of linear terms | ajj | coefficients of quadratic terms |
| B | constant | ci | concentration of component i |
| cNaCl | NaCl concentration | upper limit of NaCl concentration for microbial growth | |
| optimum NaCl concentration for microbial growth | cs | concentration of the solid phase | |
| csucrose | sucrose concentration | D | decimal reduction time |
| Dj | decimal time of the j-th step | Dji | component dispersibility (j = w, o, g) |
| E | history matching error | Ea | activation energy |
| Ea,j | activation energy of the j-th step | Fdiv | division factor |
| FFreq | frequency factor | FFreq,p | frequency factor for each pressure step |
| K | transmissibility | dispersion tensor | |
| k | changed permeability | k50 | mean permeability at 50% probability |
| k81.4 | permeability at 84.1% probability | kmul | multiplier factor |
| ko | initial permeability | N | number of surviving microbes |
| n | constant | NC | number of components (or ) |
| nf | number of neighboring regions or grid block faces | No | initial number of microbes |
| NP | number of phases | Merr | measurement error |
| mi | mass fraction of component i | pj | pressure of the j-th step |
| qik | phase rates in layer k (j = w, o, g) | R | gas constant |
| r | kinetic reaction rate | rdecay | reaction rate for bacterial decay |
| rdecay, p | reaction rate as affected by pressure | rNaCl | reaction rate as a function of NaCl |
| rNaCl,backward | backward reaction rate by NaCl | ropt | optimum reaction rate |
| rtemp | reaction rate as a function of temperature | S | saturation |
| s | normalization scale | T | temperature |
| t | time | telap | elapsed time |
| tp | pressure-hold time | Tj | temperature of the j-th step |
| Tmax | upper limit for growth temperature | Tmin | lower limit for growth temperature |
| Topt | optimum temperature for microbial growth | superficial velocity | |
| VDP | Dykstra–Parsons coefficient | Vf | volume of fluid phase |
| wi | mole fraction of component i in aqueous phase | xi | mole fraction of component i in oil phase |
| xixj | interaction effects of parameters | xj | linear effect of parameters |
| quadratic effects of the parameters | y | objective function | |
| yi | mole fraction of component i in gas phase | ΔΥm | measured maximum change |
| measured result | simulated result | ||
| zp | negative reciprocal slope of log D vs. p | ||
| Subscript | |||
| g | gas phase | k | layer k |
| o | oil phase | s | solid phase |
| w | water phase | ||
| Greek | |||
| α | rate constant | ρ | density |
| Φ | potential | ɸ | porosity |
| ɸo | initial porosity | ||
Appendix A

Table A1.
Batch model for bacterial growth and dextran generation [54].
Table A1.
Batch model for bacterial growth and dextran generation [54].
 | Growth Media Composition (Mole Fraction) | |
| Sucrose | 0.000789 | |
| NH4+ | 0.000133 | |
| HCO3− | 0.000219 | |
| H2O | 0.99883 | |
| Microbes | 2.92 × 10−5 | |

Table A2.
Sandpack model for permeability reduction [54].
Table A2.
Sandpack model for permeability reduction [54].
 | Model Conditions | |
| Size (mm3) | 62.1 × 62.1 × 160 | |
| Initial Porosity | 0.38 | |
| Initial Permeability (md) | 4150 | |
| Injector Location | 1, 1, 20 (bottom block) | |
| Producer Location | 1, 1, 1 (top block) | |
| Pressure Measurement | (1, 1, 8) and (1, 1, 13) | |

Table A3.
Description of the initial conditions of the target reservoir [54].
Table A3.
Description of the initial conditions of the target reservoir [54].
| Permeability Distribution | ||
|---|---|---|
 | ||
| Initial Conditions | ||
| Size (ft3) | 200 × 10 × 100 | |
| Grid | 20 × 1 × 10 | |
| Temperature (°F) | 100 | |
| Pressure (psi) | 4000 | |
| NaCl concentration (%) | 10 | |
| Permeability (md) | High-k zone | 1000 |
| Low-k zone | 50 | |
| Porosity | 0.25 | |
| Saturation | Oil | 0.8 |
| Water | 0.2 | |
| Relative Permeability: Brooks–Corey Model | ||
| Connate water saturation | 0.3 | |
| Irreducible oil saturation | 0.2 | |
| krw at irreducible oil | 0.5 | |
| kro at connate water | 0.8 | |
| Exponent for krw | 2 | |
| Exponent for kro | 2 | |
Appendix B
The proxy model for ideal case:
The proxy model for real case:
The proxy model for King Island:
References
- Davarpanah, A. A feasible visual investigation for associative foam>/ polymer injectivity performances in the oil recovery enhancement. Eur. Polym. J. 2018, 105, 405–411. [Google Scholar] [CrossRef]
- Davarpanah, A.; Mirshekari, B. Experimental investigation and mathematical modeling of gas diffusivity by carbon dioxide and methane kinetic adsorption. Ind. Eng. Chem. Res. 2019, 58, 12392–12400. [Google Scholar] [CrossRef]
- Davarpanah, A.; Mirshekari, B. Numerical simulation and laboratory evaluation of alkali-surfactant-polymer and foam flooding. Int. J. Environ. Sci. Technol. 2019, 17, 1123–1136. [Google Scholar] [CrossRef]
- Davarpanah, A. Parametric study of polymer-nanoparticles-assisted injectivity performance for axisymmetric two-phase flow in EOR processes. Nanomaterials 2020, 10, 1818. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Xie, J.; Cai, W.; Wang, R.; Davarpanah, A. Thermodynamic effects of cycling carbon dioxide injectivity in shale reservoirs. J. Pet. Sci. Eng. 2020, 195, 107717. [Google Scholar] [CrossRef]
- Pan, F.; Xhang, Z.; Zhang, X.; Davarpanah, A. Impact of anionic and cationic surfactants interfacial tension on the oil recovery enhancement. Powder Technol. 2020, 373, 93–98. [Google Scholar] [CrossRef]
- Hu, Y.; Cheng, Q.; Yang, J.; Zhang, L.; Davarpanah, A. A laboratory approach on the hybrid-enhanced oil recovery techniques with different saline brines in sandstone reservoirs. Processes 2020, 8, 1051. [Google Scholar] [CrossRef]
- Belyaev, S.S.; Borzenkov, I.A.; Nazina, T.N.; Rozanova, E.P.; Glumov, I.F.; Ibatullin, R.R.; Ivanov, M.V. Use of microorganisms in the biotechnology for the enhancement of oil recovery. Microbiology 2004, 73, 590–598. [Google Scholar] [CrossRef]
- Lazar, I.; Petrisor, I.G.; Yen, T.F. Microbial enhanced oil recovery (MEOR). Pet. Sci. Technol. 2007, 25, 1353–1366. [Google Scholar] [CrossRef]
- Guo, H.; Li, Y.; Yiran, Z.; Wang, F.; Wang, Y.; Yu, Z.; Haicheng, S.; Yuanyuan, G.; Chuyi, J.; Xian, G. Progress of Microbial Enhanced Oil Recovery in China. In Proceedings of the SPE Asia Pacific Enhanced Oil Recovery Conference, Kuala Lumpur, Malaysia, 11–13 August 2015. [Google Scholar]
- Jenneman, G.E.; Moffitt, P.D.; Young, G.R. Application of a microbial selective-plugging process at the North Burbank Unit: Prepilot tests. SPE Prod. Facil. 1996, 11, 11–17. [Google Scholar] [CrossRef]
- Brown, L.R.; Vadie, A.A.; Stephens, J.O. Slowing production decline and extending the economic life of an oil field: New MEOR technology. SPE Reserv. Eval. Eng. 2002, 5, 33–41. [Google Scholar] [CrossRef]
- Cusack, F.M.; Singh, S.; Novosad, J.; Chmilar, M.; Blenknsopp, S.A.; Costerton, J.W. The Use of Ultramicrobacteria for Selective Plugging in Oil Recovery by Waterflooding. In Proceedings of the International Meeting on Petroleum Engineering, Beijing, China, 24–27 March 1992. [Google Scholar]
- Jack, T.R.; Stehmeier, L.G.; Islam, M.R.; Ferris, F.G. Microbial selective plugging to control water channeling. In Microbial Enhancement of Oil Recovery–Recent Advances; Donaldson, E.C., Ed.; Elsevier: Amsterdam, The Netherland, 1991; pp. 433–440. [Google Scholar]
- Hitzman, D.O. Review of Microbial Enhanced Oil Recovery Field Tests. In Proceedings of the Symposium on Applications of Microorganisms to Petroleum Technology, Bartlesville, OK, USA, 12–13 August 1987. [Google Scholar]
- Patel, J.; Borgohain, S.; Kumar, M.; Rangarajan, V.; Somasundaran, P.; Sen, R. Recent developments in microbial enhanced oil recovery. Renew. Sustain. Energy Rev. 2015, 52, 1539–1558. [Google Scholar] [CrossRef]
- Safdel, M.; Anbaz, M.A.; Daryasafar, A.; Jamialahmadi, M. Microbial enhanced oil recovery, a critical review on worldwide implemented field trials in different countries. Renew. Sustain. Energy Rev. 2017, 74, 159–172. [Google Scholar] [CrossRef]
- Wang, J.; Xu, H.; Guo, S. Isolation and characteristics of a microbial consortium for effectively degrading phenanthrene. Pet. Sci. 2007, 4, 68–75. [Google Scholar] [CrossRef]
- Sen, R. Biotechnology in petroleum recovery: The microbial EOR. Prog. Energy Combust. Sci. 2008, 34, 714–724. [Google Scholar] [CrossRef]
- Darvishi, P.; Ayatollahi, S.; Mowla, D.; Niazi, A. Biosurfactant production under extreme environmental conditions by efficient microbial consortium, ERCPPI-2. Colloids Surf. B Biointerfaces 2011, 84, 292–300. [Google Scholar] [CrossRef]
- Stanier, R.Y.; Doudoroff, M.; Adelberg, E.A. The Microbial World, 3rd ed.; Prentice-Hall: Englewood Cliffs, NJ, USA, 1970. [Google Scholar]
- Brock, T.D.; Brock, K.M.; Belly, R.T.; Weiss, R.L. Sulfolobus: A new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Microbiol. 1972, 84, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Brock, T.D. Thermophilic Microorganisms and Life at High Temperatures; Springer: New York, NY, USA, 1978. [Google Scholar]
- Corliss, J.B.; Dymond, J.; Gordon, L.I.; Edmond, J.M.; von Herzen, R.P.; Ballard, R.D.; Green, K.; Williams, D.; Bainbridge, A.; Crane, K.; et al. Submarine thermal springs on the Galápagos Rift. Science 1979, 203, 1073–1083. [Google Scholar] [CrossRef]
- Karl, D.M.; Wirsen, C.O.; Jannasch, H.W. Deep-sea primary production at the Galápagos hydrothermal vents. Science 1980, 207, 1345–1347. [Google Scholar] [CrossRef]
- Jannash, H.W.; Wirsen, C.O. Morphological survey of microbial mats near deep-sea thermal vents. Appl. Environ. Microbiol. 1981, 41, 528–538. [Google Scholar] [CrossRef]
- Baross, J.A.; Liley, M.D.; Gordon, L.I. Is the CH4, H2 and CO venting from submarine hydrothermal systems produced by thermophilic bacteria? Nature 1982, 298, 366–368. [Google Scholar] [CrossRef]
- Yayanos, A.A.; Dietz, A.S. Thermal inactivation of a deep-sea barophilic bacterium, isolate CNPT-3. Appl. Environ. Microbiol. 1982, 43, 1481–1489. [Google Scholar] [CrossRef]
- Leigh, J.A.; Jones, J.W. A New Extremely Thermophilic Methanogen from a Submarine Hydrothermal Vent. In Proceedings of the 83rd Annual Meeting of the American Society for Microbiology, New Orleans, LA, USA, 6–11 March 1983. [Google Scholar]
- Villadsen, J.; Nielsen, J.; Lidén, G. Bioreaction Engineering Principles; Springer Science and Business Media: Berlin, Germany, 2011. [Google Scholar]
- Zobell, C.E. Pressure effects on morphology and life processes of bacteria. In High Pressure Effects on Cellular Processes; Zimmerman, A.M., Ed.; Academic Press: New York, NY, USA, 1970; pp. 85–130. [Google Scholar]
- Morita, R.Y. Pressure: Bacteria, fungi and blue-green algae. In Marine Ecology; Kinne, Ed.; Wiley: New York, NY, USA, 1972; pp. 1361–1388. [Google Scholar]
- Marquis, R.E.; Matsumura, P. Microbial life under pressure. In Microbial Life in Extreme Environments; Kushner, D.J., Ed.; Academic Press: London, UK, 1978; pp. 105–158. [Google Scholar]
- Marquis, R.E. Barobiology of Deep Oil Formations. In Proceedings of the International Conference on Microbial Enhancement of Oil Recovery, Afton, OK, USA, 16 May 1983. [Google Scholar]
- Basak, S.; Ramaswamy, H.S.; Piette, J.P.G. High pressure destruction kinetics of Leuconostoc mesenteroides and Saccharomyces cerevisiae in single strength and concentrated orange juice. Innov. Food Sci. Emerg. Technol. 2002, 3, 223–231. [Google Scholar] [CrossRef]
- Stanley, S.O.; Morita, R.Y. Salinity effect on the maximal growth temperature of some bacteria isolated from marine environments. J. Bacteriol. 1968, 95, 169–173. [Google Scholar] [CrossRef]
- Bilsky, A.Z.; Armstrong, J.B. Osmotic reversal of temperature sensitivity in Escherichia coli. J. Bacteriol. 1973, 113, 76–81. [Google Scholar] [CrossRef]
- Boyer, E.W.; Ingle, M.B.; Mercer, G.D. Bacillus alcalophilus subsp. halodurans subsp. nov.: An alkaline-amylase-producing, alkalophilic organism. Int. J. Syst. Bacteriol. 1973, 23, 238–242. [Google Scholar]
- Novitsky, T.J.; Kushner, D.J. Influences of temperature and salt concentration on the growth of a facultative halophilic “Micrococcus” sp. Can. J. Microbiol. 1975, 21, 107–110. [Google Scholar] [CrossRef]
- Kushner, D.J. Life in high salt and solute concentrations: Halophilic bacteria. In Microbial Life in Extreme Environments; Kushner, D.J., Ed.; Academic Press: London, UK, 1978; pp. 317–368. [Google Scholar]
- Belyaev, S.S.; Wolkin, R.; Kenealy, W.R.; DeNiro, M.J.; Epstein, S.; Zeikus, J.G. Methanogenic bacteria from the Bondyuzhskoe oil field: General characterization and analysis of stable-carbon isotopic fractionation. Appl. Environ. Microbiol. 1983, 45, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Paterek, J.R.; Smith, P.H. Isolation of a Halophilic Methanogenic Bacterium from the Sediments of Great Salt Lake and a San Francisco Bay Saltern. In Proceedings of the 83rd Annual Meeting of the American Society for Microbiology, New Orleans, LA, USA, 6–11 March 1983. [Google Scholar]
- Yu, I.K.; Hungate, R.E. Isolation and Characterization of an Obligately Halophilic Methanogenic Bacterium. In Proceedings of the 83rd Annual Meeting of the American Society for Microbiology, New Orleans, LA, USA, 6–11 March 1983. [Google Scholar]
- Park, C.; Raines, R.T. Quantitative analysis of the effect of salt concentration on enzymatic catalysis. J. Am. Chem. Soc. 2001, 123, 11472–11479. [Google Scholar] [CrossRef]
- Gran, K.; Bjørlykke, K.; Aagaard, P. Fluid salinity and dynamics in the North Sea and Haltenbanken Basins derived from well log data. In Geological Applications of Wireline Logs II; Hurst, A., Griffiths, C.M., Worthington, P.F., Eds.; Geological Society: London, UK, 1992; pp. 327–338. [Google Scholar]
- Maudgalya, S.; Knapp, R.M.; McInerney, M. Microbially Enhanced Oil Recovery Technologies: A Review of the Past, Present and Future. In Proceedings of the Production and Operations Symposium, Oklahoma City, OK, USA, 31 March–3 April 2007. [Google Scholar]
- Islam, M.R. Mathematical Modeling of Microbial Enhanced Oil Recovery. In Proceedings of the SPE Annual Technical Conference and Exhibition, New Orleans, LA, USA, 23–26 September 1990. [Google Scholar]
- Chang, M.M.; Chung, F.T.H.; Bryant, R.S.; Gao, H.W.; Burchfield, T.E. Modeling and Laboratory Investigation of Microbial Transport Phenomena in Porous Media. In Proceedings of the SPE Annual Technical Conference and Exhibition, Dallas, TX, USA, 6–9 October 1991. [Google Scholar]
- Zhang, X.; Knapp, R.M.; McInerney, M.J. A Mathematical Model for Microbially Enhanced Oil Recovery Process. In Proceedings of the SPE/DOE Enhanced Oil Recovery Symposium, Tulsa, OK, USA, 22–24 April 1992. [Google Scholar]
- Delshad, M.; Asakawa, K.; Pope, G.A.; Sepehrnoori, K. Simulations of Chemical and Microbial Enhanced Oil Recovery Methods. In Proceedings of the SPE/DOE Improved Oil Recovery Symposium, Tulsa, OK, USA, 13–17 April 2002. [Google Scholar]
- Stewart, T.L.; Kim, D. Modeling of biomass-plug development and propagation in porous media. Biochem. Eng. J. 2004, 17, 107–119. [Google Scholar] [CrossRef]
- Vilcáez, J.; Li, L.; Wu, D.; Hubbard, S.S. Reactive transport modeling of induced selective plugging by Leuconostoc mesenteroides in carbonate formations. Geomicrobiol. J. 2013, 30, 813–828. [Google Scholar] [CrossRef]
- Surasani, V.K.; Li, L.; Ajo-Franklin, J.B.; Hubbard, C.; Hubbard, S.S.; Wu, Y. Bioclogging and permeability alteration by L. mesenteroides in a sandstone reservoir: A reactive transport modeling study. Energy Fuels 2013, 27, 6538–6551. [Google Scholar] [CrossRef]
- Jeong, M.S.; Noh, D.; Hong, E.; Lee, K.S.; Kwon, T. Systematic modeling approach to selective plugging using in situ bacterial biopolymer production and its potential for microbial-enhanced oil recovery. Geomicrobiol. J. 2019, 36, 468–481. [Google Scholar] [CrossRef]
- Hong, E.; Jeong, M.S.; Lee, K.S. Optimization of nonisothermal selective plugging with a thermally active biopolymer. J. Pet. Sci. Eng. 2019, 173, 434–446. [Google Scholar] [CrossRef]
- Lake, L.W. Enhanced Oil Recovery; Prentice-Hall: Englewood Cliffs, NJ, USA, 1989. [Google Scholar]
- Kwon, T.; Ajo-Franklin, J.B. High-frequency seismic response during permeability reduction due to biopolymer clogging in unconsolidated porous media. Geophysics 2013, 78, 117–127. [Google Scholar] [CrossRef]
- Noh, D.; Ajo-Franklin, J.B.; Kwon, T.; Muhunthan, B. P and S wave responses of bacterial biopolymer formation in unconsolidated porous media. J. Geophys. Res. Biogeosciences 2016, 121, 1–20. [Google Scholar] [CrossRef]
- Santos, M.; Texeira, J.; Rodrigues, A. Production of dextransucrase, dextran and fructose from sucrose using Leuconostoc mesenteroides NRRL B512(f). Biochem. Eng. J. 2000, 4, 177–188. [Google Scholar] [CrossRef]
- Bültemeier, H.; Alkan, H.; Amro, M. A New Modeling Approach to MEOR Calibrated by Bacterial Growth and Metabolite Curves. In Proceedings of the SPE EOR Conference at Oil and Gas West Asia, Muscat, Oman, 31 March–2 April 2014. [Google Scholar]
- Rosso, L.; Lobry, J.R.; Bajard, S.; Flandrois, J.P. Convenient model to describe the combined effects of temperatures and pH on microbial growth. Appl. Environ. Microbiol. 1995, 61, 610–616. [Google Scholar] [CrossRef]
- CMG (Computer Modelling Group). CMOST User Guide: Version 2017; CMG (Computer Modelling Group): Calgary, AB, Canada, 2017. [Google Scholar]
- Leroi, F.; Fall, P.A.; Pilet, M.F.; Chevalier, F.; Baron, R. Influence of temperature, pH and NaCl concentration on the maximal growth rate of Brochothrix thermosphacta and a bioprotective bacteria Lactococcus piscium CNCM I-4031. Food Microbiol. 2012, 31, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Antunes, A.; Rainey, F.A.; Nobre, F.; Schumann, P.; Margarida Ferreira, A.; Ramos, A.; Santos, H.; da Costa, M.S. Leuconostoc ficulneum sp. nov., a novel lactic acid bacterium isolated from a ripe fig, and reclassification of Lactobacillus fructosus as Leuconostoc fructosum comb. nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 647–655. [Google Scholar] [CrossRef]
- Drosinos, E.H.; Mataragas, M.; Nasis, P.; Galiotou, M.; Metaxopoulos, J. Growth and bacteriocin production kinetics of Leuconostoc mesenteroides E131. J. Appl. Microbiol. 2005, 99, 1314–1323. [Google Scholar] [CrossRef]
- Cardamone, L.; Quiberoni, A.; Mercanti, D.J.; Fornasari, M.E.; Reinheimer, J.A.; Guglielmotti, D.M. Adventitious dairy Leuconostoc strains with interesting technological and biological properties useful for adjunct starters. Dairy Sci. Technol. 2011, 91, 457–470. [Google Scholar] [CrossRef]
- Cioppa, T.M. Efficient Nearly Orthogonal and Space-Filling Experimental Designs for High-Dimensional Complex Models. Ph.D. Thesis, Naval Postgraduate School, Monterey, CA, USA, September 2002. [Google Scholar]
- Peters, E.J. Advanced Petrophysics: Volume 1: Geology, Porosity, Absolute Permeability, Heterogeneity, and Geostatistics; Live Oak Book Company: Austin, TX, USA, 2012. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).