Abstract
Understanding of the metabolic pathways connected with a removal of micropollutant bisphenol A (BPA) may help to better design effective wastewater treatment processes. The aim of this study was to determine changes in gene expression in an aerobic granular sludge (AGS) community exposed to BPA. In the study, AGS adapted to BPA degradation was used. In this sludge, BPA was dosed; as a control sample, granules without BPA addition were used. mRNA was isolated from both samples and sequenced using the Illumina platform. Metatranscriptome analysis of AGS exposed to BPA indicated direct biodegradation as the main mechanism of BPA removal from wastewater. High expression of genes coding pilus and flagellin proteins in the BPA-exposed biomass indicated that exposition to BPA stimulated aggregation of microbial cells and formation of biofilm. In the BPA-exposed biomass, nitrogen was mainly used as an energy source, as indicated by the presence of genes coding nitrification enzymes and urease. Moreover, exposition to BPA stimulated expression of genes coding proteins responsible for xenobiotic degradation, including enzymes responsible for benzoate degradation. These results increase knowledge about BPA metabolism in complex microbial communities in wastewater treatment systems and indicate that AGS is suitable for efficiently removing BPA from wastewater.
1. Introduction
Accelerated urbanization and significant industrial development in recent years have increased the occurrence of micropollutants in aquatic environments. Many wastewater treatment plants (WWTPs) are not adapted for the elimination of those toxic compounds, and as a result, many of these micropollutants pass through the treatment process, enter surface waters, and become a danger to wildlife and a problematic issue for the drinking water industry [1].
Bisphenol A (BPA) is an anthropogenic compound used for the manufacturing of plastic or epoxy and polycarbonate resins. BPA is also commonly used for the production of electronic and electric equipment, layers of metal cans for food and beverages, flame retardants, and many other products [2]. Research by Bolz et al. [3] detected BPA in German surface waters at concentrations up to 0.4 µg/L. Another study showed that in the United States, 41% of examined streams were polluted with BPA in concentrations ranging from 0.14 µg/L to 12 µg/L [4]. Even small BPA concentrations of 1–10 µg/L cause serious toxicity to aquatic organisms—fish, algae, and invertebrates [5]. Matsumura et al. [6] indicated that low BPA concentrations in soil (1 mg BPA/g) do not affect the microbial community, but high BPA concentrations (100 mg/g) significantly suppress microbial activity and growth.
Microorganisms present in the biological reactors of WWTP decompose complex compounds in wastewater and use simple constituents as energy resources for metabolic activities or building materials for cell synthesis. Treatment of BPA by biomass is possible due to biodegradation and biosorption by the microorganisms [7]. Biodegradation of BPA by bacteria proceeds via complicated metabolic routes and creates several kinds of biodegradation products (BDPs). Known BPA-degrading bacteria can grow on BPA as a sole carbon source—about 70–85% of the total organic carbon is mineralized to CO2 or used to build bacterial cells, while the remaining 15–30% is accumulated in the culture medium as soluble BDPs [8]. According to the metabolic pathways of BPA degradation (Figure 1) proposed by Spivack et al. [9], the major pathway (80%) is the cleavage of BPA to form p-hydroxyacetophenone and phydroxybenzaldehyde, followed by further degradation of both BDPs via p-hydroxybenzoic acid. In addition, about 20% of the BPA is converted to 2,3-bis-(4-hydroxyphenyl)-1,2-propanediol via bis(4-hydroxyphenyl)-1-propanol in the minor pathway.
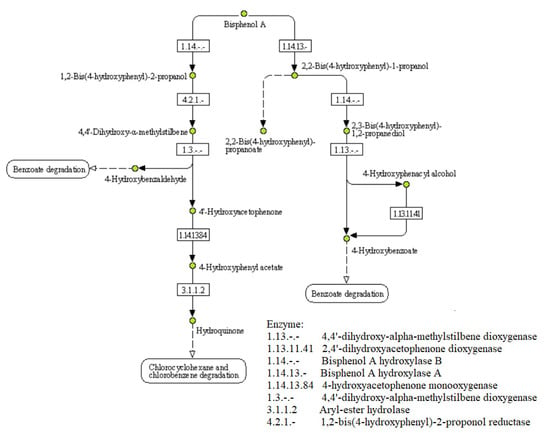
Figure 1.
Pathways of BPA biodegradation [10].
In biological reactors, different types of biomass can be used. The constant need for improving existing treatment technologies has led to the cultivation of aerobic granular sludge (AGS), a promising alternative to conventional activated sludge for micropollutant removal [11]. AGS has better resistance to toxins than conventional activated sludge because of its high content of extracellular polymers (EPS) that protect bacterial cells. Also, the presence of zones with different oxic conditions in the granule structure favors high species variety and the occurrence of many metabolic pathways enabling micropollutant degradation [12].
Effective bacterial BPA degraders belong to the genera Sphingomonas sp., Pandoraea sp., Pseudomonas sp., Bordetella sp., Achromobacter sp., and Nitrosomonas sp. [13,14,15]. Oh and Choi [16] observed that during the first 2 weeks of the acclimation period, microbial communities from activated sludge rapidly increased their efficiency of removal of 500–5000 μg BPA/L from 23–29% to 89–99%, after which removal rates remained stable at >90% for 3 months. Biochemical assays demonstrated that BPA was removed by biodegradation, rather than abiotic removal routes (e.g., adsorption and volatilization). 16S rRNA gene-based community analysis revealed that exposure to 500–5000 μg BPA/L selected for three Sphingomonadaceae genera (Sphingobium, Novosphingobium, and Sphingopyxis). The Sphingomonadaceae-enriched communities that had acclimated to BPA removed BPA at a rate of 7.0 mg VSS−1 day−1, which was higher than that of other potential BPA degraders.
The cooperation of bacteria in complex technical biocenoses is crucial for effective BPA removal. A meta-omics analysis that correlated the BPA biodegradation performance of the microbial community with bacterial activities identified specific substrates, such as 4-1,2-bis(4-hydroxyphenyl)-2-propanol, hydroxy-acetophenone, and 4-hydroxybenzoate, that were involved in a synergistic interaction of cross-feeding between BPA-degrading Sphingonomas species and intermediate users such as Pseudomonas sp. and Pusillimonas sp. [17]. This proposed synergistic interaction was supported by the observation that a co-culture of Sphingonomas sp. and Pseudomonas sp. isolates demonstrated better BPA biodegradation than an isolate of Sphingonomas sp. alone.
Understanding microbial interactions in engineering bioprocesses is important for improving and optimizing performance outcomes. Despite the recognized effect of BPA on the microbial structure in wastewater treatment systems, very little is known about the effect of BPA on microbial activity in the complex microbial systems in WWTPs. Therefore, the objective of this study was to use metatranscriptomic analysis to determine how exposure to 12 mg/L BPA changes gene expression in a microbial community in aerobic granular sludge.
2. Materials and Methods
2.1. Experiment Setup
In the first step of the experiment, a sequencing batch reactor with aerobic granular sludge (GSBR) with a height of 100 cm and diameter of 10 cm was set up. GSBR was supplied with synthetic municipal wastewater according to [18]. Synthetic wastewater was used because real wastewater contains other micropollutants that would affect the study results. To prepare a stock solution, 3.2 g of BPA (purity > 99%, Sigma-Aldrich, Saint Lous, MO, USA) were dissolved in 50 mL of methanol (99% purity, Chempur, Piekary Śląskie, Poland). The stock solution was added to wastewater to obtain a concentration of 12 mg BPA/L. BPA concentrations in a range of 10–12 mg/L are typical of highly contaminated wastewater [19]. The GSBR was operated at a cycle length of 8 h and a volumetric exchange rate of 50% for over 100 cycles as described in [12]. Air was supplied continuously at a rate of 4 L/min through a fine-bubble diffuser. The cycle consisted of 5 min of settling, 5 min of decantation, 5 min of feeding, and 465 min of aeration in the reaction phase. The concentration of biomass in the GSBR was about 4–5 g MLSS/L. The efficiency of COD, total nitrogen, and BPA removal exceeded 90%. In the influent and effluent, total nitrogen (TN), COD (HACH cuvette tests, HACH Lange, Dusseldorf, Germany), total suspended solids (TSS), and BPA were analyzed. BPA concentrations were determined using HPLC (Varian, Palo Alto, Australia) with a UV–Vis detector.
In the second step of the experiment, the biomass from GSBR was first kept at 4 °C for 2 weeks (removal of all BPA sorbed on the biomass) and then separated into two flasks (2 g MLSS/L). In the control flask (control), distilled water was added, while a single dose of 10 mg BPA/L was added to the second flask (BPA) [19]. Two hours after BPA addition, the biomass samples were taken from both flasks and embedded in 800 μL of fenozol. The second step was repeated, and the biomass was once again embedded in fenozol.
2.2. Metatranscriptome Analysis
The biomass from both experimental repetitions was mixed, and RNA was then isolated from this mixture using a Total RNA Mini kit (A&A Biotechnology, Gdańsk, Poland). The quality and quantity of RNA were measured using Qubit 3.0 Fluorometer (ThermoFisher Scientific, Waltham, USA). From the isolated RNA, rRNA was removed using MICROBExpress™ (Life Technologies, Carlsbad, CA, USA) to enrich bacterial mRNA from purified total RNA. For the removal of genomic DNA, a reaction mixture was prepared (1 μL of 10x concentrated dsDNase Buffer, 1 μL of dsDNase (Promega, Madison, USA), 5 μg of RNA, nuclease free water to 10 μL). Purified RNA (1 μg-volume calculated based on the spectrophotometric measurement of mRNA concentration) was incubated with the genomic DNA removal buffer and with the addition of nuclease-free water at 37 °C for 2 min. Then, samples were immediately placed on ice. To obtain cDNA, the 2 μL remaining RNA was subjected to a reverse transcription using the Thermo Scientific™ Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Roche, Basel, Switzerland); in the reaction, random hexamer primers were used. The obtained cDNA was sent to the Research and Testing Laboratory (USA). For amplification, random hexamers were used. High-throughput sequencing was performed using the MiSeq Illumina platform.
All bioinformatics analyses for community diversity and gene expression were performed by the MG-RAST (MetaGenomics Rapid Annotation using Subsystem Technology) server at the Argonne National Library. Raw sequences as FASTQ files were uploaded to the platform. For metatranscriptomic analyses, sequences shorter than 75 bp were discharged. The remaining sequences were paired-ended and forwarded for further analysis. Pipeline quality-control options were chosen for the removal of artificial replicate sequences produced by sequencing artifacts. The taxonomic profiles used the NCBI taxonomy. Then, using hierarchical classification, the functional profiles were generated using KEGG Orthologs (KO) and SEED Subsystems. To get information about all general metabolic pathways, data were annotated with default settings: alignment length of 15 bp, e-value: e-5, and percent identity 60%. For general gene expression analysis, Levels 1 and 2 of KO were used. To study in detail genes responsible for metabolic pathways, Level 3 of KO and KEGG mapper internal tool was used. Sequence libraries of cDNA were archived in the MG-RAST database under accession numbers: control-4790864.3, BPA-4790863.3 (R1 and R2 Illumina paired-end reads, respectively).
3. Results
The dataset from the control biomass contained 14,884,214 sequences, totaling 1,843,195,647 bp, with an average length of 124 bp. Of the tested sequences, 13,311,779 (89.44%) failed to pass the QC pipeline. Of those, dereplication identified 101,824 sequences as artificial duplicate reads. Of the sequences that passed QC, 12,748 sequences (1.00%) contained ribosomal RNA (rRNA) genes, 4929 sequences (0.49%) coded for predicted proteins with known functions, and 997,708 sequences (98.26%) coded for predicted proteins with unknown functions.
The dataset from the BPA-exposed biomass contained 16,212,696 sequences, totaling 2,013,941,187 bp, with an average length of 124 bp. Of the tested sequences, 14,896,760 sequences (91.88%) failed to pass the QC pipeline. Of those, dereplication identified 66 432 sequences as artificial duplicate reads. Of the sequences that passed QC, 22,475 sequences (3.00%) contained ribosomal RNA genes, 2944 sequences (0.33%) coded for predicted proteins with known functions, and 861,726 sequences (97.13%) coded for predicted proteins with unknown function.
In the examined microbial communities of the AGS, five general metabolic pathways were annotated using KO at Level 1 (Figure 2). In the control sample, about 39% of the identified sequences were responsible for metabolism, and in the BPA sample, this value increased to 50%. Sequences related to human diseases, cellular processes, genetic information processing, and environmental information processing constituted 3%, 7%, 39%, and 12% of the control biomass, and 5%, 5%, 29%, and 11% of the BPA-exposed biomass, respectively. A more detailed analysis was conducted using KO at Level 2 (Figure 2), which allowed for a more precise description of the functions identified at Level 1.
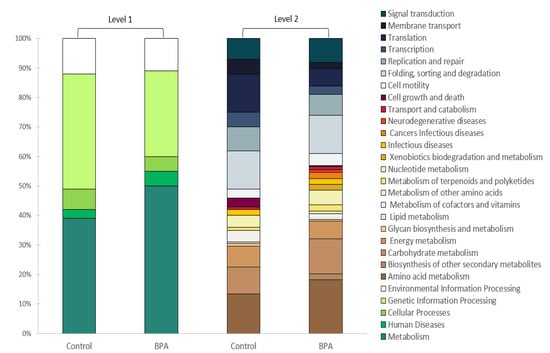
Figure 2.
Gene expression classification of control and BPA samples based on KEGG Orthologs in MG-RAST. Samples were annotated at Level 1 in five categories (metabolism, human diseases, cellular processes, genetic information processing, and environmental information processing) and the sub-functions that correspond to the Level-1 categories are shown at Level 2 (default settings were assumed: alignment length: 15 bp, e-value: e-5, percent identity: 60%, minimum abundance: 3).
Regarding particular functions in the metagenome, the largest shares of expressed genes in the control sample consisted of those responsible for amino acid metabolism (13%), protein folding, sorting and degradation (13%), translation (13%), carbohydrate metabolism (9%), replication and repair (8%), signal transduction (7%), and energy metabolism (7%). Similarly, the largest shares of genes expressed in the BPA-exposed sample also consisted of genes responsible for the same functions. However, the percentage of genes responsible for translation processes was two times higher in the BPA-exposed sample than in the control sample. The shares of genes responsible for carbohydrate metabolism and amino acid metabolism also increased noticeably (to 12% and 17%, respectively). The exposition of the biomass to BPA also led to the expression of genes responsible for biosynthesis of other secondary metabolites (2%); metabolism of other amino acids (1%), including those in a pathway of streptomycin biosynthesis; biodegradation and metabolism of xenobiotics (2%); and genes responsible for cancerogenic pathways (2%) and neurodegenerative diseases (1%) in humans. In contrast, genes coding for enzymes responsible for cell growth and death (3%) and glycan biosynthesis and metabolism (1%) were expressed only in the control sample.
To supplement the information about genes responsible for metabolic pathways, a more detailed analysis of differences in expression between the control and BPA-exposed samples was carried out manually using Level 3 of KO and the KEGG mapper visualization tool. Genes coding for alanine, aspartate, glutamate, valine, leucine, and isoleucine were expressed in both samples. In the BPA-exposed biomass, genes coding enzymes involved in the metabolism of cysteine, methionine, histidine, phenylalanine, tyrosine, and tryptophan were expressed. Glutamine synthetase, which plays an essential role in the metabolism of nitrogen by catalyzing the condensation of glutamate and ammonia to form glutamine, was expressed in the control sample, while in the BPA sample, glutamate dehydrogenase demonstrated higher expression (converts glutamate to α-ketoglutarate with a by-product of ammonia). Also, in the BPA-exposed sample, the expression of genes encoding 3-isopropylmalate connected with protein biosynthesis was observed [20].
The most important differences between the control and BPA-exposed samples were noticeable in the expression of genes of cellular respiratory pathways. The expression of genes of glycolysis and citrate cycle was almost two times higher in the control sample than in the BPA-exposed sample (47% vs. 24% and 27% vs. 6%, respectively). On the other hand, in the BPA-exposed sample, the expression of genes coding acetylo-coA synthetase, which takes part in anaerobic respiration in the glycolysis pathway, was two times higher than in the control sample. Also in the BPA-exposed sample, a high expression of genes encoding fumarate enzyme, which participates in two metabolic pathways i.e., citric acid cycle and reductive citric acid cycle (CO2 fixation), was observed.
The expression of genes related to the last stage of cellular respiration, which is oxidative phosphorylation, was much higher in the control sample than in the BPA-exposed sample. In the BPA-exposed sample, genes involved in nitrogen metabolism were expressed (67% of overall energy metabolism expression) including periplasmic nitrate reductase (NapA), which takes part in denitrification.
Within the category of folding, sorting, and degradation, in the control sample, the genes coding protein processing in the endoplasmic reticulum, heat shock proteins, as well as genes corresponding to RNA degradation processes were expressed. In the BPA-exposed sample, only genes corresponding to RNA degradation (90% of identified sequences) and genes connected with sulphur-rely systems (10%) were expressed. In both samples, DNA replication genes were active, but in the BPA-exposed sample, their expression was two times higher. In the control sample, only the DNA ligase genes were expressed, while in the BPA-exposed sample, genes coding DNA polymerase III subunits α and ε were active.
In the control and BPA-exposed sample, genes responsible for signal transduction cell motility were expressed in both sampled. In the BPA-exposed sample, high expression of the gene coding flagellin protein and gene coding protein responsible for pilus assembly was observed. In the control sample, genes coding glutamine synthetase and coenzyme Q-cytochrome c reductase were active—glutamine synthetase participates in nitrogen assimilation during low availability of this element (Figure 3).
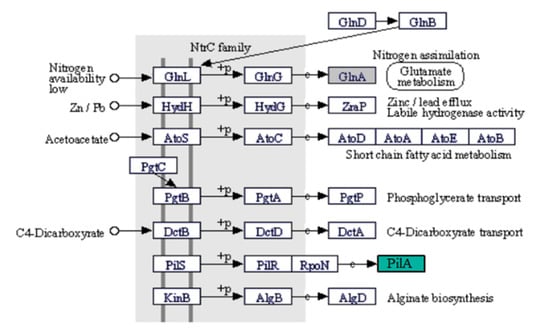
Figure 3.
Two-component system pathway fragment from KEGG mapper, showing the expression of type IV pilus assembly protein (PilA) in the BPA sample (green) and glutamine synthetase (GlnA) in the control sample (grey).
The expression of genes coding urease, converting urea into NH3, was two times higher in the BPA-exposed sample than in the control sample (Figure 4).
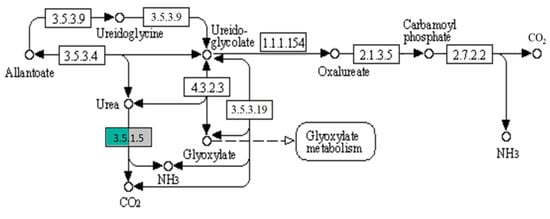
Figure 4.
Purine metabolism pathway fragment from KEGG mapper, showing the active expression of urease in the BPA sample (green) and the control sample (grey).
The microbial community of the BPA-exposed sample synthesized genes coding secondary metabolite streptomycin and took part in xenobiotic metabolism and degradation by the production of enzymes responsible for benzoate degradation. In the control sample, the activity of these enzymes was not observed.
4. Discussion
In both control and BPA-exposed biomass, the highest expression of genes coding heat shock chaperone proteins (HSP) was observed; in the BPA-exposed sample, the expression of genes coding chaperonin GroEL predominated. Those molecular systems are activated to protect newly synthesized or stress-denaturated proteins from misfolding and aggregation [21]. It was proved that expression of GroEL and DnaK genes also increased in cells exposed to osmotic stress, oxidative stress, changes in pH, UV radiation, and toxic compounds, including aromatic ones [22]. Chaperonin GroEL was found in activated sludge treated with toxic compounds (cadmium, pentachlorophenol, acetone) [23]. The GroEL was up-regulated in Acinetobacter sp. EDP3 grown at 1000 mg/L of phenol. This protein functions as an enzyme that prevents misfolding and promotes the refolding and proper assembly of unfolded polypeptides generated under stress conditions. This way, it stabilizes the protein and increases the resistance of the bacteria to deleterious chemicals or heat shock stress [24]. In our study, an increased expression of genes coding chaperone DnaK was observed in the control sample. Some chaperones are activated as starvation stress proteins [23]. To the control sample, no substrate was added, so this can be a reason for the increased expression of this HSP.
The exposition of biomass to BPA triggers the expression of genes coding enzymes involved in xenobiotic metabolism and biodegradation. Cydzik-Kwiatkowska et al. [25] used the bisdA gene coding for ferredoxin as a molecular marker to investigate BPA degrader activity in AGS from a batch reactor supplied with municipal wastewater spiked with up to 12 mg BPA/L. In the first hour of the cycle in the reactor with 2 mg BPA/L in the influent, the expression of bisdA was 14-fold higher than in the control reactor during this time. After this time, bisdA expression gradually decreased. In reactors with 6 and 12 mg BPA/L in the influent, bisdA expression peaked twice during the cycle (0.5 h and 4 h of the cycle) relative to that in the control reactor. There was a linear correlation between the concentration of BPA in wastewater introduced to the reactors and the averaged expression levels of bisdA in AGS. Ferredoxin was probably involved in an initial hydroxylation of BPA. Ferrodoxin converts BPA to 1,2-bis(hydroxyphenyl)-2-propanol and 2,2-bis(4-hydroxyphenyl)-1-propanol catalyzing the first step of two main BPA degradation pathways [26].
The high expression of genes coding pilus and flagellin proteins in BPA-exposed samples indicated that an exposition to BPA stimulated aggregation of microbial cells and formation of biofilm. Flagellin belongs to intermediate filament proteins taking part in the stress response and promotion of regeneration after injury, by providing scaffolding for the cell and binding to signaling proteins and to HSPs [27], while the pilus assembly protein is related to biofilm formation in bacteria [28]. Biofilms create multispecies communities, held together by a self-produced EPS that protects bacteria. The various metabolic capabilities of bacteria in biofilm increase the chances of successfully degrading toxins like BPA [29]. Li et al. [30] investigated the effect of exposition to BPA on the content of proteins (PN) and polysaccharides (PS) in loosely- and tightly-bound EPS in AGS. PS and PN contents changed from 17.7 and 11.3 mg/g VSS to 12.4 and 31.7 mg/g VSS in loosely-bound EPS, and from 37.7 and 35.7 to 24.3 and 88.1 mg/g VSS in tightly-bound EPS, respectively. The results implied that the increased in EPS content resulted from changes in PN concentrations, thus indicating the toxic effect of BPA on the production of PN.
Research shows that the use of immobilized biomass in the form of a biofilm or granular sludge allows for the effective removal of BPA, as high concentrations of EPS in the biomass protect microorganisms against its negative influence. In the studies by Cydzik-Kwiatkowska et al. [12], an AGS containing EPS at the level of 104.3 mg/g MLSS removed BPA with an efficiency of 99.3%. Zielińska et al. [7], in reactors with immobilized biomass, achieved a BPA removal efficiency of 87.1–92.9%. For comparison, in reactors with activated sludge, where the EPS concentration is lower [31], the BPA removal efficiency was only 84% [32].
Nitrogen metabolism in the control sample was mostly related to nitrogen assimilation, while in the BPA-exposed sample, nitrogen was mainly used as an energy source. In the control sample, a high expression of genes coding glutamine synthetase was observed indicating that microorganisms actively assimilated nitrogen. Glutamine synthetase plays an essential role in nitrogen metabolism and catalyzes the reaction of glutamate and ammonia to form glutamine [33]. In the BPA-exposed biomass, a higher expression of genes coding glutamate dehydrogenase was observed, which converts glutamate to α-ketoglutarate with ammonia as a by-product.
Zheng et al. [34], based on the metabolic map, found nitrogen genes involved with glutamate metabolism and arginine biosynthesis. The glutamate can be used either to replenish the pool of glutamate for glutamine synthetase catalysis or to donate its amino group to form other nitrogen-containing compounds. The microbial synthesis of glutamine can be regarded as the first step in a highly branched pathway, which leads to the biosynthesis of a great number of different compounds. Furthermore, glutamate synthase (gltB) and GDH are important for the incorporation of ammonia (NH3) into glutamate to form glutamine. Jiang et al. [35] observed that the relative abundance of the gltB gene in sludge, which is involved in glutamate synthesis, was 1.7-fold higher in a BPA-exposed reactor than that in a control reactor. Expression of genes coding the α subunit of urease occurred in both samples, but in the BPA-exposed sample, this expression was two times higher than in the control sample. Many ammonia-oxidizing bacteria (AOB) produce urease including Nitrosococcus sp. and Nitrosospira sp. For AOB, the hydrolysis of urea is a source of ammonia (as an energy source by nitrification or protein biosynthesis) and carbon dioxide [36]. In the BPA-exposed sample, transcripts of genes coding many enzymes needed for nitrogen metabolism were present such as urease, ammonia monooxygenase (AMO), hydroxylamine oxidoreductase, or nitrogen regulatory proteins (NtrC). It can be hypothesized that the energy and nitrogen demand may be higher during BPA degradation. In the BPA-exposed sample, the expression of signal transduction genes involved in nitrogen or carbon metabolism was two times higher.
Margot et al. [37] observed that BPA was removed better in a reactor with efficient nitrification because AOB co-metabolized BPA as a result of oxidation by AMO. The bacteria synthesize AMO, which creates OH radicals that can break down resistant, non-specific substrates such as hydrocarbons [38]. In the present study, genes coding AMO were active in both samples, but in trace amounts, indicating that BPA removal resulted mainly in heterotrophic activity. Similar conclusions were drawn by Zielińska and co-workers [7]. The authors proved that in aerobic biofilms, BPA removal was related mainly with an activity of heterotrophic bacteria. Specific oxygen uptake rates in samples with AMO inhibitor indicated that ammonium was not removed from wastewater, although BPA was. This implies that BPA removal was connected with heterotrophic bacteria activity or with BPA adsorption on biomass particles.
The presence of valine, leucine, and isoleucine in the growth medium is essential for achieving the high activity of proteins involved in signal transduction processes and for efficient regulation of the target genes [39]. This confirms the increased expression in the BPA-exposed biomass genes coding 3-isopropylmalate, which is an intermediate in the biosynthesis of those three proteins.
In the BPA-exposed biomass, transcripts of genes coding proteins responsible for xenobiotic degradation, including enzymes responsible for benzoate degradation, were observed. This might be connected with the structure of BPA, which contains hydroxyphenyl groups and acetate. One of the highly expressed genes coding enzymes from this category was phenylacetate-coA ligase, which is involved in the catabolism of phenylacetic acid in Pseudomonas putida [40].
5. Conclusions
The study of changes in gene expression in aerobic granular sludge allows to obtain information about mechanisms responsible for BPA removal, indirectly indicating the dominant microbiological changes leading to effective wastewater treatment.
Metatranscriptomic analysis of aerobic granular sludge exposed to BPA indicated direct biodegradation as the main mechanism of BPA removal from wastewater. In the BPA-exposed biomass, genes coding chaperon proteins and proteins involved in biofilm formation were expressed. In the control sample, mainly nitrogen assimilation occurred (glutamine synthetase), while in the BPA-exposed sample, nitrogen was mainly used as an energy source as indicated by the presence of genes coding nitrification enzymes and urease. In the BPA-exposed biomass, genes were synthesized coding proteins responsible for xenobiotic degradation including enzymes responsible for benzoate degradation. This might be connected with the chemical structure of BPA.
Author Contributions
Conceptualization, A.C.-K.; methodology, A.C.-K. and M.G.; software, M.G. and P.J.; validation, M.G. and A.C.-K.; formal analysis, A.C.-K. and P.J.; investigation, A.C.-K. and M.G.; resources, A.C.-K.; data curation, A.C.-K. and M.G. and P.J.; writing—original draft preparation, P.J. and A.C.-K.; writing—review and editing, A.C.-K. and P.J.; visualization, M.G. and P.J.; supervision, A.C.-K.; project administration, A.C.-K.; funding acquisition, A.C.-K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Science Centre, Poland (grant number 2013/09/B/NZ9/01811 and grant number 2016/21/B/NZ9/03627).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Sequences libraries of cDNA were archived in MG-RAST database under accession numbers: Control–4790864.3, BPA–4790863.3 (R1 and R2 Illumina paired-end reads, respectively).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Luo, Y.; Guo, W.; Ngo, H.H.; Nghiem, L.D.; Hai, F.I.; Zhang, J.; Hai, F.I.; Zhang, J.; Liang, S.; Wang, X.C. A review on the occurrence of micropollutants in the aquatic environment and their fate and removal during wastewater treatment. Sci. Total Environ. 2014, 473, 619–641. [Google Scholar] [CrossRef] [PubMed]
- Flint, S.; Markle, T.; Thompson, S.; Wallace, E. Bisphenol A exposure, effects, and policy: A wildlife perspective. J. Environ. Manag. 2012, 104, 19–34. [Google Scholar] [CrossRef]
- Bolz, U.; Hagenmaier, H.; Korner, W. Phenolic xenoestrogens in surface water, sediments, and sewage sludge from Baden–Wurttemberg, south-west Germany. Environ. Pollut. 2001, 115, 291–301. [Google Scholar] [CrossRef]
- Kolpin, D.W.; Furlong, E.T.; Meyer, M.T.; Thurman, E.M.; Zaugg, S.D.; Barber, L.B.; Buxton, H.T. Pharmaceuticals, hormones, and other organic wastewater contaminants in US streams, 1999–2000: A national reconnaissance. Environ. Sci. Technol. 2002, 36, 1202–1211. [Google Scholar] [CrossRef] [PubMed]
- Alexander, H.C.; Dill, D.C.; Smith, L.W.; Guiney, P.D.; Dorn, P. Bisphenol A: Acute aquatic toxicity. Environ. Toxicol. Chem. 1988, 7, 19–26. [Google Scholar] [CrossRef]
- Matsumura, Y.; Akahira-Moriya, A.; Sasaki-Mori, M. Bioremediation of Bisphenol-A polluted soil by Sphingomonas bisphenolicum AO1 and the microbial community existing in the soil. Biocontrol Sci. 2015, 20, 35–42. [Google Scholar] [CrossRef]
- Zielińska, M.; Cydzik-Kwiatkowska, A.; Bernat, K.; Bułkowska, K.; Wojnowska-Baryła, I. Removal of bisphenol A (BPA) in a nitrifying system with immobilized biomass. Bioresour. Technol. 2014, 171, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Lobos, J.H.; Leib, T.K.; Su, T.M. Biodegradation of bisphenol A and other bisphenols by a gram-negative aerobic bacterium. Appl. Environ. Microbiol. 1992, 58, 1823–1831. [Google Scholar] [CrossRef]
- Spivack, J.; Leib, T.K.; Lobos, J.H. Novel pathway for bacterial metabolism of bisphenol A. Rearrangements and stilbene cleavage in bisphenol A metabolism. J. Biol. Chem. 1994, 269, 7323–7329. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Bengtsson, S.; De Blois, M.; Wilén, B.M.; Gustavsson, D. Treatment of municipal wastewater with aerobic granular sludge. Crit. Rev. Environ. Sci. Technol. 2018, 48, 119–166. [Google Scholar] [CrossRef]
- Cydzik-Kwiatkowska, A.; Bernat, K.; Zielińska, M.; Bułkowska, K.; Wojnowska-Baryła, I. Aerobic granular sludge for bisphenol A (BPA) removal from wastewater. Int. Biodeter. Biodegr. 2017, 122, 1–11. [Google Scholar] [CrossRef]
- Matsumura, Y.; Hosokawa, C.; Sasaki-Mori, M.; Akahira, A.; Fukunaga, K.; Ikeuchi, T.; Oshiman, K.; Tsuchido, T. Isolation and characterization of novel bisphenol-A-degrading bacteria from soils. Biocontrol Sci. 2009, 14, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Roh, H.; Subramanya, N.; Zhao, F.; Yu, C.P.; Sandt, J.; Chu, K.H. Biodegradation potential of wastewater micropollutants by ammonia-oxidizing bacteria. Chemosphere 2009, 77, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zeng, G.; Yuan, L.; Yu, J.; Li, J.; Huang, G.; Xi, B.; Liu, H. Aerobic degradation of bisphenol A by Achromobacter xylosoxidans strain B-16 isolated from compost leachate of municipal solid waste. Chemosphere 2007, 68, 181–190. [Google Scholar] [CrossRef]
- Oh, S.; Choi, D. Microbial community enhances biodegradation of bisphenol a through selection of Sphingomonadaceae. Microb. Ecol. 2019, 77, 631–639. [Google Scholar] [CrossRef]
- Yu, K.; Yi, S.; Li, B.; Guo, F.; Peng, X.; Wang, Z.; Wu, Y.; Alvarez-Cohen, L.; Zhang, T. An integrated meta-omics approach reveals substrates involved in synergistic interactions in a bisphenol A (BPA)-degrading microbial community. Microbiome 2019, 7, 16. [Google Scholar] [CrossRef]
- Coelho, M.A.; Russo, C.; Araujo, O.Q. Optimization of sequencing batch reactor for biological nitrogen removal. Water Res. 2000, 34, 2809–2817. [Google Scholar] [CrossRef]
- Yamamoto, T.; Yasuhara, A.; Shiraishi, H.; Nakasugi, O. Bisphenol A in hazardous waste landfill leachates. Chemosphere 2001, 42, 415–418. [Google Scholar] [CrossRef]
- Miflin, B.J.; Habash, D.Z. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 2002, 53, 979–987. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, J.; Zhao, J.; Yang, N.; Zhang, F.; Jiang, Z. The influence of SBR parameters on the sludge toxicity of synthetic wastewater containing bisphenol A. Environ. Sci. Pollut. Res. 2014, 21, 9287–9296. [Google Scholar] [CrossRef]
- Susin, M.F.; Baldini, R.L.; Gueiros-Filho, F.; Gomes, S.L. GroES/GroEL and DnaK/DnaJ have distinct roles in stress responses and during cell cycle progression in Caulobacter crescentus. J. Bacteriol. 2006, 188, 8044–8053. [Google Scholar] [CrossRef]
- Bott, C.B.; Love, N.G. The immunochemical detection of stress proteins in activated sludge exposed to toxic chemicals. Water Res. 2001, 35, 91–100. [Google Scholar] [CrossRef]
- Geng, A.; Lim, C.J. Proteome Analysis of the Adaptation of a Phenol-Degrading Bacterium Acinetobacter sp. EDP3 to the Variation of Phenol Loadings. Chin. J. Chem. Eng. 2007, 15, 781–787. [Google Scholar] [CrossRef]
- Cydzik-Kwiatkowska, A.; Zielińska, M.; Bernat, K.; Bułkowska, K.; Wojnowska-Baryła, I. Insights into mechanisms of bisphenol A biodegradation in aerobic granular sludge. Bioresour. Technol. 2020, 315, 123806. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Akahira, A.; Oshiman, K.I.; Tsuchido, T.; Matsumura, Y. Purification of cytochrome P450 and ferredoxin, involved in bisphenol A degradation, from Sphingomonas sp. strain AO1. Appl. Environ. Microbiol. 2005, 71, 8024–8030. [Google Scholar] [CrossRef]
- Toivola, D.M.; Strnad, P.; Habtezion, A.; Omary, M.B. Intermediate filaments take the heat as stress proteins. Trends Cell Biol. 2010, 20, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Matsumoto, S. Bacterial adhesion: From mechanism to control. Biochem. Eng. J. 2010, 48, 424–434. [Google Scholar] [CrossRef]
- López, D.; Vlamakis, H.; Kolter, R. Biofilms. Cold Spring Harb. Perspect. Biol. 2010, 2, a000398. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wei, D.; Zhang, G.; Shi, L.; Wang, Y.; Wang, B.; Wang, X.; Du, B.; Wei, Q. Toxicity of bisphenol A to aerobic granular sludge in sequencing batch reactors. J. Mol. Liq. 2015, 209, 284–288. [Google Scholar] [CrossRef]
- Guo, H.; Felz, S.; Lin, Y.; van Lier, J.B.; de Kreuk, M. Structural extracellular polymeric substances determine the difference in digestibility between waste activated sludge and aerobic granules. Water Res 2020, 181, 115924. [Google Scholar] [CrossRef]
- Melcer, H.; Klečka, G. Treatment of wastewaters containing bisphenol A: State of the science review. Water. Environ. Res. 2011, 83, 650–666. [Google Scholar] [CrossRef] [PubMed]
- Spodenkiewicz, M.; Diez-Fernandez, C.; Rüfenacht, V.; Gemperle-Britschgi, C.; Häberle, J. Minireview on glutamine synthetase deficiency, an ultra-rare inborn error of amino acid biosynthesis. Biology 2016, 5, 40. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhao, Z.; Lv, F.; Wang, R.; Gong, Q.; Zhai, B.; Wang, Z.; Zhao, Z.; Li, Z. Metagenomic exploration of the interactions between N and P cycling and SOM turnover in an apple orchard with a cover crop fertilized for 9 years. Biol. Fertil. Soils 2019, 55, 365–381. [Google Scholar] [CrossRef]
- Jiang, X.; Yan, Y.; Feng, L.; Wang, F.; Guo, Y.; Zhang, X.; Zhang, Z. Bisphenol A alters volatile fatty acids accumulation during sludge anaerobic fermentation by affecting amino acid metabolism, material transport and carbohydrate-active enzymes. Bioresour. Technol. 2021, 323, 124588. [Google Scholar] [CrossRef]
- Koper, T.E.; El-Sheikh, A.F.; Norton, J.M.; Klotz, M.G. Urease-encoding genes in ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 2004, 70, 2342–2348. [Google Scholar] [CrossRef] [PubMed]
- Margot, J.; Lochmatter, S.; Barry, D.A.; Holliger, C. Role of ammonia-oxidizing bacteria in micropollutant removal from wastewater with aerobic granular sludge. Water Sci. Technol. 2016, 73, 564–575. [Google Scholar] [CrossRef]
- Chang, S.W.; Hyman, M.R.; Williamson, K.J. Cooxidation of naphthalene and other polycyclic aromatic hydrocarbons by the nitrifying bacterium, Nitrosomonas europaea. Biodegradation 2002, 13, 373–381. [Google Scholar] [CrossRef]
- Belitsky, B.R. Role of branched-chain amino acid transport in Bacillus subtilis CodY activity. J. Bacteriol. 2015, 197, 1330–1338. [Google Scholar] [CrossRef]
- Martinez-Blanco, H.; Reglero, A.; Rodriguez-Aparicio, L.B.; Luengo, J.M. Purification and biochemical characterization of phenylacetyl-CoA ligase from Pseudomonas putida. A specific enzyme for the catabolism of phenylacetic acid. J. Biol. Chem. 1990, 265, 7084–7090. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).